Abstract
Background & Aims
Inherited mutations in the BRCA2 tumor suppressor have been associated with an increased risk of pancreatic cancer. To establish the contribution of Brca2 to pancreatic cancer we developed a mouse model of pancreas-specific Brca2 inactivation. Since BRCA2 inactivating mutations cause defects in repair of DNA double-strand breaks that result in chromosomal instability, we evaluated whether Brca2 inactivation induced instability in pancreatic tissue from these mice and whether associated pancreatic tumors were hypersensitive to DNA damaging agents.
Methods
We developed mouse models that combined pancreas-specific Kras activation and Trp53 deletion with Brca2 inactivation. Development of pancreatic cancer was assessed; tumors and non-malignant tissues were analyzed for chromosomal instability and apoptosis. Cancer cell lines were evaluated for sensitivity to DNA damaging agents.
Results
In the presence of disrupted Trp53, Brca2 inactivation promoted development of premalignant lesions and pancreatic tumors that reflected the histology of the human disease. Cancer cell lines derived from these tumors were hypersensitive to specific DNA damaging agents. In contrast, in the presence of KrasG12D, Brca2 inactivation promoted chromosomal instability and apoptosis and unexpectedly inhibited growth of premalignant lesions and tumors.
Conclusions
Trp53 signaling must be modified before inactivation of the Brca2 wild-type allele, irrespective of Kras status, for Brca2-deficient cells to form tumors.
Keywords: pancreas, cancer genetics, oncogene, neoplasia
Introduction
The BRCA2 gene encodes a 3418 amino acid protein that is a key component of the homology-directed DNA repair pathway1,2 and is involved in maintenance of structural and numerical chromosomal stability. While germline mutations in BRCA2 predispose to breast and ovarian cancer3, an increased risk (relative risk, 5.54) of pancreatic ductal adenocarcinoma (PDAC) was also observed among BRCA2 mutation carriers from 173 breast and ovarian cancer families3. In addition, germline mutations in BRCA2 are among the most common genetic lesions associated with familial pancreatic cancer4,5 and somatic BRCA2 mutations have been associated with 10% of sporadic pancreatic cancers6,7. Mutations in PALB2 and the FANCC and FANCG Fanconi anemia complex components that interact with BRCA2 have also been implicated in pancreatic cancer8. While these studies suggest a role for BRCA2 in pancreatic cancer, direct evidence of the contribution of BRCA2 to pancreatic cancer development remains to be elucidated.
Histological analysis of PDAC has identified a common pattern of disease progression from pancreatic intraepithelial neoplasia (PanIN) precursor lesions with increasingly severe stages of cellular atypia to invasive tumors9. Common genetic lesions associated with the development and progression of this disease have been identified including KRAS activating mutations in >90% of PDAC cases10 that occur early in tumor development and TP53 mutations in 50–70% of tumors11 that occur late in tumor development12, similarly to BRCA2 mutations9. Conditional activation of Kras alone and in combination with inactivation of tumor suppressors such as Trp53 in the pancreas of mouse models has been shown to promote formation of PanINs and invasive tumors13. Here we utilized these models in combination with a conditional knockout mouse model of Brca2 to demonstrate a direct role for BRCA2 in pancreatic cancer.
Results
To evaluate the role of BRCA2 in pancreatic cancer we used a mouse model expressing a functional wild type Brca2 gene, in which exon 11 of Brca2 is flanked by loxP sites (B2F11)14. Conditional rearrangement of this allele in the developing pancreas in response to pdx-1-cre(C) expression resulted in deletion of Brca2 exon 11, and the generation of a functionally null brca2 allele (B2Δ11)14 (Figure 1A). TP53 is frequently mutated in BRCA2 associated breast and ovarian tumors15, and mutations in Brca2 and Trp53 act synergistically to promote tumorigenesis in mouse mammary glands14. Therefore, we crossed CB2Δ11/Δ11 mice with conditional Trp53F2-10/F2-10 (P) mice, in which exons 2 and 10 were flanked by loxP sites14 (Figure 1A), to ultimately generate Trp53-null CPB2Δ11/Δ11, CPB2wt/Δ11 and CPB2wt/wt mice. Allele-specific PCR of DNA extracted from tail snip and pancreas DNA demonstrated that the floxed alleles of Brca2 and Trp53 were present in the tail and that these alleles were efficiently rearranged by Cre recombinase in the pancreas (Figure 1B).
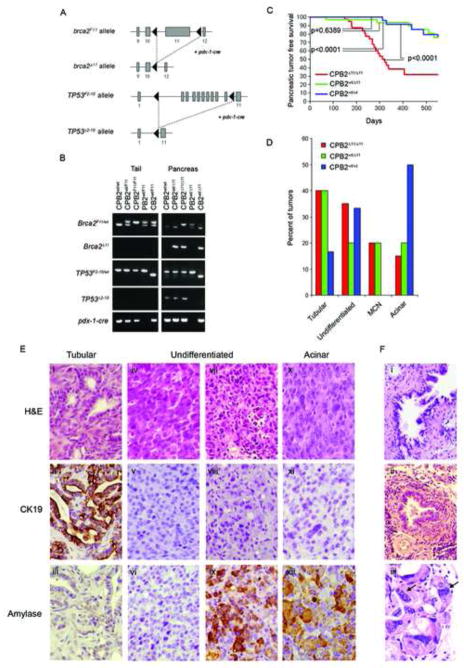
Figure 1. Brca2 inactivation promotes pancreatic cancer when combined with Trp53 inactivation.
(A) Schematic representation of the Brca2F11 allele and the Trp53F2-10 allele before and after pdx-1-cre dependent recombination. (B) PCR analysis demonstrating rearrangement of the Brca2 and Trp53 alleles in response to pdx-1-cre expression using DNA from mouse tail and pancreas. (C) Kaplan-Meier plots showing pancreatic cancer free survival of aged CPB2Δ11/Δ11 (n=47), CPB2wt/Δ11 (n=41), and CPB2wt/wt (n=34) mice. p- values were determined by a log rank test. (D) Frequency of histological subtypes of tumors detected in CPB2Δ11/Δ11, CPB2wt/Δ11 and CPB2wt/wt mice. (E) Representative images from tumors derived from CPB2Δ11/Δ11 mice stained with H&E (i,iv,vii,x), cytokeratin 19 (ii,v,viii,xi) or amylase (iii,vi,ix,xii). (F) PanIN lesions (i,ii) and multinucleated cells (iii) from a CPB2Δ11/Δ11 pancreas.
