Abstract
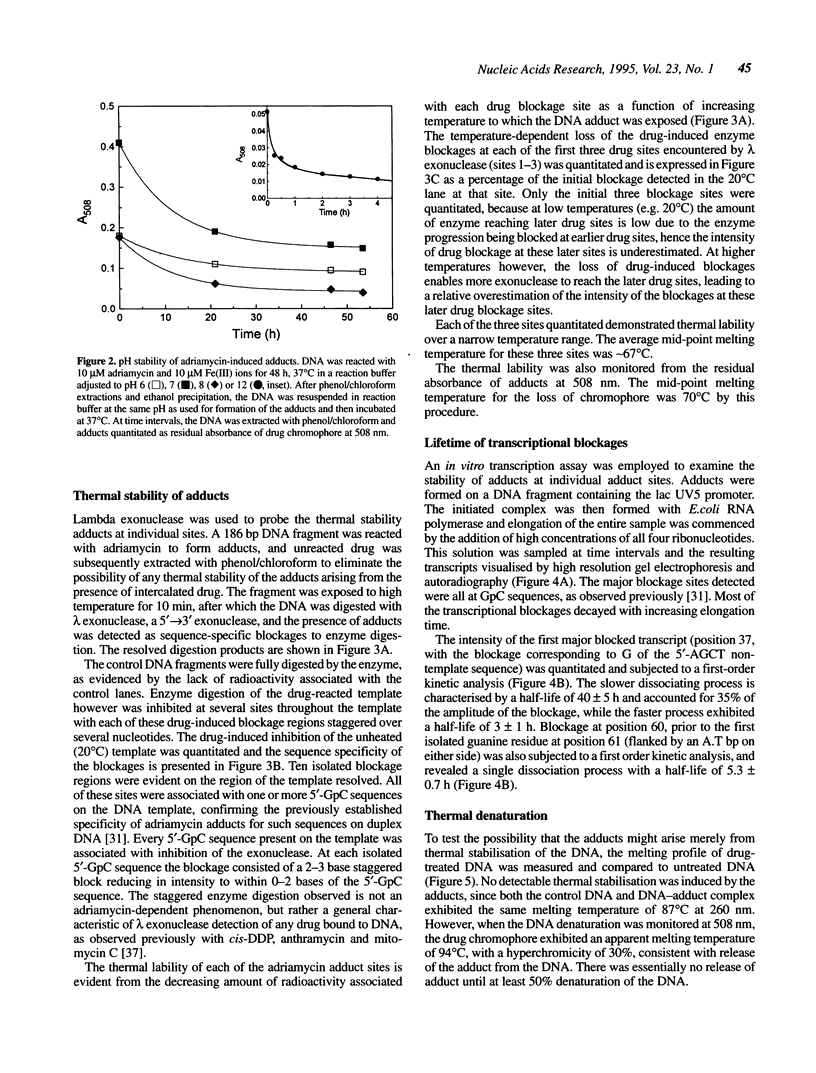
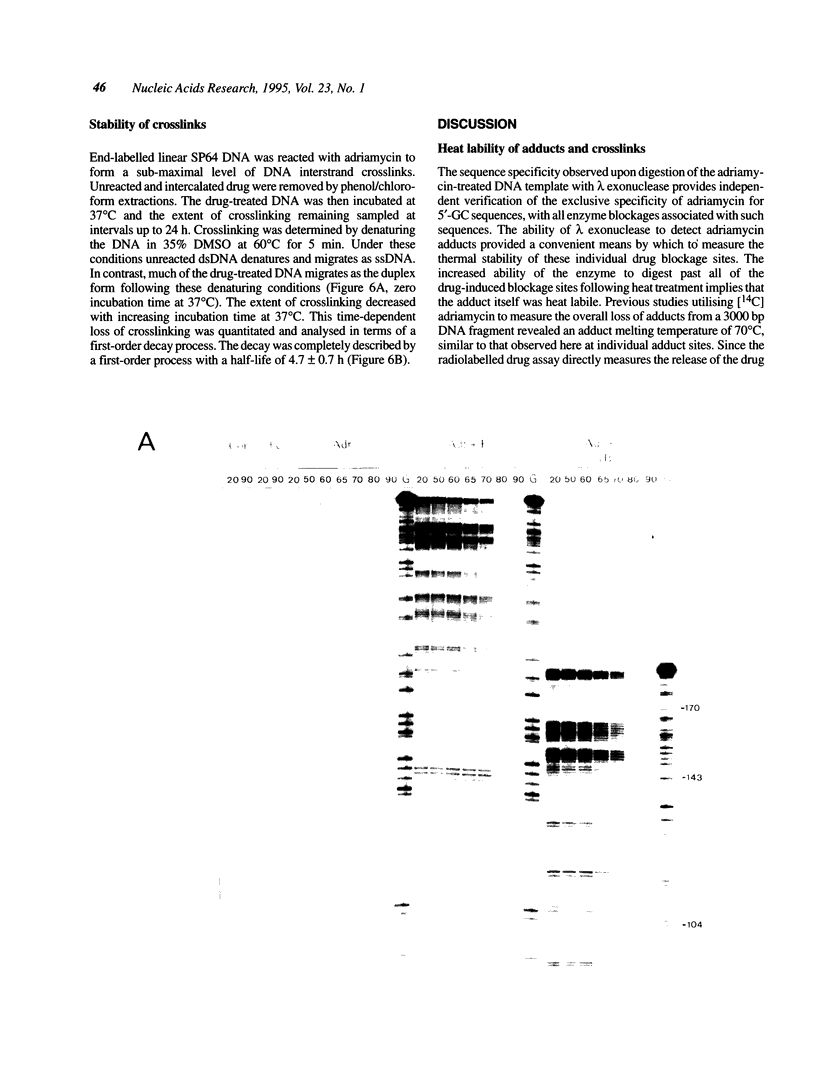
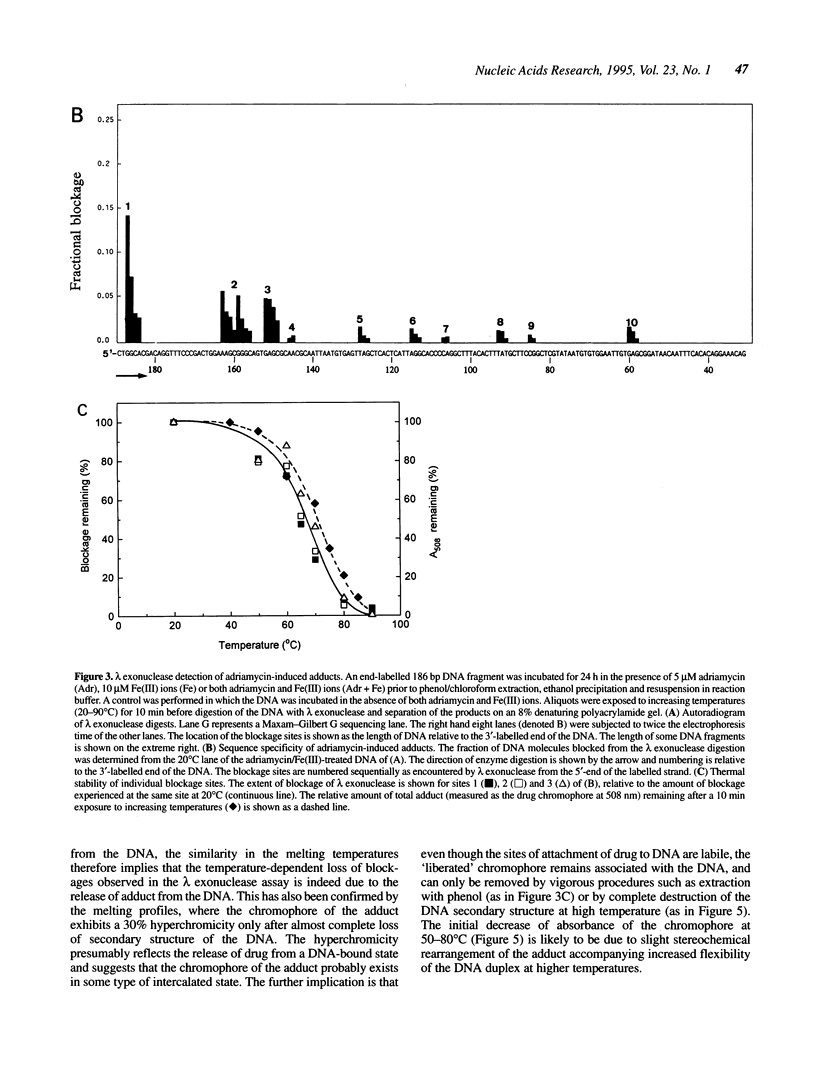
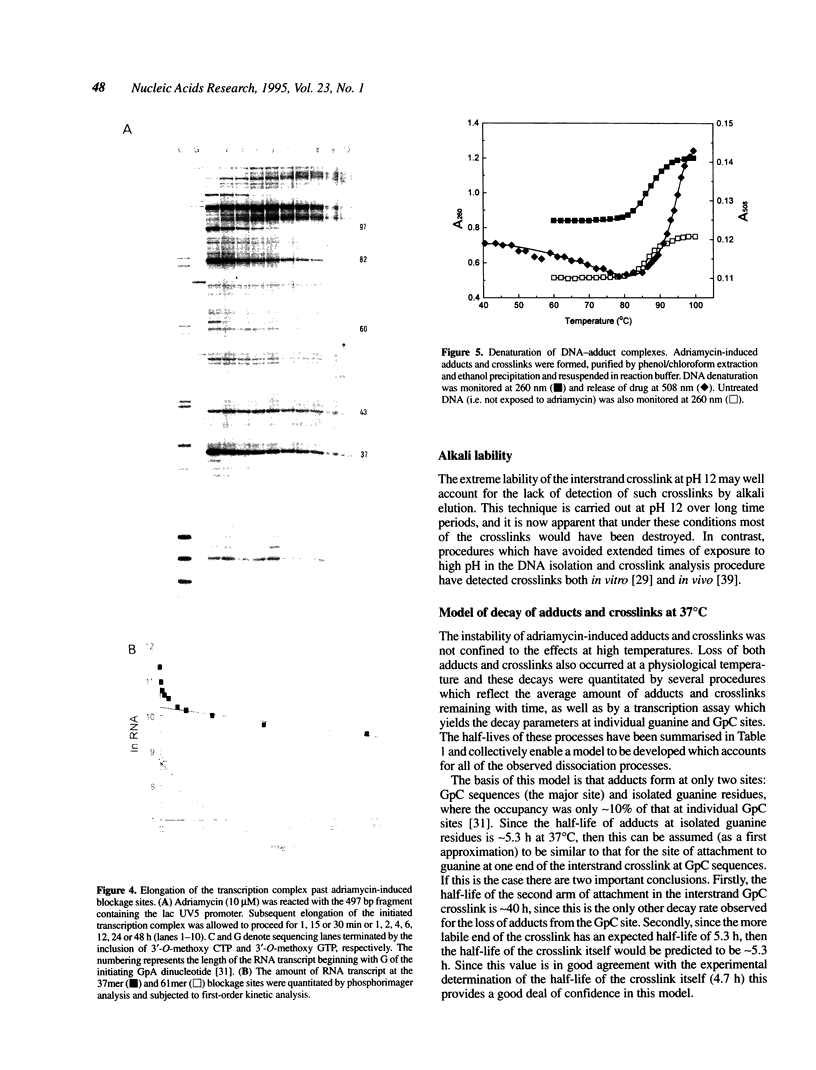
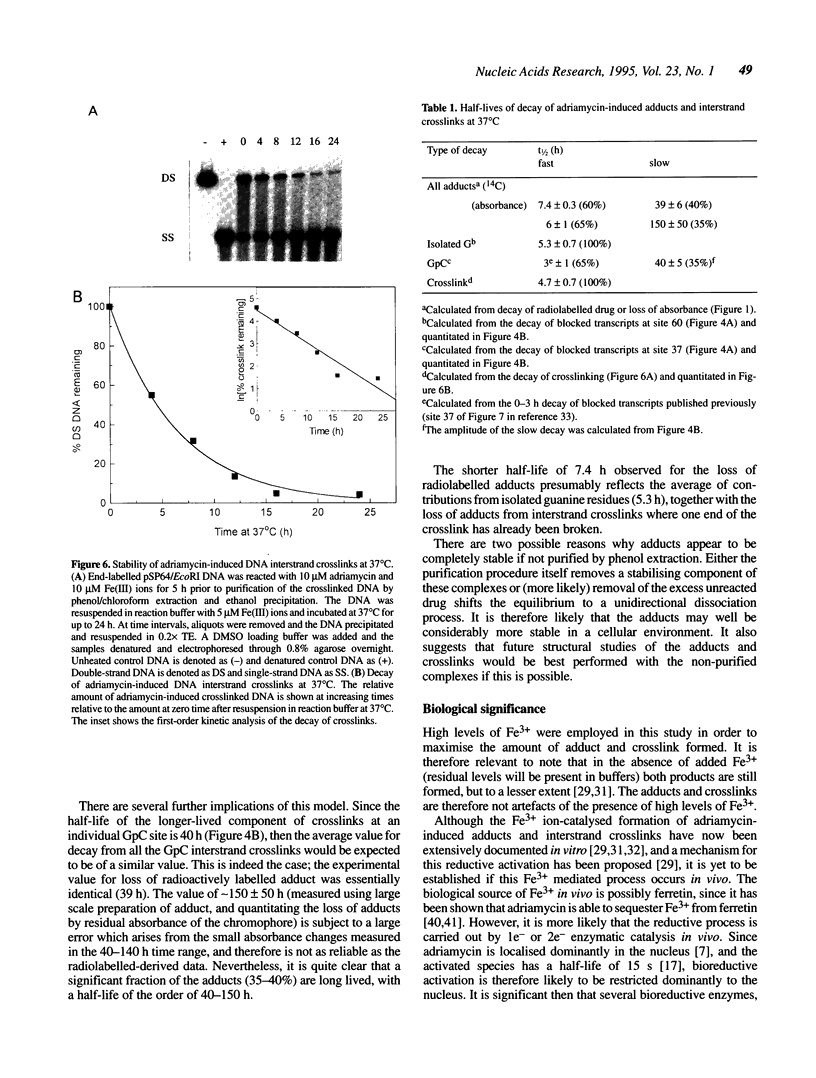
The stability of adriamycin-induced DNA adducts and interstrand crosslinks was measured at 37 degrees C by three independent procedures. The loss of [14C]-labelled adducts was described by two first-order decays with half-lives of 7.4 h (60% amplitude) and 39 h (40%). The loss of the drug chromophore also exhibited a biphasic character, with half-lives of 6 h (65%) and approximately 150 h (35%). The decay of transcriptional blockages at an isolated, apparent interstrand GpC crosslinking site was described by two first-order processes, with half-lives of 3 h (65%) and 40 h (35%), whereas the decay of transcriptional blockages at an isolated guanine residue (apparent site of monoadduct) was completely described by a first-order decay with a half-life of 5.3 h. The loss of interstrand crosslinks was measured using a gel electrophoresis assay, and the decay was characterised by a single first-order process with a half-life of 4.7 h. Collectively, these values serve to define a model of the interstrand crosslink with unstable sites of attachment at both ends of the crosslink, with half-lives at either end being approximately 5 and 40 h. The adducts exhibited increasing lability with increasing pH, and were particularly unstable at pH 12, with a half-life of approximately 0.5 h. The adducts were also heat labile, with an overall melting temperature of 67 degrees C (10 min exposure) and this was also the thermal lability measured at three individual adduct sites probed by lambda exonuclease.
