Abstract
This article introduces a hypometabolic convergence index (HCI) for the assessment of Alzheimer’s disease (AD), compares it to other biological, cognitive and clinical measures, and demonstrate its promise to predict clinical decline in mild cognitive impairment (MCI) patients using data from the AD Neuroimaging Initiative (ADNI). The HCI is intended to reflect in a single measurement the extent to which the pattern and magnitude of cerebral hypometabolism in an individual’s fluorodeoxyglucose positron emission tomography (FDG PET) image corresponds to that in probable AD patients, and is generated using a fully automated voxel-based image analysis algorithm. HCIs, magnetic resonance imaging (MRI) hippocampal volume measurements, cerebrospinal fluid (CSF) assays, memory test scores, and clinical ratings were compared in 47 probable AD patients, 21 MCI patients who converted to probable AD within the next 18 months, 76 MCI patients who did not, and 47 normal controls (NCs) in terms of their ability to characterize clinical disease severity and predict conversion rates from MCI to probable AD. HCIs were significantly different in the probable AD, MCI converter, MCI stable and NC groups (p = 9e-17) and correlated with clinical disease severity. Using retrospectively characterized threshold criteria, MCI patients with either higher HCI’s or smaller hippocampal volumes had the highest hazard ratios (HRs) for 18-month progression to probable AD (7.38 and 6.34, respectively), and those with both had an even higher HR (36.72). In conclusion, the HCI, alone or in combination with certain other biomarker measurements, have the potential to help characterize AD and predict subsequent rates of clinical decline. More generally, our conversion index strategy could be applied to a range of imaging modalities and voxel-based image-analysis algorithms.
Keywords: hypometabolic convergence index, Alzheimer’s disease, FDG, PET, MCI, hippocampal volume
Introduction
Alzheimer’s disease (AD) is the most common form of dementia in older adults, affecting approximately 10% of adults over age 65 and almost 50% of adults over age 85 (Corrada et al., 2008; Evans et al., 1989). Mild cognitive impairment (MCI) (Petersen et al., 1999) is associated with an increased rate of progression to AD dementia and affects about 15–20% of adults over age 65 (Lopez et al., 2003). There is growing interest in using brain imaging and other biomarkers for differential diagnosis, detection, and tracking of AD, especially in its earliest non-symptomatic and symptomatic stages. Brain imaging may also help identify genetic and non-genetic risk factors, and evaluate putative AD-slowing treatments in a cost-effective way (Reiman and Langbaum, 2009).
To date, the best established biomarkers of clinical AD progression are fluorodeoxyglucose positron emission tomography (FDG-PET) measurements of the regional cerebral metabolic rate for glucose (CMRgl) decline (Alexander et al., 2002; Reiman and Langbaum, 2009; Reiman et al., 2001) and volumetric magnetic resonance imaging (MRI) measurements of hippocampal or whole brain shrinkage, regional gray matter loss and cortical thinning (Fleisher et al., 2008; Fox et al., 2000; Huang et al., 2010; Jack, Jr. et al., 1999; Jack, Jr. et al., 2008b; Whitwell et al., 2007). The best established biomarkers of AD pathology are fibrillar amyloid-β (Aβ) PET measurements using Pittsburgh Compound B (PiB) (Ikonomovic et al., 2008; Klunk et al., 2004) or other recently developed radioligand (Wong et al., 2010) and cerebrospinal fluid (CSF) amyloid-beta1–42 (Aβ1–42) levels alone or in combination with total tau (t-tau) or phosphorylated tau levels (p-tau181) (Fagan et al., 2007; Hansson et al., 2006; Li et al., 2007; Shaw et al., 2009a).
When analyzing FDG-PET or any other brain images, it may be helpful to capitalize on as much of the data in the image as possible (rather than one or more preselected regions-of-interest [ROIs]), while overcoming the problem of inflated Type I error due to multiple regional comparisons. We developed a voxel-based data analysis method that capitalizes on all data in person’s image and captures the extent to which the pattern and magnitude of a person’s brain alterations, relative to a normal control (NC) group, correspond to the pattern and magnitude of the brain alterations in AD patients.
Here we used FDG-PET data from the multi-center AD Neuroimaging Initiative (ADNI), and an automated brain-mapping algorithm (SPM) to compute an “AD-related hypometabolic convergence index (HCI)” for each person. As described below, the HCI provides a single measurement of the extent to which a person’s pattern and magnitude of cerebral hypometabolism corresponds to that in a specific group - in this case clinically diagnosed AD patients. We compared the HCI to the more widely used measurements, namely, MRI measures of hippocampal volume, CSF assays, memory test scores and clinical ratings in their ability to distinguish probable AD patients, MCI patients who converted to probable AD in the next 18 months, MCI patients who remained stable during that time, and NCs. Finally, we examined biomarker, memory test score, and clinical rating thresholds that had optimal specificity and sensitivity to predict progression from MCI to probable AD within 18 months. We compared their ability to predict rates of progression from MCI to probable AD over the same time-period.
Material and Methods
Participants
To date, ADNI has enrolled 819 adults 55–90 years of age, including 192 patients with mild probable AD, 398 patients with amnestic MCI (Petersen et al., 2001), and 229 NCs from 58 clinical sites in the United States and Canada. Mild AD patients had Mini-Mental State Examination (MMSE) (Folstein et al., 1975) scores of 20–26, a Clinical Dementia Rating (CDR) global scores (Morris, 1993) of 0.5 or 1.0, and met NINCDS/ADRDA criteria for probable AD (McKhann et al., 1984). MCI patients had MMSE scores of at least 24, a subjective memory complaint, objective memory loss measured by education adjusted scores on the Wechsler Memory Scale Logical Memory II, a CDR global score of 0.5, absence of significant levels of impairment in other cognitive domains, preserved activities of daily living (ADLs), and an absence of dementia. NCs had MMSE scores of at least 24, a CDR global core of 0, and no diagnosis of depression, MCI, or dementia. Additional inclusion and exclusion criteria, including lists of approved medications, can be found on the ADNI website (www.loni.ucla.edu/ADNI).
At the time of this analysis, an ADNI database search identified 44 AD, 97 MCI (21 of whom converted to probable AD within 18 months after baseline) and 47 NC participants with both baseline FDG-PET scans and MRI hippocampal volume data available for downloading from the ADNI Laboratory on Neuroimaging (LONI) website (www.loni.ucla.edu/ADNI/). Note that four of the NCs had converted to MCI after the baseline scan, one at month 6 (who turned to AD at month 36) and three at month 24.