CPB2Δ11/Δ11(n=47), CPB2wt/Δ11(n=41), and CPB2wt/wt (n=34) mice were aged and evaluated for pancreatic tumor development. CPB2Δ11/Δ11 mice developed pancreatic cancer at high frequency (median survival of 300 days) and exhibited substantially reduced pancreatic cancer-free survival relative to CPB2wt/Δ11 mice (p<0.0001) and CPB2wt/wt mice (p<0.0001) (Figure 1C). In contrast, CB2Δ11/Δ11(n=12), CB2wt/Δ11(n=21), and CB2wt/wt (n=18) mice expressing wildtype Trp53 alleles failed to develop PDAC. Efficient rearrangement of alleles in these mice was verified by PCR. Histological evaluation of serial sections from all pancreas glands from the CB2Δ11/Δ11, CB2wt/Δ11 and CB2wt/wt mice at 24 months of age confirmed the absence of precursor lesions or PDAC. In addition, immunohistochemistry (IHC) with antibodies against cytokeratin 19 (CK19), amylase and insulin identified normal ductal, acinar, and islet cell components of the pancreas (Figure S1A). These findings suggest that inactivation of Brca2 alone does not promote pancreatic cancer development, but that disruption of Trp53 signaling in combination with inactivation of Brca2 significantly enhances pancreatic tumor formation. In addition, the results show that disruption of Trp53, by deletion of exons 2–10, can promote pancreatic cancer with long latency.
The pancreatic tumors observed in the CPB2Δ11/Δ11 mice were histologically similar to human pancreatic cancers. Over 40% resembled human tubular PDAC (Figure 1D) and stained positive for CK19 and negative for amylase by IHC (Figure 1E, panel ii–iii), suggesting a ductal origin. Another 15% of tumors were acinar carcinomas that stained positive for amylase and negative for CK19 (Figure 1E, panel xi–xii). A further 35% were high grade undifferentiated carcinomas. Since 50% were negative for CK19 and amylase (Figure 1E, panels v–vi) and 50% were negative for CK19 but positive for amylase (Figure 1E, panels viii–ix), the cell of origin of these tumors is uncertain. The final 20% were mucinous tumors. There was no evidence of significant desmoplastic stroma in any of these tumors. The proportion of tumors from CPB2wt/Δ11 mice in each histological subgroup was remarkably consistent with those from CPB2Δ11/Δ11 mice. However, tumors forming in CPB2wt/wt mice were predominantly acinar and undifferentiated. Since both the B2wt and B2Δ11 alleles were expressed in cell lines derived from tumors in CPB2wt/Δ11 mice (data not shown), it appears that the similarity in histology of tumors from CPB2wt/Δ11 and CPB2Δ11/Δ11 mice was not the result of somatic loss of the wildtype allele in the pancreas tissue from CPB2wt/Δ11 mice. Alternatively, since Brca2 may exhibit haploinsufficiency in murine pancreatic tissue16, it is possible that the inactivation of a single allele of Brca2 may influence the tumor histology but not tumor frequency in these mice.
Next we evaluated pancreas glands from 8 month-old mice without invasive pancreatic cancer for the presence of premalignant lesions. CPB2Δ11/Δ11 mice displayed severe acinar cell dysplasia and reduced numbers of islets (72%) (Figure 1F, panel i–ii). The pancreata were severely atrophic (60%) with acini replaced by mature adipose tissue. Mild focal acute and chronic inflammatory infiltrate (67%) with little evidence of fibrosis was also evident. In contrast, dysplasia (<8%), atrophy (<9%), and chronic inflammatory infiltrate (<14%) was less severe and less frequent in age matched CPB2wt/Δ11 and CPB2wt/wt mice. Similar evaluation of pancreatic tissue from CPB2Δ11/Δ11 mice harvested during resection of tumors or at time of death identified PanIN lesions in 66% and flat epithelial high grade dysplasia in 72% of the pancreas glands. In contrast, PanINs were observed in <6% of pancreas glands from the aged CPB2wt/Δ11 and CPB2wt/wt mice. Thus, combined disruption of Brca2 and Trp53, but not disruption of Brca2 or Trp53 alone, causes extensive remodeling of the pancreas and rapid development of premalignant and malignant lesions.
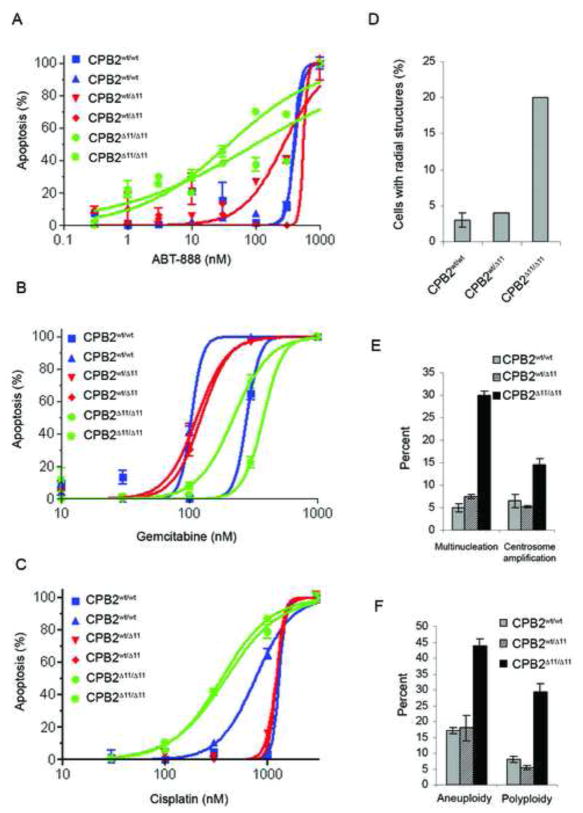
To confirm that the CPB2Δ11/Δ11 tumors displayed a BRCA2 null phenotype we characterized a series of early passage tumor cell lines (Figure S2A) from CPB2Δ11/Δ11, CPB2wt/Δ11, and CPB2wt/wt mice. Cells with defects in BRCA2 and other HR DNA repair pathway proteins display chromosomal aberrations and defective Rad51 focus formation in response to DNA damage1. Here we showed that cells from CPB2Δ11/Δ11 tumors displayed increased inter-chromosomal radial structures relative to CPB2wt/Δ11 and CPB2wt/wt cells, in response to mitomycin-c (MMC) treatment (Figure 2D and S2B). Similarly, CPB2Δ11/Δ11 cells exhibited decreased Rad51 foci, but not γH2AX foci (Figure S2C). Recently, it has been shown that cells deficient in BRCA2 are hypersensitive to poly-ADP-ribose polymerase (PARP) inhibitors17,18 and DNA cross-linking agents such as cisplatin19. Consistent with these observations, we found that CPB2Δ11/Δ11 tumor cell lines displayed increased sensitivity to the PARP inhibitor ABT-888 and to cisplatin, but not to gemcitabine (Figure 2A–C). These results suggest that these and agents that promote replication defects may be useful in treating pancreatic tumors with BRCA2 mutations.