Full text
PDF

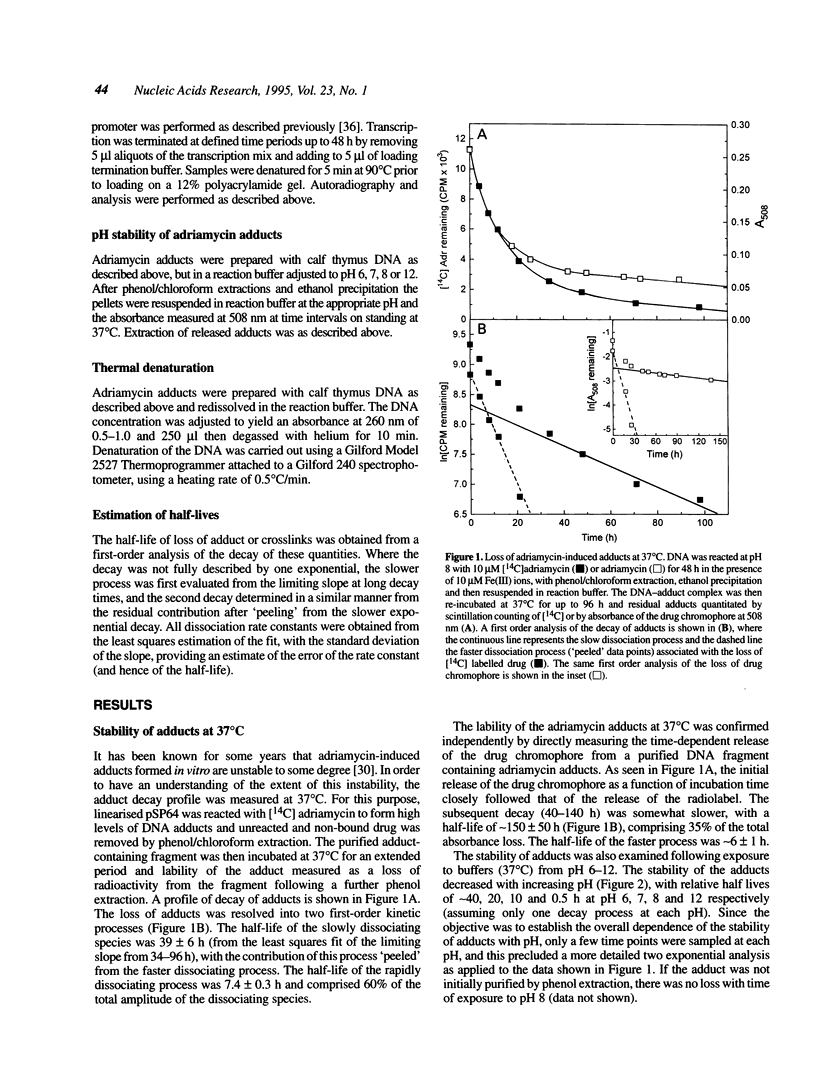

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Butler J., Hoey B. M. Are reduced quinones necessarily involved in the antitumour activity of quinone drugs? Br J Cancer Suppl. 1987 Jun;8:53–59. [PMC free article] [PubMed] [Google Scholar]
- Cullinane C., Cutts S. M., van Rosmalen A., Phillips D. R. Formation of adriamycin--DNA adducts in vitro. Nucleic Acids Res. 1994 Jun 25;22(12):2296–2303. doi: 10.1093/nar/22.12.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullinane C., Phillips D. R. Induction of stable transcriptional blockage sites by adriamycin: GpC specificity of apparent adriamycin-DNA adducts and dependence on iron(III) ions. Biochemistry. 1990 Jun 12;29(23):5638–5646. doi: 10.1021/bi00475a032. [DOI] [PubMed] [Google Scholar]
- Cullinane C., Phillips D. R. Thermal stability of DNA adducts induced by cyanomorpholinoadriamycin in vitro. Nucleic Acids Res. 1993 Apr 25;21(8):1857–1862. doi: 10.1093/nar/21.8.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullinane C., van Rosmalen A., Phillips D. R. Does adriamycin induce interstrand cross-links in DNA? Biochemistry. 1994 Apr 19;33(15):4632–4638. doi: 10.1021/bi00181a025. [DOI] [PubMed] [Google Scholar]
- Cummings J., Anderson L., Willmott N., Smyth J. F. The molecular pharmacology of doxorubicin in vivo. Eur J Cancer. 1991;27(5):532–535. doi: 10.1016/0277-5379(91)90209-v. [DOI] [PubMed] [Google Scholar]
- Cummings J., Bartoszek A., Smyth J. F. Determination of covalent binding to intact DNA, RNA, and oligonucleotides by intercalating anticancer drugs using high-performance liquid chromatography. Studies with doxorubicin and NADPH cytochrome P-450 reductase. Anal Biochem. 1991 Apr;194(1):146–155. doi: 10.1016/0003-2697(91)90162-m. [DOI] [PubMed] [Google Scholar]
- Cummings J., Willmott N., Hoey B. M., Marley E. S., Smyth J. F. The consequences of doxorubicin quinone reduction in vivo in tumour tissue. Biochem Pharmacol. 1992 Dec 1;44(11):2165–2174. doi: 10.1016/0006-2952(92)90343-h. [DOI] [PubMed] [Google Scholar]
- Demant E. J. Transfer of ferritin-bound iron to adriamycin. FEBS Lett. 1984 Oct 15;176(1):97–100. doi: 10.1016/0014-5793(84)80919-9. [DOI] [PubMed] [Google Scholar]
- Favaudon V. On the mechanism of reductive activation in the mode of action of some anticancer drugs. Biochimie. 1982 Jul;64(7):457–475. doi: 10.1016/s0300-9084(82)80162-4. [DOI] [PubMed] [Google Scholar]