Cognitive and clinical evaluations
Clinical ratings acquired at the time of each scan were used to help track the progression of cognitive impairment in each subject. They included the AD Assessment Scale-Cognitive Subscale (ADAS-Cog) (Rosen et al., 1984), Auditory Verbal Learning Test (AVLT) Total and Long-Term Memory (LTM) scores (Rey, 1941), CDR sum of boxes (SB), MMSE, Boston Naming Test-Total Score (BNT), Categorical Fluency (Animal) (Animals), Categorical Fluency (Vegetable) (Vegetables), Clock Drawing Test – Total Score (Clock), Digit Span-Forward (Digits-Forward), Digit Span-Backward (Digits-Backwards), Digit Span-Total Score (Digits-Total), Trail Making Test Part A-Time to Complete (TMT-A), and Trail Making Test Part B-Time to Complete (TMT-B). More detailed information can be found at www.adni-info.org.
FDG-PET brain imaging
Data Acquisition and Pre-processing
Each participating site acquired and reconstructed the FDG-PET data with the use of measured-attenuation correction and the specified reconstruction algorithm for each scanner type according to a standardized protocol (www.loni.ucla.edu/ADNI/Data/ADNI_Data.shtml). All images were pre-processed by ADNI PET Coordinating Center investigators at the University of Michigan and uploaded to the LONI ADNI website. These images were downloaded by investigators at Banner Alzheimer’s Institute for additional pre-processing using SPM5 (http://www.fil.ion.ucl.ac.uk/spm). Briefly, FDG-PET images were deformed into a standard space of the Talairach atlas and spatially re-smoothed with a 3D Gaussian kernel with 12-mm FWHM.
Hippocampal Volume
1.5T structural MRI scans were acquired at multiple ADNI sites using a standardized protocol described elsewhere (Jack, Jr. et al., 2008a), and corrected for image artifacts (image inhomogeneity, distortion correction, geometrical scaling) at the Mayo Clinic (Rochester, MN, USA). Bilateral hippocampal volumes were computed by ADNI investigators at University of California, San Francisco (UCSF) using Surgical Navigation Technologies (SNT, Medtronic, Louisville, CO). Anatomical boundaries were characterized using a semi-automated brain mapping method based on a high-dimensional fluid transformation algorithm (Christensen et al., 1997). This algorithm uses initially coarse and subsequently fine transformations to deform a manually drawn hippocampal MRI template onto each person’s MRI (Schuff et al., 2009a).
CSF biomarkers
Baseline CSF samples were obtained for the analysis of Aβ1–42, total tau (t-tau), and phospho-tau181 (p-tau181) levels, as well as t-tau181/ Aβ1–42 and p-tau181/ Aβ1–42 ratios in a subset of these participants, including 26 probable AD, 12 converter MCI, 42 stable MCI, and 27 NC. CSF assays were conducted at the University of Pennsylvania AD Biomarker Fluid Bank Laboratory as previously described (Shaw et al., 2009b).
Hypometabolic Convergence Index (HCI)
Normal Control (NC) Database
FDG PET images from the NCs provided the normative database needed to characterize the extent of regional CMRgl reductions in each person and compare it to the extent of regional CMRgl reductions in the probable AD group.
Map of Cerebral Hypometabolism in the Probable AD Group
The t-score map of AD-related cerebral hypometabolism was generated by comparing the FDG PET scans in the probable AD patients with those of NCs using SPM5, normalizing each image for the individual variation in whole brain measurements using proportionate scaling, and using the voxel-wise general linear model. The t-score map was transformed to a z-score map referred to as the AD hypometabolic map.
Map of Cerebral Hypometabolism in each Subject
Similarly, the t-score map of cerebral hypometabolism in each subject was generated by comparing the person’s FDG PET scan to the NCs’ scans (excluding the subject of interest) using SPM5, normalizing each image for the individual variation in whole brain measurements using proportionate scaling, and using the general linear model. In this case, the t-statistic was the ratio of difference of the subject’s CMRgl and the NC mean CMRgl over the pooled standard errors, containing only the NC group standard deviation and the available degree of freedom. Again, the resulting t-score map was transformed to a z-score map.
Computation of each Subject’s HCI
To calculate the HCI summary statistic for each subject, a new map was formed which was the product of the subject’s hypometabolic map and the AD hypometabolic map (voxel-by-voxel multiplication). The HCI was computed as the voxel-wise summation across all the voxels at which z-scores from both maps were negative, divided by 10,000 (to reduce the HCI to be in two-digit range). Thus,
where ZPi is the z-score at voxel i for person P, ZAi is the z-score of at voxel i for AD group (A), and n is the total number of voxels associated with hypometabolism in both the person of interest and the probable AD group within the whole brain volume. The HCI thus reflects the proportion of brain voxels which are both: a) hypometabolic in the subject compared to the NC group and; b) whose locations overlap with that of voxels which are hypometabolic in pAD subjects compared to the NC group, both being weighted by the product of their degree of hypometabolism in comparison to the NC group. In other words, the HCI measures how strongly the pattern of cerebral hypometabolism in the individual of interest corresponds to that in the AD group. In this way, voxels with z-scores close zero have a minor influence on the HCI, but voxels with higher hypometabolic z-scores in both the person of interest and the AD group would make a larger contribution to the HCI. The construction of individual and AD group hypometabolic maps did not involve any statistical inferences and the subsequent statistical analyses for HCI, a single numeric summary for each person, are without the need to address multiple comparisons. Also note that dividing by a scalar value 10000 does not affect any subsequent analyses of HCI, since all subjects’ HCI were scaled by the same constant value.
Statistical Analysis
Group Comparisons
We initially compared the probable AD, 18-month MCI converters, 18-month MCI stable, and NC groups in their demographic characteristics, proportion of apolipoprotein E (APOE) ε4 carriers, HCIs, other biomarker measurements, cognitive test scores and clinical ratings. We analyzed the relationship between the HCI and categorical measures of clinical AD severity (NC<MCI stable<MCI converter<probable AD) using Analysis of Variance (ANOVA) with linear trend. We also assessed HCI correlations with continuous measures of clinical AD severity (MMSE, ADAS-cog, AVLT-Total, AVLT-LTM, and CDR-SB, BNT, Animals, Vegetables, Clock, Digits-Forward, Digits-Backwards, Digits-Total, TMT-A, and TMT-B).