Figure 2. Inactivation of Brca2 in pancreatic cancer cell lines induces sensitivity to DNA damage and promotes chromosomal instability.
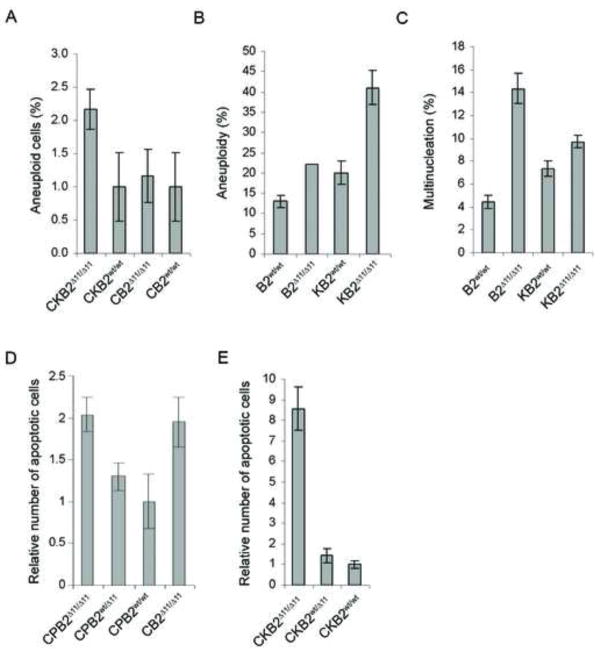
(A–C) Apoptosis in cell lines in response to ABT-888, gemcitabine, and cisplatin. (D) Quantification of radial structures in tumor cell lines treated with 100nM MMC. (E) Multinucleation and centrosome amplification in murine pancreatic tumor cell lines. (F) Aneuploidy and polyploidy in tumor cell lines with and without BRCA2.
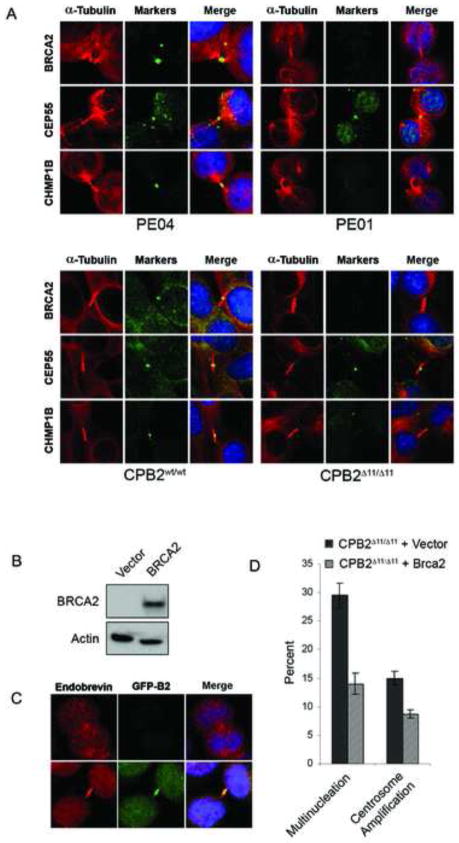
BRCA2 deficient tumors display numerical as well as structural chromosomal instability. Aneuploid cells may derive from impaired DNA damage repair and/or aberrant chromosomal segregation, whereas polyploidy cells may result from failure of cytokinesis20,21. Here immunofluorescence microscopy showed that CPB2Δ11/Δ11 tumor cell lines exhibited elevated levels of multinucleation and centrosome amplification (Figure 2E). Similarly, metaphase spreads verified increased aneuploidy and polyploidy in these cells (Figure 2F). Furthermore, multinucleated cells were frequently detected in H&E stained sections of CPB2Δ11/Δ11 tumors (Figure 1F, panel iii). Because of the significantly elevated levels of polyploidy in CPB2Δ11/Δ11 cells we investigated the influence of Brca2 on cytokinesis. We verified the absence of Brca2, but not CEP55, from the midbody in brca2Δ11/Δ11 cells by immunofluorescence staining. Similarly, endosomal membrane resorting complex (ESCRT) proteins, such as CHMP1B, that are involved in the final stage of cytokinesis, were reduced or absent from the midbodies of BRCA2 null cells (CPB2Δ11/Δ11 and PEO1 cells), relative to their wildtype counterparts (CPB2wt/wt and PEO4) (Figure 3A). Reconstitution of CPB2Δ11/Δ11 cells with GFP-tagged wildtype BRCA2 (Figure 3B and 3C), enhanced recruitment of membrane-associated endobrevin to the midbody and substantially reduced the levels of multinucleation and centrosome amplification over a 72 hr period (Figure 3D), suggesting a direct role for BRCA2 in regulation of numerical chromosomal instability.
Figure 3. Inactivation of Brca2 in pancreatic cancer cell lines disrupts localization of midbody proteins during cytokinesis.
(A) Immunofluorescence images of intercellular bridges and midbody structures in tumor cell lines stained with antibodies against α-tubulin, BRCA2, CEP55, and CHMP1B. (B) Expression of BRCA2 in a reconstituted CPB2Δ11/Δ11 cell line. (C) Immunofluorescence images showing the presence of endobrevin (red) and BRCA2 (green) at the midbody of a CPB2Δ11/Δ11 cell line (top) reconstituted with GFP-BRCA2 (bottom). (D) Multinucleation and centrosome amplification measured by immunofluorescence in a CPB2Δ11/Δ11 cell line reconstituted with empty vector or wild type BRCA2. Error bars represent SEM.