- Fisher J., Abdella B. R., McLane K. E. Anthracycline antibiotic reduction by spinach ferredoxin-NADP+ reductase and ferredoxin. Biochemistry. 1985 Jul 2;24(14):3562–3571. doi: 10.1021/bi00335a026. [DOI] [PubMed] [Google Scholar]
- Fu L. X., Waagstein F., Hjalmarson A. A new insight into adriamycin-induced cardiotoxicity. Int J Cardiol. 1990 Oct;29(1):15–20. doi: 10.1016/0167-5273(90)90267-9. [DOI] [PubMed] [Google Scholar]
- Gigli M., Doglia S. M., Millot J. M., Valentini L., Manfait M. Quantitative study of doxorubicin in living cell nuclei by microspectrofluorometry. Biochim Biophys Acta. 1988 May 6;950(1):13–20. doi: 10.1016/0167-4781(88)90068-1. [DOI] [PubMed] [Google Scholar]
- Kappus H. Overview of enzyme systems involved in bio-reduction of drugs and in redox cycling. Biochem Pharmacol. 1986 Jan 1;35(1):1–6. doi: 10.1016/0006-2952(86)90544-7. [DOI] [PubMed] [Google Scholar]
- Konopa J. Adriamycin and daunomycin induce interstrand DNA crosslinks in Hela S3 Cells. Biochem Biophys Res Commun. 1983 Feb 10;110(3):819–826. doi: 10.1016/0006-291x(83)91035-5. [DOI] [PubMed] [Google Scholar]
- Land E. J., Mukherjee T., Swallow A. J., Bruce J. M. One-electron reduction of adriamycin: properties of the semiquinone. Arch Biochem Biophys. 1983 Aug;225(1):116–121. doi: 10.1016/0003-9861(83)90013-9. [DOI] [PubMed] [Google Scholar]
- Mattes W. B. Lesion selectivity in blockage of lambda exonuclease by DNA damage. Nucleic Acids Res. 1990 Jul 11;18(13):3723–3730. doi: 10.1093/nar/18.13.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore H. W. Bioactivation as a model for drug design bioreductive alkylation. Science. 1977 Aug 5;197(4303):527–532. doi: 10.1126/science.877572. [DOI] [PubMed] [Google Scholar]
- Nielsen D., Skovsgaard T. P-glycoprotein as multidrug transporter: a critical review of current multidrug resistant cell lines. Biochim Biophys Acta. 1992 Jul 7;1139(3):169–183. doi: 10.1016/0925-4439(92)90131-6. [DOI] [PubMed] [Google Scholar]
- Pan S. S., Bachur N. R. Xanthine oxidase catalyzed reductive cleavage of anthracycline antibiotics and free radical formation. Mol Pharmacol. 1980 Jan;17(1):95–99. [PubMed] [Google Scholar]
- Ramakrishnan K., Fisher J. 7-Deoxydaunomycinone quinone methide reactivity with thiol nucleophiles. J Med Chem. 1986 Jul;29(7):1215–1221. doi: 10.1021/jm00157a017. [DOI] [PubMed] [Google Scholar]