Predicting Rates of Progression from MCI to probable AD, Retrospective Analyses
We next compared HCIs, hippocampal volumes, CSF analytes, memory test scores and clinical ratings in their ability to predict rates of conversion from MCI to probable AD within 18 months after baseline. We initially used Receiver Operating Characteristic (ROC) analyses to determine the measurement cut-offs with maximal sensitivity and specificity in distinguishing between the MCI converters and those who were stable. We used the cut-off to compare the MCI patients with or without a positive test score (i.e., those with or without a higher HCI, smaller hippocampal volume, lower CSF Aβ1–42, a higher t-tau, p-tau181, t-tau181/ Aβ1–42, p-tau181/ Aβ1–42, a lower AVLT-Total, AVLT-LTM, or MMSE, or a higher ADAS-cog or CDR-SB, respectively) in the hazard ratio (HR) of converting from MCI to probable AD within 18 months after baseline. Thus, for each measure of interest, univariate and multivariate Cox proportional hazard models were constructed to determine the hazard ratio for progression to AD within 18 months in MCI patients who were classified into MCI patients with scores above the cut-off value in comparison with those with scores below. Since the CSF biomarkers were strongly correlated with each other, only p-tau181 was incorporated into the multivariate proportional hazard model, as it had the highest predictive value in the univariate model. Kaplan-Meier plots were used to illustrate the ability of the most significant predictors to distinguish between those MCI patients with and without a positive test.
To clarify the consistency of our HR measurements and minimize their inflation due to the use of the same subjects in both the ROC and hazard model analyses, we repeated this approach after randomly assigning 60% of the MCI patients to a training data set to compute the optimal measurement cut-offs using ROC analyses. We then computed the HRs using the empirically derived thresholds in the remaining 40% of MCI patients. The partitioning process was repeated 10 times using a random sampling method. Statistical analyses were conducted using Stata 11.0 (College Station, TX, StataCorp LP) and Statistica (Tulsa, OK, StatSoft Inc).
Results
The probable AD, MCI converter, MCI stable, and NC groups’ demographic characteristics, clinical ratings, and memory test scores, and their proportion of APOE ε4 homozygotes, heterozygotes and non-carriers are shown in Table 1. The groups did not differ in their mean age, gender distribution or educational level. As expected, the groups differed in their clinical ratings, memory test scores, and proportion of number APOE ε4 alleles.
Table 1.
Subject Characteristics, Clinical Ratings, Memory Test Scores and HCIs
| NC (n=47) |
Stable MCI (n=76) |
MCI Converter (n=21) |
pAD (n=44) |
P-value* | |
|---|---|---|---|---|---|
| Age | 75.4±4.1 | 74.9±7.5 | 77.5±6.9 | 75.4±5.7 | 0.44 |
| Gender (%F/M) | 68/32 | 71/29 | 57/43 | 59/41 | 0.45 |
| Years of Education | 15.7±3.5 | 15.8±2.9 | 16.3±2.8 | 14.7±2.8 | 0.14 |
| MMSE | 28.9±1.2 | 27.2±1.8 | 26.7±1.7 | 23.3±1.9 | 5.1e-36 |
| ADAS-cog | 7.0±3.2 | 10.1±3.8 | 13.9±4.8 | 17.8±5.6 | 4.17e-25 |
| CDR-SB | 0.0±0.1 | 1.4±0.7 | 1.9±1.1 | 4.6±1.5 | 3.4e-55 |
| AVLT-LTM | 6.9±4.3 | 3.1±3.1 | 1.6±2.5 | 0.4±1.4 | 3.3e-18 |
| APOE ε4 gene dose (%) | 0.002a | ||||
| Homozygotes | 4.3 | 13.2 | 23.8 | 25.0 | |
| Hetrozygotes | 23.4 | 35.5 | 33.3 | 47.8 | |
| Non-carriers | 72.3 | 51.3 | 42.9 | 27.3 | |
| HCI | 5.4±2.0 | 7.2±2.4 | 10.2±2.9 | 10.9±4.4 | 8.6e-17 |
Group differences were computed using one-way analysis of variance (ANOVA) or Chi-square (χ2) tests. Scheffé-multiple comparison test was used to compare the differences between each pair of means.
all pairwise differences are significant except that of MCI converter and MCI stable subjects.
As shown in Table 1 and Figure 1, HCIs were different in the four subject groups (p=8.6e-17) and were associated with categorical clinical disease severity (linear trend p=3.1e-18). In the post hoc pair-wise comparisons, the probable AD, MCI converter and MCI stable groups each had higher HCIs than the NC group (P=5e-13, 2e-8 and 0.01, respectively), the probable AD and MCI converter groups each had higher HCIs than the MCI non-converter group (P=8e-10 and 2e-4,), and HCIs in the probable AD and MCI converter groups were not different (P=0.74). As shown in Figure 2A, the locations of the voxels that contributed to a high HCI score for an AD patient (and therefore contributed to the between-group differences) are in temporal, occipital and parietal area which is consistent of the areas known to be affected by AD.
Figure 1.
Hypometabolic Convergence Index (Mean±SD) in normal controls, MCI stable subjects, MCI converters, and probable AD patients
Figure 2.
An example of the brain regions where A) an individual AD patient and B) a normal control subject shows the presence/absence of consistent hypometabolic pattern with AD. For illustration purposes, the overlap display was created with a cut-off of the ZPi • ZAi≥ 8.37. Note that this method uses all cerebral voxels without having to specify a preselected region of interest, so the search area is over the whole brain.
As shown in Table 2, HCIs were correlated with ratings of clinical disease severity and cognitive test scores (even after Bonferroni correction), with the exception of Digit Span Forward and Backward. In addition, HCIs were correlated with smaller baseline hippocampal volumes and each CSF measurement of Aβ and/or tau pathology.
Table 2.