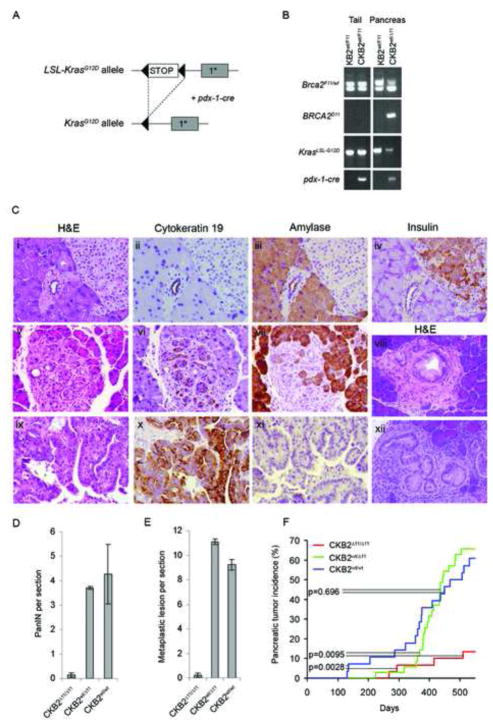
Since Brca2 deficiency in combination with inactivation of Trp53 promoted pancreatic cancer in mice, we further evaluated whether disruption of Brca2 also enhanced pancreatic tumor formation in a pdx-1-cre dependent activated KrasG12D (CK) mouse model13 (Figure 4A). Allele-specific PCR verified the presence of floxed Brca2F11 and LSL-KrasG12D alleles in the tail and cre recombinase-dependent rearranged alleles in the pancreas (Figure 4B). CKB2Δ11/Δ11, CKB2wt/Δ11 and CKB2wt/wt mice displayed normal pancreatic development and normal ductal, acinar, and islet cell architecture (Figure 4C, panels i–iv), although 20% of CKB2Δ11/Δ11 mice exhibited pancreatic insufficiency due to replacement of acinar tissue with adipose tissue at young age. Histological evaluation of serial sections from pancreas glands of 8 month old CKB2wt/Δ11 and CKB2wt/wt mice detected PanINs (Figure 4C, panel viii and xii) and metaplastic lesions (Figure 4C, panels v–vii), with an average of 4.3 and 3.7 PanIN lesions per section (Figure 4D) and an average of 10.2 and 11.1 transdifferentiation/metaplastic lesions per section22 (Figure 4E). In contrast, only 0.14 PanIN lesions and 0.24 metaplastic lesions per section were observed in CKB2Δ11/Δ11 mice (Figure 4D and 4E). Consistent with these findings, only 13% (4/30) of CKB2Δ11/Δ11 mice developed tumors (Figure 4F), whereas 66% (23/35) of CKB2wt/Δ11 (p=0.0095) and 61% (17/28) of CKB2wt/wt (p=0.0028) mice developed pancreatic tumors with an average latency of 366 and 406 days, respectively. The rate of tumor development did not differ between CKB2wt/Δ11 and CKB2wt/wt mice (p=0.696). The majority of the tumors (90%) in CKB2wt/Δ11 and CKB2wt/wt mice, and the four tumors arising in CKB2Δ11/Δ11 mice, were CK19-positive and amylase-negative pancreatic ductal adenocarcinomas (Figure 4C, panels ix–xi). Thus, loss of the Brca2 tumor suppressor gene inhibits the development of premalignant lesions and pancreatic tumors that are induced by activated Kras.
Figure 4. Brca2 mutant alleles prevent development of premalignant lesions and pancreatic tumors in LSL-KrasG12D mice.
(A) Schematic representation of the LSL-KrasG12D allele before and after pdx-1-cre dependent rearrangement. (B) PCR analysis of the Brca2F11 and KrasG12D alleles in response to pdx-1-cre expression in the mouse tail and pancreas. (C) (i)H&E, (ii)cytokeratin 19, (iii)amylase and (iv)insulin staining of a normal pancreas from a CKB2Δ11/Δ11 mouse. (v)H&E, (vi)cytokeratin 19 and (vii)amylase staining of a metaplastic lesion from a CKB2wt/wt mouse. (viii, xii)H&E staining of PanIN lesions from an 8-month old KrasG12D mouse. (ix)H&E (x)cytokeratin 19 and (xi)amylase staining of a ductal tumor from a CKB2Δ11/Δ11 mouse. (D,E) Quantitation of PanINs (D) and metaplastic lesions (E) from the pancreata of CKB2wt/wt, CKB2wt/Δ11 and CKB2Δ11/Δ11 mice. Error bars represent SEM. (F) Tumor incidence in CKB2Δ11/Δ11 mice (n=28), CKB2wt/Δ11 mice (n=35) and CKB2wt/wt mice (n=30). p- values were determined by a log rank test.
Since inactivated Brca2 inhibited KrasG12D associated pancreatic cancer but acted synergistically with disrupted Trp53 to promote pancreatic cancer, we evaluated whether Kras activation and Trp53 disruption co-occurred in tumors derived from these animal models. The four tumors from CKB2Δ11/Δ11 mice stained strongly positive for Trp53 expression suggesting the presence of Trp53 mutations. In addition, we successfully PCR amplified and sequenced all Trp53 exons from DNA extracted from one paraffin-embedded tumor and identified an alteration encoding a C239R missense mutation (Figure S3B) that was predicted by sequence conservation analysis (http://agvgd.iarc.fr) to disrupt Trp53 activity. Thus, KrasG12D tumors arising in the absence of Brca2 appeared to require inactivation of Trp53 signaling pathways. In contrast, sequencing of the Kras gene in six ductal, five undifferentiated, and two acinar tumors from CPB2Δ11/Δ11 mice yielded activating Kras mutations (G12D) in only one ductal and one undifferentiated tumor (Figure S3A), indicating that Kras activation was rarely associated with Brca2 associated pancreatic cancer.