- Sinha B. K. Binding specificity of chemically and enzymatically activated anthracycline anticancer agents to nucleic acids. Chem Biol Interact. 1980 Apr;30(1):67–77. doi: 10.1016/0009-2797(80)90115-5. [DOI] [PubMed] [Google Scholar]
- Sinha B. K., Chignell C. F. Binding mode of chemically activated semiquinone free radicals from quinone anticancer agents to DNA. Chem Biol Interact. 1979 Dec;28(2-3):301–308. doi: 10.1016/0009-2797(79)90170-4. [DOI] [PubMed] [Google Scholar]
- Sinha B. K., Gregory J. L. Role of one-electron and two-electron reduction products of adriamycin and daunomycin in deoxyribonucleic acid binding. Biochem Pharmacol. 1981 Sep 15;30(18):2626–2629. doi: 10.1016/0006-2952(81)90594-3. [DOI] [PubMed] [Google Scholar]
- Sinha B. K., Katki A. G., Batist G., Cowan K. H., Myers C. E. Differential formation of hydroxyl radicals by adriamycin in sensitive and resistant MCF-7 human breast tumor cells: implications for the mechanism of action. Biochemistry. 1987 Jun 30;26(13):3776–3781. doi: 10.1021/bi00387a006. [DOI] [PubMed] [Google Scholar]
- Sinha B. K., Sik R. H. Binding of [14C]-adriamycin to cellular macromolecules in vivo. Biochem Pharmacol. 1980 Jun 15;29(12):1867–1868. doi: 10.1016/0006-2952(80)90156-2. [DOI] [PubMed] [Google Scholar]
- Sinha B. K., Trush M. A., Kennedy K. A., Mimnaugh E. G. Enzymatic activation and binding of adriamycin to nuclear DNA. Cancer Res. 1984 Jul;44(7):2892–2896. [PubMed] [Google Scholar]
- Skladanowski A., Konopa J. Interstrand DNA crosslinking induced by anthracyclines in tumour cells. Biochem Pharmacol. 1994 Jun 15;47(12):2269–2278. doi: 10.1016/0006-2952(94)90265-8. [DOI] [PubMed] [Google Scholar]
- Skladanowski A., Konopa J. Relevance of interstrand DNA crosslinking induced by anthracyclines for their biological activity. Biochem Pharmacol. 1994 Jun 15;47(12):2279–2287. doi: 10.1016/0006-2952(94)90266-6. [DOI] [PubMed] [Google Scholar]
- Skorobogaty A., White R. J., Phillips D. R., Reiss J. A. Elucidation of the DNA sequence preferences of daunomycin. Drug Des Deliv. 1988 Jul;3(2):125–151. [PubMed] [Google Scholar]
- Weiss R. B. The anthracyclines: will we ever find a better doxorubicin? Semin Oncol. 1992 Dec;19(6):670–686. [PubMed] [Google Scholar]
- Winterbourn C. C., Vile G. F., Monteiro H. P. Ferritin, lipid peroxidation and redox-cycling xenobiotics. Free Radic Res Commun. 1991;12-13 Pt 1:107–114. doi: 10.3109/10715769109145774. [DOI] [PubMed] [Google Scholar]