Biomarker correlations with HCIs, clinical ratings, and memory test scores
| HCI | Hipp vol | p-tau181 | t-tau | Aβ1–42 | t-tau/Aβ 1–42 | p-tau181/Aβ 1–42 |
APOEε4 gene dose |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R | P value | r | P value | R | P value | r | P value | r | P value | R | P value | R | P value | r | P value | |
| HCI | −0.27* | 1.50e-4 | 0.37* | 7.20e-5 | 0.24 | 0.013 | −0.38* | 3.70e-5 | 0.27 | 0.004 | 0.37* | 6.30e-5 | 0.21 | 0.04 | ||
| ADAS-cog | 0.53* | 5.40e-15 | −0.42* | 1.70e-9 | 0.28 | 0.003 | 0.22 | 0.022 | −0.4* | 1.50e-5 | 0.29 | 0.003 | 0.33* | 3.60e-4 | 0.05 | 0.654 |
| CDR-SB | 0.54* | 1.10e-15 | −0.45* | 1.60e-10 | 0.26 | 0.007 | 0.36* | 1.30e-4 | −0.42* | 5.00e-6 | 0.39* | 2.60e-5 | 0.31 | 0.001 | −0.19 | 0.059 |
| MMSE | −0.48* | 4.70e-12 | 0.38* | 7.00e-8 | −0.26 | 0.006 | −0.32 | 0.001 | 0.35* | 1.70e-4 | −0.35* | 1.70e-4 | −0.31 | 0.001 | 0.02 | 0.871 |
| AVLT-Total | −0.43* | 6.20e-10 | 0.24 | 1.00e-3 | −0.31 | 0.001 | −0.27 | 0.005 | 0.3 | 0.002 | −0.3 | 0.002 | −0.33 | 4.60e-4 | 0.05 | 0.634 |
| AVLT-LTM | −0.36* | 2.90e-7 | 0.34* | 2.40e-6 | −0.33 | 5.00e-4 | −0.34* | 2.80e-4 | 0.32 | 0.001 | −0.36* | 1.20e-4 | −0.35* | 1.90e-4 | 0.12 | 0.242 |
| BNT | −0.34* | 2.10e-6 | 0.31* | 1.20e-5 | −0.14 | 0.154 | −0.18 | 0.062 | 0.2 | 0.04 | −0.18 | 0.067 | −0.15 | 0.13 | 0.08 | 0.42 |
| Animals | −0.29* | 5.60e-5 | 0.2 | 0.007 | −0.18 | 0.057 | −0.1 | 0.282 | 0.28 | 0.003 | −0.19 | 0.051 | −0.25 | 0.01 | 0.15 | 0.136 |
| Vegetables | −0.37* | 1.50e-7 | 0.21 | 0.004 | −0.25 | 0.01 | −0.16 | 0.103 | 0.36* | 1.10e-4 | −0.25 | 0.009 | −0.31 | 0.001 | −0.15 | 0.132 |
| Clock | −0.40* | 1.50e-8 | 0.18 | 0.015 | −0.05 | 0.617 | −0.14 | 0.135 | 0.21 | 0.032 | −0.16 | 0.099 | −0.08 | 0.43 | −0.03 | 0.792 |
|
Digits- Forward |
−0.08 | 2.90e-2 | 0.14 | 0.052 | −0.06 | 0.554 | −0.07 | 0.492 | 0.24 | 0.011 | −0.14 | 0.158 | −0.12 | 0.218 | −0.07 | 0.498 |
|
Digits- Backwards |
−0.25 | 4.80e-4 | 0.06 | 0.422 | −0.06 | 0.537 | −0.06 | 0.54 | 0.26 | 0.006 | −0.09 | 0.33 | −0.11 | 0.279 | −0.01 | 0.961 |
|
Digits- Total |
−0.44* | 3.40e-10 | 0.22 | 0.003 | −0.24 | 0.011 | −0.27 | 0.005 | 0.35* | 2.50e-4 | −0.22 | 0.02 | −0.21 | 0.029 | 0.13 | 0.194 |
| TMT-A | 0.44* | 3.40e-10 | 0.01 | 0.853 | 0.16 | 0.095 | 0.28 | 0.003 | −0.18 | 0.057 | 0.2 | 0.039 | 0.12 | 0.232 | −0.18 | 0.081 |
| TMT-B | 0.45* | 5.50e-11 | −0.18 | 0.014 | 0.18 | 0.066 | 0.3 | 0.002 | −0.33 | 0.001 | 0.25 | 0.01 | 0.14 | 0.137 | 0.05 | 0.644 |
indicates the correlation coefficient remain significant after Bonferroni correction
Correlations were computed using Pearson’s correlation
HCI, Hypometabolic Convergence Index; Hipp vol, Hippocampal volume; ADAS-cog, Alzheimer’s Disease Assessment Scale-Cognitive Subscale; CDR-SB, Clinical Dementia Rating Scale-Sum of Boxes; MMSE, Mini Mental State Exam; ALVT-Total, Auditory Verbal Learning Test - Total Learning; AVLT-LTM, Auditory Verbal Learning Test-Long Term Memory, BNT, Boston Naming Test-Total Score; Animals, Categorical Fluency (Animal); Vegetables, Categorical Fluency (Vegetable); Clock, Clock Drawing Test – Total Score; Digits-Forward, Digit Span-Forward; Digits-Backwards, Digit Span-Backward; Digits-Total, Digit Span-Total Score; TMT-A, Trail Making Test Part A-Time to Complete; TMT-B, Trail Making Test Part B-Time to Complete
As shown in Table 3, MCI patients who converted to probable AD within 18 months after baseline were distinguished from the stable MCI patients by significantly higher HCIs, smaller hippocampal volumes, higher ADAS-cog, CDR-SB ratings and lower AVLT-LTM scores. While there were fewer MCI patients with CSF assays, there were non-significant trends for lower Aβ1–42 levels and higher p-tau181/Aβ1–42 ratios in the MCI converters than in the MCI stable group. After controlling for higher ADAS-cog scores, the MCI converters continued to be distinguished from the stable MCI by higher HCIs (p=0.0001) and hippocampal volume (p=0.02), but not by any of the other measurements (p-value range 0.09 to 0.9).
Table 3.