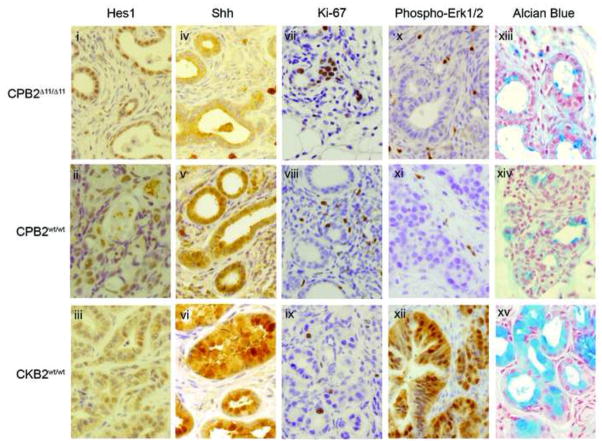
Next we evaluated biomarkers for signaling pathways commonly altered in pancreatic cancers in the tumors from the CPB2wt/wt, CPB2Δ11/Δ11, CKB2wt/wt, and CKB2Δ11/Δ11 mice. The Notch ligand and the Notch target, Hes1, have been implicated in PanIN development through induction of transdifferentiation of acinar cells to ductal-like cells13. Additionally, Sonic hedgehog (Shh) is upregulated in early PanIN lesions, and is often associated with Kras mutations in PDAC23. Hes1 expression levels in the tumors did not differ (Figure 5, panels i–iii), whereas Shh levels were higher in CKB2 tumors than in CPB2 tumors (Figure 5, panels iv–vi). The status of the brca2 gene appeared to have no effect on either Hes1 or Shh expression levels. β-catenin has been shown to inhibit Kras dependent transdifferentiation of acinar cells into PanIN lesions24. Here β-catenin expression was elevated but did not differ among the various tumors. In contrast, the neuroendocrine marker synaptophysin displayed low expression, suggesting that the tumors did not originate among islet cells (data not shown). Proliferation measured by Ki-67 staining was markedly increased in CPB2 tumors compared to CKB2 tumors, presumably due to the loss of p53 dependent cell cycle control (Figure 5, panels vii–ix). Also, CKB2 but not CPB2 tumors displayed high levels of phospho-Erk1/2, consistent with the effects of activated Kras (Figure 5, panels x–xii). Finally, alcian blue staining confirmed that the tumors and PanIN lesions in CKB2 mice but not CPB2 mice were highly mucinous (Figure 5, panels xiii–xv). These results suggest that tumors involving disruption of the Trp53 gene follow different developmental pathway from tumors associated with Kras activation.
Figure 5. Tumors from CKB2wt/wt, CPB2wt/wt and CPB2Δ11/Δ11 mice display differences in biomarker expression.
Representative images of pancreatic tumors from CPB2Δ11/Δ11, CPB2wt/wt, and CKB2wt/wt stained by IHC for (i–iii)Hes1, (iv–vi)Shh, (vii–ix)Ki-67, (x–xii)phospho-Erk1/2 and (xiii–xv)alcian blue dye.
Given the role of BRCA2 in regulation of chromosomal instability and the increased numerical chromosomal instability in CPB2Δ11/Δ11 mice, we evaluated the influence of Brca2 on instability in the presence of KrasG12D. Fluorescent in situ hybridization (FISH) studies of pancreas tissue from 8 month old mice using murine chromosome 9 and 12 centromeric probes detected elevated chromosome copy number in pancreas glands of CKB2Δ11/Δ11 mice relative to CKB2wt/wt mice (Figure 6A and S4A). This suggests that inactivation of Brca2 significantly enhanced levels of numerical chromosomal instability in vivo. Similarly, mouse embryonic fibroblasts (MEFs) from CKB2Δ11/Δ11 mice, infected with adenoviral-cre to rearrange the Brca2 and Kras loci (Figure S4B), displayed elevated levels of aneuploidy and multinucleation relative to MEFs from CKB2wt/wt mice, in both the presence and absence of KrasG12D (Figure 6B and 6C). To evaluate whether the structural and numerical chromosomal instability resulting from Brca2 deficiency resulted in elevated levels of cell death in the presence of Trp53 disruption and activated Kras, we measured in vivo apoptosis by cleaved caspase 3 staining of acinar and ductal cells in the pancreas glands of 4 month old mice. Levels of apoptosis were increased 2-fold in CPB2Δ11/Δ11 mice relative to CPB2wt/wt mice (Figure 6D), suggesting that the instability caused by absence of Brca2 enhances apoptosis. However, the levels of apoptosis were equivalent in CPB2Δ11/Δ11 and CB2Δ11/Δ11 pancreata. Thus, apoptosis resulting from Brca2 deficiency in vivo may not be dependent on Trp53 status. In contrast, 4 month old CKB2Δ11/Δ11 mice displayed 8.6-fold higher levels of in vivo apoptosis than CKB2wt/Δ11 and CKB2wt/wt mice (Figure 6E and S4C), suggesting that activated Kras and inactive Brca2 co-operate to promote cell death.
Figure 6. Brca2 mutant alleles promote chromosomal instability and apoptosis in mouse pancreatic tissue and MEFs.
(A) FISH analysis of pancreatic tissue from mice showing the percentage of cells with aneuploidy. (B) Metaphase spreads of MEFs scored for the presence of aneuploidy. (C) Percentage of MEFs with multinucleation measured by immunofluorescence microscopy after staining with α-tubulin antibody. (D,E). Quantitation of cleaved caspase 3 expression measured by IHC in the pancreas of 4 month old CPB2 and CB2 mice (D) and CKB2 (E) mice. Error bars represent SEM.
Discussion
Germline mutations in the BRCA2 gene have been observed in pancreatic cancer families and BRCA2 mutations have been detected in unselected adenocarcinomas from the pancreas, suggesting a role for BRCA2 in the development of pancreatic cancer. Here we show, using a pancreas specific knockout mouse model, that disruption of Brca2 promotes the development and progression of pancreatic cancer when combined with Trp53 inactivation, but not in the presence of active Trp53 signaling. Based on our findings we suggest a model, whereby disruption of Trp53 signaling occurs prior to inactivation of the second Brca2 allele. In this model, inactive Trp53 signaling allows pancreatic cells to evade the growth inhibitory or cell death14 effects caused by the extensive numerical and structural instability that develops in the absence of functional Brca2 protein (Figure 7). This is consistent with the presence of TP53 mutations in human PDACs containing BRCA2 mutations25. The model further suggests that loss of the wildtype BRCA2 allele in human carriers of germline BRCA2 mutations must occur late in the pancreatic tumor development process after the inactivation of TP53 signaling. Support for this comes from studies of human PDAC, which showed that the loss of heterozygosity (LOH) of BRCA2 appears to be a late event in tumorigenesis9,26.