Clinical Ratings, Memory Test Scores and Biomarker Measurements in MCI Converters and Stable MCIs.
| MCI Converters (n=21) |
Stable MCI (n=76) |
p-value | |
|---|---|---|---|
| ADAS-cog | 13.9±4.8 | 10.1±3.8 | 1.6e-4 |
| CDR-SB | 1.9±1.1 | 1.4±0.7 | 0.04 |
| AVLT-LTM | 1.6±2.5 | 3.1±3.1 | 0.05 |
| HCI | 10.2±2.97 | 7.2±2.4 | 4e-6 |
| Hipp Volume (mm3) | 3231±597 | 3836±766 | 0.001 |
| APOE ε4 carrier status (%) | 0.47 | ||
| Homozygotes | 23.8 | 13.2 | |
| Heterozygotes | 33.3 | 35.5 | |
| Non-carriers | 42.9 | 51.3 | |
| CSF | (n=12) | (n=42) | |
| Aβ1–42 | 148.83±45.04 | 174.61 ±55.87 | 0.08 |
| p-tau181 | 37.33±10.49 | 31.08±15.99 | 0.13 |
| t-tau | 89.78±23.81 | 89.00±41.55 | 0.94 |
| p-tau181/Aβ1–42 | 0.28±0.12 | 0.21±0.15 | 0.09 |
| t-tau/Aβ1–42 | 0.65±0.25 | 0.58±0.39 | 0.48 |
Group differences were computed using two-sample t-tests or Chi-square (χ2) tests.
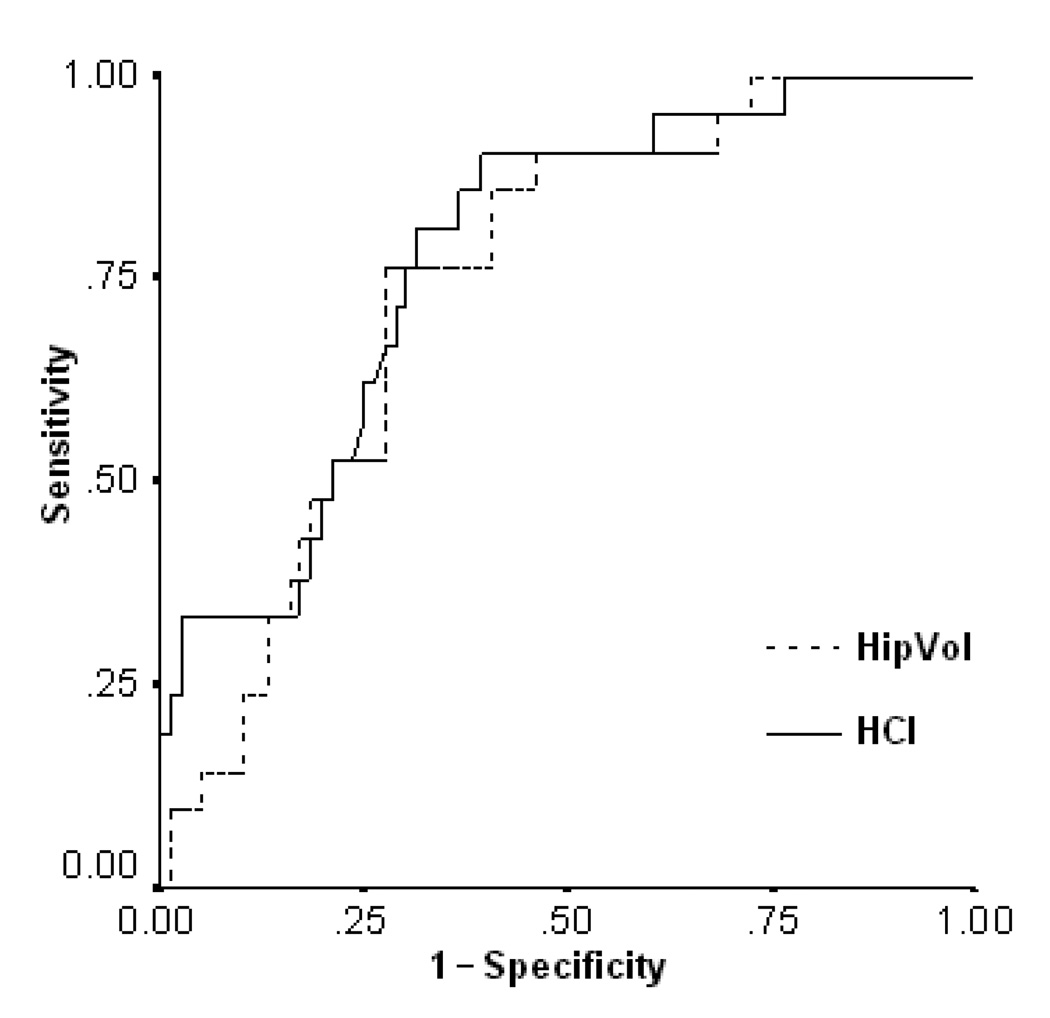
The biomarker, clinical rating, and memory test cut-off values, area-under-curves (AUC) and their 95% confidence interval found using ROC analyses to distinguish with optimal sensitivity and specificity those MCI patients who did or did not convert to probable AD within 18 months after baseline are shown in Table 4. For further performance comparison, ROCs of HCI and hippocampal volume are shown in Figure 3. Using those cut-offs to sub-divide the MCI patients into those with and without positive scores, Table 5 shows the estimated hazard risk of converting to probable AD in those MCI patients who were positive on each of the measurements. When each of the candidate predictors was analyzed independently, the MCI patients with a positive (i.e., higher) HCI or positive (i.e., smaller) hippocampal volume had the highest HRs of converting to probable AD within 18 months after baseline (7.38, 95% CI=2.48–21.98 and 6.34, 95% CI=2.32–17.36, respectively). When all of the candidate predictors were included in the same model, only the HCI and hippocampal volume remained significantly associated with a higher HR of conversion to probable AD.
Table 4.
The biomarker, clinical rating, and memory test cut-off values found using ROC analyses to distinguish with optimal sensitivity and specificity those MCI patients who did or do not convert to probable AD within 18 months
| Predictors | Cut-off values | AUC(95% CI) | Asymptotic Significance |
|---|---|---|---|
| ADAS-cog | 11.84 | 0.74 (0.62–0.86) | 7.5e-4 |
| CDR-SB | 2.25 | 0.62 (0.47–0.76) | 0.11 |
| AVLT-LTM | 1.5 | 0.66 (0.53–0.80) | 0.02 |
| HCI | 8.36 | 0.78 (0.68–0.88) | 9.4e-5 |
| Hipp Volume (mm3) | 3554.72 | 0.75 (0.64–0.85) | 6.0e-4 |
| APOE ε4 carrier status | APOE carrier | 0.56 (0.38–0.42) | 0.38 |
| CSF | |||
| Aβ1–42 | 148 | 0.59 (0.43–0.76) | 0.31 |
| p-tau181 | 32 | 0.66(0.50–0.81) | 0.10 |
| t-tau | 81 | 0.57(0.42–0.72) | 0.46 |
| p-tau181/Aβ1–42 | 0.23 | 0.65(0.50–0.81) | 0.11 |
| t-tau/Aβ1–42 | 0.56 | 0.61(0.45–0.76) | 0.27 |
Figure 3.