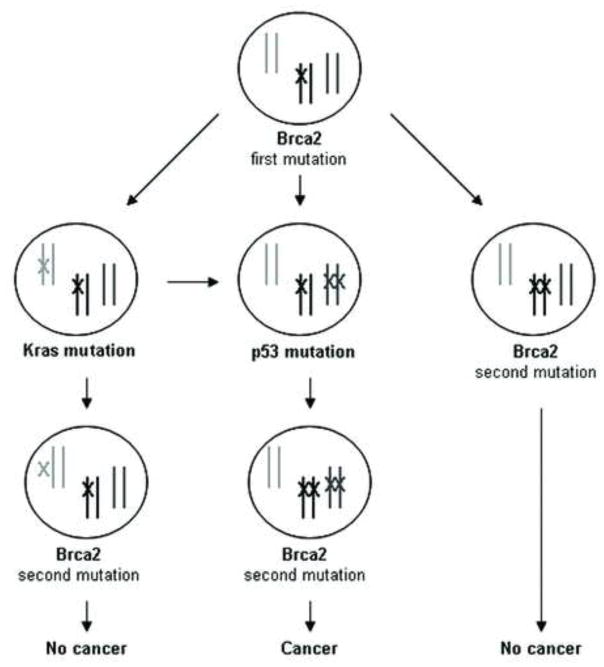
Figure 7. A model of BRCA2 deficient tumorigenesis in the pancreas.
The model reflects germline inheritance of a BRCA2 mutation. Inactivation of Tp53 signaling precedes inactivation or loss of the 2nd BRCA2 allele and facilitates cancer development. Early loss of the 2nd BRCA2 allele prior to disruption of Tp53 is inconsistent with cell growth/cell survival and tumor formation. Activation of Kras or other oncogenes prior to disruption of the 2nd BRCA2 allele is insufficient to maintain tumor formation if wildtype Tp53 signaling remains intact. Activation of oncogenes after inactivation of the 2nd BRCA2 allele and Tp53 signaling may promote tumor development.
Somewhat surprisingly our studies also showed that inactivation of Brca2 inhibits development of PanINs, metaplastic lesions and PDAC in the well-characterized pdx-1-cre;LSL-KrasG12D mouse model. This synthetic lethal effect appears to be associated with the increased chromosomal instability caused by Brca2 deficiency with some evidence suggesting a synergistic effect of Kras activation and Brca2 disruption on apoptosis (Figure 6E). Given our data suggesting that the few pancreatic tumors arising in CKB2Δ11/Δ11 mice contained Trp53 mutations, and the known presence of BRCA2, TP53 and KrasG12V mutations in the human Capan-1 pancreatic cancer cell line, the suggestion is that disruption of Trp53 signaling is again required to bypass the effects of Brca2 inactivation in cells expressing KrasG12D. While we were unable to generate sufficient numbers of CKPB2Δ11/Δ11 mice to confirm this model, a pancreas specific CKPB2Tr/Δ11 model involving a Trp53R270H allele instead of a Trp53 truncating mutation and a Brca2Tr allele that truncates Brca2 at amino acid 1492 has recently been described16. These CKPB2Tr/Δ11 mice develop pancreatic tumors at high frequency, which in part supports the model that Trp53 disruption is required for tumor formation in Brca2 deficient pancreata, both in the presence and absence of activated Kras. However, in the same report it was suggested that CKB2Tr/Δ11 mice developed pancreatic cancer in the presence of wildtype Trp53, an observation that would appear to be contrary to our proposed model. Careful examination of the presented PDAC-free Kaplan-Meier survival estimates suggests that only a small number/proportion of CKB2Tr/Δ11 mice developed pancreatic tumors16, fully consistent with the 13% tumor incidence at 500 days in our CKB2Δ11/Δ11 mice (Figure 4F). Should the tumors arising in the CKB2Tr/Δ11 mice contain Trp53 mutations or exhibit altered Trp53 signaling, similarly to the four tumors from our CKB2Δ11/Δ11 mice, then the results would further support the proposed model. Since the Trp53 status of the tumors was not reported, additional studies of pancreatic tumors arising in these mice are needed. Furthermore, whether aberrations in other regulators of apoptosis and cell cycle can rescue the effects of Brca2 deficiency remains to be determined. Taken together, our results point to critical temporal regulation of the second BRCA2 “hit” and the importance of the interplay between BRCA2 and TP53 for development of PDAC.
The variety of different tumor types observed in the CPB2Δ11/Δ11 mice suggests a high degree of plasticity among cells of the pancreas. We noted that CPB2wt/wt mice displayed predominantly acinar tumors, whereas additional inactivation of Brca2 in CPB2Δ11/Δ11 mice reduced the frequency of acinar tumors and promoted formation of ductal-like tumors. Similarly, disruption of other tumor suppressor genes in the pancreas has promoted development of other types of pancreatic tumors. In particular, pdx-1-cre;Ink4a−/−;LSL-KrasG12D mice develop tumors of spindle cell or sarcomatoid histology27, while pdx-1-cre;Smad4−/−;LSL-KrasG12D mice develop intraductal papillary mucinous neoplasia (IPMN)28. Furthermore, it is now well established that KrasG12D expression promotes transdifferentiation of acinar cells to ductal-like cells in pdx-1-cre;LSL-KrasG12D mice. Thus, the temporality of these alterations in combination with the roles of specific signaling pathways in development and differentiation may influence the histology of the resulting tumors. Alternatively, pancreatic tumors of different tumor histology may arise from effects on progenitor cells in the murine pancreas29,30. Studies have shown that expression of PyMT in the murine pancreas induces tumors with different histological features that express the pancreatic progenitor marker pdx-1 and/or markers of other cell lineages, suggesting that a progenitor cell that can differentiate into cells of different lineages may be the tumor cell of origin29.