Receiver Operating Curves showing the sensitivity and 1-specificity of the HCI and hippocampal volume
Table 5.
Estimated risk of 18-month conversion to probable AD in MCI patients with positive versus negative test measurements
| Univariate | Multivariate | ||
|---|---|---|---|
| HCI |
HR 95%CI |
7.38* 2.48–21.98 |
6.55* 2.19–19.61 |
| Hippocampal volume |
HR 95%CI |
6.34* 2.32–17.36 |
5.60* 2.04–15.41 |
|
APOE ε4 carrier status |
HR 95%CI |
1.33 0.56–3.16 |
NS |
| Aβ1–42** |
HR 95% CI |
2.31 0.69–7.67 |
|
| p-tau181** |
HR 95%CI |
4.94* 1.34–18.29 |
NS |
| t-tau** |
HR 95% CI |
2.39 0.72–7.96 |
|
| p-tau181/Aβ1–42** |
HR 95% CI |
3.81* 1.03–14.10 |
|
| t-tau/Aβ1–42** |
HR 95% CI |
1.90 0.57–6.33 |
|
| ADAS-cog |
HR 95% CI |
3.91* 1.58–9.69 |
NS |
| AVLT-LTM |
HR 95% CI |
2.78* 1.08–7.20 |
NS |
| CDR-SB |
HR 95% CI |
3.70* 1.51–9.31 |
NS |
Hazards Risks (HR) were estimated using the Cox Proportional Hazard Model
denotes statistical significance at p<0.05.
denotes that the analysis is conducted using 42 stable MCI and 12 converter MCI due to limited availability of the data.
After the non-significant predictors were removed and the multivariate model was refitted, 26 MCI patients with a higher HCI (27% of MCI patients) had an HR of 6.55 (95% CI=2.19–19.61) of conversion to probable AD within 18 months after baseline (Figure 4a), 21 MCI patients with a smaller hippocampal volume (22%) had an HR of 5.60 (95% CI=2.04–15.41) (Figure 4b), and 13 MCI patients with both a higher HCI and smaller hippocampal volume (13%) had an HR of 36.72 (95%CI=4.46–302.34) compared to those with both a lower HCI and larger hippocampal volume (20 and 38 MCI patients or 21% and 39% of the MCI patients) (Figure 4d). In comparison with all of the MCI participants, those with both a higher HCI and hippocampal volume had an HR of 10.17 times (95% CI=4.17–24.86) (Figure 4c).
Figure 4.
Kaplan-Meier curves showing the probability of an MCI patient not converting to probable AD within 18 months from baseline in a) the 26 MCI patients with a higher HCI versus the 71 MCI patients with a lower HCI, b) the 21 MCI patients with a smaller hippocampal volume versus 76 MCI patients with a larger hippocampal volume, and c) the 20 MCI patients with both a higher HCI and smaller hippocampal volume versus the 77 other MCI patients, d) the 20 MCI patients with both a higher HCI and smaller hippocampal volume versus the 38 MCI patients with both a lower HCI and larger hippocampal volume.
When the analyses were performed iteratively using training set data to characterize the cut-off values and independent test set data to characterize HRs, the findings were generally consistent with our original analysis: the MCI patients with a higher HCI (average HR=5.32) or smaller hippocampal volume (average HR=4.77) had the highest average HRs of converting to probable AD within 18 months after baseline. Average HRs were 1.54 for those with lower Aβ1–42 levels, 1.62 for t-tau levels, 3.11 for p-tau181 levels, 1.29 for t-tau/Aβ1–42 ratios, 2.44 for p-tau181/Aβ1–42 ratios, 3.58 for ADAS-cog scores, 2.40 for CDR-SB scores, 2.55 for AVLT-LTM scores, and 2.14 for APOE ε4 carriers.
Because there was concern of including subjects who subsequently converted to MCI in the normal database, we re-computed the HCIs without these 4 subjects in the normal database and compared the results to those with the 4 subjects included. The re-computed HCIs were highly correlated to the original HCI computed with the 4 subjects (Pearson correlation r>0.9999), suggesting that to some degree, the HCI is robust to changes in the normal database.
Discussion
This study introduces the concept and use of the AD-related HCI. Using data from ADNI, we showed it could distinguish between probable AD patients, MCI patients who did or did not convert within 18 months after baseline, and NCs. HCIs were closely associated with categorical measures of disease severity and significantly correlated with other AD biomarkers. Finally, we demonstrate possible advantages of using HCIs, alone or in combination with hippocampal volumes, over other promising biomarkers, to predict rates of clinical progression from MCI to probable AD.
The HCI provides a single measurement, free from multiple comparisons. It capitalizes on all of the cerebral data in the individual’s FDG PET image (without having to specify a preselected region of interest), as well as that from normative and AD-related FDG PET databases. It is generated using a fully automated voxel-based image-analysis technique, and has been shown to work well using data acquired at different imaging centers and from different laboratories. This approach could be applied to a wide variety of imaging methods and voxel-based image-analysis techniques. It could not only be used to determine the extent to which the magnitude and pattern of brain differences in a person’s image corresponds to that in AD, but the extent to which the magnitude and pattern corresponds to other pathological conditions (e.g,, that associated with frontotemporal dementia or neurological or psychiatric disorders) and other normal conditions (e.g., that associated with normal aging).
Additional studies are needed to determine the extent to which the HCI could be used, alone or in combination with other information, in the differential diagnosis, early detection and tracking of AD. The HCI may also assist with enrichment or stratification of subjects in clinical trials or as an endpoint in clinical trials. Preliminary data from our laboratory suggests it can distinguish cognitively normal late-middle-aged APOE ε4 homozygote, heterozygote, and non-carrier groups, and that it has the potential to track AD progression and evaluate AD-modifying treatments in clinical trials (unpublished data)—all free from multiple comparisons.