The role of BRCA2 in homology directed repair of DNA double strand breaks is well-established. However, a role for BRCA2 in regulation of cytokinesis and cell division has also been proposed, based on frequent multinucleation in Brca2−/− ES cells, localization of BRCA2 to intercellular bridges and abnormalities in myosin II organization at the cleavage furrow following depletion of BRCA220. Here we show that up to 30% of cells from CPB2Δ11/Δ11 tumor cell lines display multinucleation and polyploidy, whereas only 5% of CPB2wt/wt tumor cells show similar effects (Figure 2E and 2F). Similarly B2Δ11/Δ11 MEFs display increases in unresolved cytokinetic bridge structures and multinucleation relative to B2wt/wt MEFs. In addition, the localization of Brca2 to the midbody and the reduced levels of membrane remodeling complexes at the midbody in response to Brca2 inactivation suggest that disruption of Brca2 may result in delays in or failure of cytokinesis because of inefficient membrane remodeling at the midbody. Our findings suggest that disruption of BRCA2 activity at the midbody may contribute to the numerical instability observed in all BRCA2 deficient cells and may contribute to tumorigenesis.
The studies reported here may have significant therapeutic implications. Specifically, we have verified that Brca2 deficient pancreatic tumors display enhanced sensitivity to cisplatin and PARP inhibitors but not DNA damaging agents such as Gemcitabine. These effects are consistent with the response to PARP inhibitors observed in a CKPB2Tr/Δ11 murine model of Brca2 deficient pancreatic cancer16 and in breast, ovarian and prostate cancer patients with germline BRCA2 mutations31. Recent phase 2 clinical trials also suggest that PARP inhibitors can be used successfully to treat cancer patients with germline mutations in BRCA1 or BRCA232. Our findings suggest that human pancreatic tumors arising in individuals with germline BRCA2 mutations may be particularly sensitive to PARP inhibitors and other agents that induce similar replication defects. The mouse model of Brca2-associated inherited pancreatic cancer described here may also prove useful for further characterization of the in vivo response to these therapeutics.
Experimental Procedures
Transgenic mice
Conditional Brca2F11, LSL-KrasG12D/+, Trp53F2-10, and pdx-1-cre mice were as previously described13,14. Mice were housed in a specific pathogen free facility and were interbred to generate the cohorts of mice used in this study. All experiments were conducted in compliance with Mayo Clinic IACUC guidelines.
Genotyping
DNA was harvested from mouse tails using the DNeasy Blood and Tissue Kit (Qiagen) and genotyped by PCR. Reaction conditions for Brca2, Trp53, and Cre were 36 cycles of 94°C for 30 sec, 56°C for 30 sec, and 72°C for 1 min. Conditions for Kras were 40 cycles of 94°C for 30 sec, 63°C for 30 sec and 72°C for 1 min. Genotyping primers are listed in supplemental information.
Histology and immunohistochemistry
Pancreatic tissue from mice was fixed in 10% formalin overnight and embedded in paraffin. Slides were deparaffinized in xylene and rehydrated. Antigen retrieval was carried out in citrate buffer. Slides were incubated with H2O2 and blocked in Rodent Block Mouse (Biocare Medical, Walnut Creek, CA) for 20 minutes. Sections were incubated in primary antibody for 60 minutes and incubated with HRP conjugated secondary antibody. Slides were further incubated in diaminobenzidine (DAB+) (DAKO) and counterstained with Modified Schmidts’ Hematoxylin.
Chromosome enumeration of metaphase spreads
1–2×106 cells were treated with 0.05 μg/ml colcemid (GIBCO BRL) for 5–6 h at 37°C. Cells were harvested, suspended in KCl and fixed in Carnoy’s solution (75% methanol, 25% acetic acid). 10–20 μl aliquots were dropped onto glass slides, stained in 5% Giemsa solution, and analyzed on an Olympus AX70 microscope using a 100× objective. For MMC response, cells were treated with 100nM MMC for 1 hour and 48 hours later were harvested for metaphase spreads.
Immunofluorescence
Cells grown on glass slides were fixed with 3% paraformaldehyde and permeabilized with 0.2% TritonX-100. Slides were blocked in 3% non-fat milk and incubated with primary antibodies overnight. Slides were then incubated with secondary antibodies for 2–3 hours, fixed with 3% paraformaldehyde and mounted with Prolong-anti-fade containing DAPI (Molecular Probes). Slides were imaged using a Zeiss LSM510 confocal microscope. Antibodies used are listed in supplemental information.
Tumor cell culture and transfection
Mouse pancreatic tumor tissue was minced and seeded in DMEM supplemented with 10% fetal bovine serum to establish tumor cell lines. Confluent cells were trypsinized and reseeded 1:3. Transfections were carried out using Lipofectamine Plus (Invitrogen).
Drug response
Mouse tumor cell lines were grown in 6-well plates in the presence of ABT-888, Gemcitabine, and cisplatin at the indicated dose for 6 days. Cells were harvested and resuspended in hypotonic propidium iodide solution (50 μg/mL propidium iodide) and analyzed by fluorescent activated cell sorting (FACS). Apoptotic cells were quantified as the percent of sub-diploid cells in the population.
Kras and Trp53 mutation screens
Tumor tissue was macrodissected from 4 to 8 unstained 5μM paraffin sections and DNA was isolated using the QIAamp DNA mini kit (Qiagen). PCR fragments containing codons 12, 13, and 61 of Kras or all exons of Trp53 were amplified with 40 cycles of 94°C for 30 sec, 56°C for 30 sec, 72°C for 1 min. PCR products were treated with ExosapIT and subjected to bidirectional DNA sequencing in the Mayo Clinic DNA sequencing core using primers listed in supplemental information.
Isolation of MEFs
Pregnant female mice were euthanized at d12.5 and embryos were surgically removed from the abdomen. Embryos were minced, transferred to a 12 well culture plate, incubated at 37°C/5% C02 for 15 minutes, and re-homogenized for 3 cycles. Cells were collected, centrifuged at 1000rpm for 5 minutes and plated in DMEM. Experiments were performed on passage 3 to 6 cells.
FISH
Centrosomal DNA probe working solution was hybridized to 5μm paraffin sections previously deparaffinized in CitriSolv and treated with 10mM Citric Acid. DAPI counterstain was applied and visualization of FISH signals was accomplished using a fluorescent microscope.
Supplementary Material
Acknowledgments
Grant Support: This study was supported by an NCI Specialized Program of Research Excellence (SPORE) grant (CA102701) in pancreatic cancer at Mayo Clinic.