Limitations of this study include the relatively small number of subjects for whom both FDG PET and MRI hippocampal volume were available, the even smaller sample with CSF measures, the absence of PET measures of Aβ burden, and the retrospective HR analysis. As we used the same MCI patients to characterize the thresholds used to subdivide the MCI patients based on “positive” or “negative” biomarker, clinical rating and memory test values and then used these values to compute HRs. On the other hand, the study capitalized on the same strategy to compare HRs using these different candidate predictors of clinical progression. We confirmed this when the threshold values and HRs were computed in independent training and test data sets. Still, additional studies are needed to clarify the extent to which the HCI predicts rates of conversion, not only within the 18-month but over an extended period of time, and how it compares to other biomarker and psychometric predictors. Moreover, additional studies in longitudinally assessed and neuropthologically verified subjects are needed to clarify further the extent to which the HCI, alone or in combination with clinical, cognitive, or other image-based biomarker endpoints, predicts subsequent cognitive decline and distinguishes AD from other neuropathologically verified causes of cognitive impairment.
As we have previously noted, FDG PET changes in AD could be related to a reduction in the density or activity of terminal neuronal fields, peri-synaptic astroglial cells, metabolic dysfunction, the combined effects of brain atrophy and partial-volume averaging, or a combination of these factors. Based on findings from previous studies, these changes do not appear to be solely attributable to the combined effects of atrophy and partial volume averaging (Bokde et al., 2001; Ibanez et al., 1998; Reiman et al., 2004; Reiman et al., 2005; Sakamoto et al., 2003). No attempt was made to further address the contribution of brain atrophy to the HCI in this study, as our primary aim was to provide a single measurement for the detection and future tracking of AD rather than to clarify the biological contributions to this measurement.
Please note our ability to find such a strong association between the HCI, based in regional-to-whole brain FDG count ratios in each cerebral voxel, and AD. While the well established association between whole brain CMRgl reductions and clinical severity would cause us to underestimate reductions in regional-to-whole brain FDG count ratios (Borghammer et al., 2008; Borghammer et al., 2009a; Borghammer et al., 2009b; Yakushev et al., 2008), the HCI was still able to characterize the preferential pattern of CMRgl reductions in AD-affected brain regions.
This study supports the complementary value of hippocampal volumes in the detection of AD and the prediction of subsequent clinical decline in MCI patients (Beckett et al., 2010; Chupin et al., 2009; Hua et al., 2008; Kohannim et al., 2010; Landau et al., 2010; Leung et al., 2010; Lorenzi et al., 2010; Morra et al., 2008; Risacher et al., 2009; Schott et al., 2010; Schuff et al., 2009b), even though smaller hippocampal volumes may also be found in patients with other neurodegenerative disorders (e.g., van de Pol et al., 2006). Please note, however, that all hippocampal measurements are not alike. It has previously been shown that Freesurfer measurements of cortical thinning in the hippocampus are superior to SNT measurements of hippocampal volume in the longitudinal tracking of AD and, thus, may be more suitable as an endpoint in clinical trials of AD-modifying treatments. In contrast, a post hoc analysis of more recently uploaded data suggests that freesurfer-based hippocampal measurements are associated with a lower HR for 18-month progression to probable AD than the SNT hippocampal volumes used here (HR=4.12 versus 6.34).
In addition to relating and comparing FDG-PET-based HCI with MRI-based volume measurements of hippocampus and others investigated in this study, it will also be informative to examine the relationship between amyloid PET(such as PIB-PET) and FDG-PET. However, given that only 2 of the subjects (1 AD and 1 MCI patient) in our study had PIB-PET data, we must await the acquisition from ongoing ADNI Grand Opportunity and ADNI2 studies, which will provide florbetapir F18 PET measurements of fibrillar amyloid in every subject.
The proposed HCI is similar with the FDG PET indicator of AD described by Herholz et al (Herholz et al., 2002). They computed the sum of hypometabolic T-scores exceeding 95% of an age-adjusted normal distribution in those voxels shown to be hypometabolic in AD using the same threshold and was shown to distinguish between AD patients and NCs with high sensitivity and specificity. Unlike the HCI, it does not include information on the CMRgl reduction in voxels outside of this threshold. Direct comparison between these two approaches requires separate studies.
In conclusion, the automatically generated HCI characterizes the extent to which the pattern and magnitude of cerebral hypometabolism in a person’s FDG PET image corresponds to that in AD patients. It offers promise for the differential diagnosis, early detection and tracking, the prediction of clinical decline in MCI patients, and in the cost-effective evaluation of AD-modifying treatments.
Acknowledgements
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI; Principal Investigator: Michael Weiner; NIH grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering (NIBIB), and through generous contributions from the following: Pfizer Inc., Wyeth Research, Bristol-Myers Squibb, Eli Lilly and Company, GlaxoSmithKline, Merck & Co. Inc., AstraZeneca AB, Novartis Pharmaceuticals Corporation, Alzheimer’s Association, Eisai Global Clinical Development, Elan Corporation plc, Forest Laboratories, and the Institute for the Study of Aging, with participation from the U.S. Food and Drug Administration. Industry partnerships are coordinated through the Foundation for the National Institutes of Health. The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory of NeuroImaging at the University of California, Los Angeles.
This work was also partly supported by grants from the National Institute on Aging (R01AG031581, P30AG19610, R01AG025526 ), the National Institute of Mental Health (R01MH057899), the Evelyn G. McKnight Brain Institute (GEA), the state of Arizona (EMR, RJC, GEA, KC), and contributions from the Banner Alzheimer’s Foundation and Mayo Clinic Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (www.loni.ucla.edu/ADNI). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in the analyses or writing of this report. The ADNI investigator list is at www.loni.ucla.edu/ADNI/Data/ADNI_Authorship_List.pdf.