Abbreviations
- B2
Brca2
- C
Cre
- K
Kras
- P
Trp53, PanIN, pancreatic intraepithelial neoplasia
- PDAC
pancreatic ductal adenocarcinoma
Footnotes
Author contributions:
MR: acquisition of data, analysis and interpretation of data, statistical analysis, drafting of the manuscript
AO, GM, LM, LY, LZ, RS, MM, TS, VS: acquisition of data; critical revision of the manuscript for important intellectual content
FJC: study concept and design, analysis and interpretation of data, critical revision of the manuscript
Disclosures: The authors have no conflicts to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7:263–72. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 2.Tutt A, Bertwistle D, Valentine J, et al. in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. Embo J. 2001;20:4704–16. doi: 10.1093/emboj/20.17.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Consortium TBCL. Cancer risks in BRCA2 mutation carriers. J Natl Cancer Inst. 1999;91:1310–6. doi: 10.1093/jnci/91.15.1310. [DOI] [PubMed] [Google Scholar]
- 4.Couch FJ, Johnson MR, Rabe K, et al. Germ line Fanconi anemia complementation group C mutations and pancreatic cancer. Cancer Res. 2005;65:383–6. [PubMed] [Google Scholar]
- 5.Murphy KM, Brune KA, Griffin C, et al. Evaluation of candidate genes MAP2K4, MADH4, ACVR1B, and BRCA2 in familial pancreatic cancer: deleterious BRCA2 mutations in 17% Cancer Res. 2002;62:3789–93. [PubMed] [Google Scholar]
- 6.Goggins M, Schutte M, Lu J, et al. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res. 1996;56:5360–4. [PubMed] [Google Scholar]
- 7.Ozcelik H, Schmocker B, Di Nicola N, et al. Germline BRCA2 6174delT mutations in Ashkenazi Jewish pancreatic cancer patients. Nat Genet. 1997;16:17–8. doi: 10.1038/ng0597-17. [DOI] [PubMed] [Google Scholar]
- 8.Jones S, Hruban RH, Kamiyama M, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hruban RH, Wilentz RE, Kern SE. Genetic progression in the pancreatic ducts. Am J Pathol. 2000;156:1821–5. doi: 10.1016/S0002-9440(10)65054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Almoguera C, Shibata D, Forrester K, et al. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53:549–54. doi: 10.1016/0092-8674(88)90571-5. [DOI] [PubMed] [Google Scholar]
- 11.Redston MS, Caldas C, Seymour AB, et al. p53 mutations in pancreatic carcinoma and evidence of common involvement of homocopolymer tracts in DNA microdeletions. Cancer Res. 1994;54:3025–33. [PubMed] [Google Scholar]
- 12.Maitra A, Adsay NV, Argani P, et al. Multicomponent analysis of the pancreatic adenocarcinoma progression model using a pancreatic intraepithelial neoplasia tissue microarray. Mod Pathol. 2003;16:902–12. doi: 10.1097/01.MP.0000086072.56290.FB. [DOI] [PubMed] [Google Scholar]
- 13.Hingorani SR, Petricoin EF, Maitra A, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–50. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 14.Jonkers J, Meuwissen R, van der Gulden H, et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–25. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 15.Crook T, Brooks LA, Crossland S, et al. p53 mutation with frequent novel condons but not a mutator phenotype in BRCA1- and BRCA2-associated breast tumours. Oncogene. 1998;17:1681–9. doi: 10.1038/sj.onc.1202106. [DOI] [PubMed] [Google Scholar]
- 16.Skoulidis F, Cassidy LD, Pisupati V, et al. Germline Brca2 Heterozygosity Promotes Kras(G12D) -Driven Carcinogenesis in a Murine Model of Familial Pancreatic Cancer. Cancer Cell. 18:499–509. doi: 10.1016/j.ccr.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 17.Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 18.Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 19.Yuan SS, Lee SY, Chen G, et al. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 1999;59:3547–51. [PubMed] [Google Scholar]
- 20.Daniels MJ, Wang Y, Lee M, et al. Abnormal cytokinesis in cells deficient in the breast cancer susceptibility protein BRCA2. Science. 2004;306:876–9. doi: 10.1126/science.1102574. [DOI] [PubMed] [Google Scholar]
- 21.Tutt A, Gabriel A, Bertwistle D, et al. Absence of Brca2 causes genome instability by chromosome breakage and loss associated with centrosome amplification. Curr Biol. 1999;9:1107–10. doi: 10.1016/s0960-9822(99)80479-5. [DOI] [PubMed] [Google Scholar]
- 22.Zhu L, Shi G, Schmidt CM, et al. Acinar cells contribute to the molecular heterogeneity of pancreatic intraepithelial neoplasia. Am J Pathol. 2007;171:263–73. doi: 10.2353/ajpath.2007.061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thayer SP, di Magliano MP, Heiser PW, et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–6. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris JPt, Cano DA, Sekine S, et al. Beta-catenin blocks Kras-dependent reprogramming of acini into pancreatic cancer precursor lesions in mice. J Clin Invest. 120:508–20. doi: 10.1172/JCI40045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rozenblum E, Schutte M, Goggins M, et al. Tumor-suppressive pathways in pancreatic carcinoma. Cancer Res. 1997;57:1731–4. [PubMed] [Google Scholar]
- 26.Goggins M, Hruban RH, Kern SE. BRCA2 is inactivated late in the development of pancreatic intraepithelial neoplasia: evidence and implications. Am J Pathol. 2000;156:1767–71. doi: 10.1016/S0002-9440(10)65047-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aguirre AJ, Bardeesy N, Sinha M, et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17:3112–26. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bardeesy N, Cheng KH, Berger JH, et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20:3130–46. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis BC, Klimstra DS, Varmus HE. The c-myc and PyMT oncogenes induce different tumor types in a somatic mouse model for pancreatic cancer. Genes Dev. 2003;17:3127–38. doi: 10.1101/gad.1140403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gidekel Friedlander SY, Chu GC, et al. Context-dependent transformation of adult pancreatic cells by oncogenic K-Ras. Cancer Cell. 2009;16:379–89. doi: 10.1016/j.ccr.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fong PC, Boss DS, Yap TA, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 32.Tutt A, Robson M, Garber JE, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 376:235–44. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.