References
- Alexander GE, Chen K, Pietrini P, Rapoport SI, Reiman EM. Longitudinal PET evaluation of cerebral metabolic decline in dementia: A potential outcome measure in Alzheimer’s disease treatment studies. Am.J.Psychiatry. 2002;159:738–745. doi: 10.1176/appi.ajp.159.5.738. [DOI] [PubMed] [Google Scholar]
- Beckett LA, Harvey DJ, Gamst A, Donohue M, Kornak J, Zhang H, Kuo JH. The Alzheimer’s Disease Neuroimaging Initiative: Annual change in biomarkers and clinical outcomes. Alzheimers.Dement. 2010;6:257–264. doi: 10.1016/j.jalz.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokde AL, Pietrini P, Ibanez V, Furey ML, Alexander GE, Graff-Radford NR, Rapoport SI, Schapiro MB, Horwitz B. The effect of brain atrophy on cerebral hypometabolism in the visual variant of Alzheimer disease. Arch.Neurol. 2001;58:480–486. doi: 10.1001/archneur.58.3.480. [DOI] [PubMed] [Google Scholar]
- Borghammer P, Aanerud J, Gjedde A. Data-driven intensity normalization of PET group comparison studies is superior to global mean normalization. Neuroimage. 2009a;46:981–988. doi: 10.1016/j.neuroimage.2009.03.021. [DOI] [PubMed] [Google Scholar]
- Borghammer P, Cumming P, Aanerud J, Forster S, Gjedde A. Subcortical elevation of metabolism in Parkinson’s disease--a critical reappraisal in the context of global mean normalization. Neuroimage. 2009b;47:1514–1521. doi: 10.1016/j.neuroimage.2009.05.040. [DOI] [PubMed] [Google Scholar]
- Borghammer P, Jonsdottir KY, Cumming P, Ostergaard K, Vang K, Ashkanian M, Vafaee M, Iversen P, Gjedde A. Normalization in PET group comparison studies--the importance of a valid reference region. Neuroimage. 2008;40:529–540. doi: 10.1016/j.neuroimage.2007.12.057. [DOI] [PubMed] [Google Scholar]
- Christensen GE, Joshi SC, Miller MI. Volumetric transformation of brain anatomy. IEEE Trans.Med.Imaging. 1997;16:864–877. doi: 10.1109/42.650882. [DOI] [PubMed] [Google Scholar]
- Chupin M, Gerardin E, Cuingnet R, Boutet C, Lemieux L, Lehericy S, Benali H, Garnero L, Colliot O. Fully automatic hippocampus segmentation and classification in Alzheimer’s disease and mild cognitive impairment applied on data from ADNI. Hippocampus. 2009;19:579–587. doi: 10.1002/hipo.20626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrada MM, Brookmeyer R, Berlau D, Paganini-Hill A, Kawas CH. Prevalence of dementia after age 90. Results from The 90+ Study. Neurology. 2008;71:337–343. doi: 10.1212/01.wnl.0000310773.65918.cd. [DOI] [PubMed] [Google Scholar]
- Evans DA, Funkenstein HH, Albert MS, Scherr PA, Cook NR, Chown MJ, Hebert LE, Hennekens CH, Taylor JO. Prevalence of Alzheimer’s disease in a community population of older persons. Higher than previously reported. JAMA. 1989;262:2551–2556. [PubMed] [Google Scholar]
- Fagan AM, Roe CM, Xiong C, Mintun MA, Morris JC, Holtzman DM. Cerebrospinal fluid tau/beta-amyloid(42) ratio as a prediction of cognitive decline in nondemented older adults. Arch.Neurol. 2007;64:343–349. doi: 10.1001/archneur.64.3.noc60123. [DOI] [PubMed] [Google Scholar]
- Fleisher AS, Sun S, Taylor C, Ward CP, Gamst AC, Petersen RC, Jack CR, Jr, Aisen PS, Thal LJ. Volumetric MRI vs clinical predictors of Alzheimer disease in mild cognitive impairment. Neurology. 2008;70:191–199. doi: 10.1212/01.wnl.0000287091.57376.65. [DOI] [PubMed] [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR. "Mini-mental state". A practical method for grading the cognitive state of patients for the clinician. J Psychiatr.Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Fox NC, Cousens S, Scahill R, Harvey RJ, Rossor MN. Using serial registered brain magnetic resonance imaging to measure disease progression in Alzheimer disease: power calculations and estimates of sample size to detect treatment effects. Arch.Neurol. 2000;57:339–344. doi: 10.1001/archneur.57.3.339. [DOI] [PubMed] [Google Scholar]
- Hansson O, Zetterberg H, Buchhave P, Londos E, Blennow K, Minthon L. Association between CSF biomarkers and incipient Alzheimer’s disease in patients with mild cognitive impairment: a follow-up study. Lancet Neurol. 2006;5:228–234. doi: 10.1016/S1474-4422(06)70355-6. [DOI] [PubMed] [Google Scholar]
- Herholz K, Salmon E, Perani D, Baron JC, Holthoff V, Frolich L, Schonknecht P, Ito K, Mielke R, Kalbe E, Zundorf G, Delbeuck X, Pelati O, Anchisi D, Fazio F, Kerrouche N, Desgranges B, Eustache F, Beuthien-Baumann B, Menzel C, Schroder J, Kato T, Arahata Y, Henze M, Heiss WD. Discrimination between Alzheimer dementia and controls by automated analysis of multicenter FDG PET. Neuroimage. 2002;17:302–316. doi: 10.1006/nimg.2002.1208. [DOI] [PubMed] [Google Scholar]
- Hua X, Leow AD, Parikshak N, Lee S, Chiang MC, Toga AW, Jack CR, Jr, Weiner MW, Thompson PM. Tensor-based morphometry as a neuroimaging biomarker for Alzheimer’s disease: an MRI study of 676 AD, MCI, and normal subjects. Neuroimage. 2008;43:458–469. doi: 10.1016/j.neuroimage.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Li J, Sun L, Ye J, Fleisher A, Wu T, Chen K, Reiman E. Learning brain connectivity of Alzheimer’s disease by sparse inverse covariance estimation. Neuroimage. 2010;50:935–949. doi: 10.1016/j.neuroimage.2009.12.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez V, Pietrini P, Alexander GE, Furey ML, Teichberg D, Rajapakse JC, Rapoport SI, Schapiro MB, Horwitz B. Regional glucose metabolic abnormalities are not the result of atrophy in Alzheimer’s disease. Neurology. 1998;50:1585–1593. doi: 10.1212/wnl.50.6.1585. [DOI] [PubMed] [Google Scholar]
- Ikonomovic MD, Klunk WE, Abrahamson EE, Mathis CA, Price JC, Tsopelas ND, Lopresti BJ, Ziolko S, Bi W, Paljug WR, Debnath ML, Hope CE, Isanski BA, Hamilton RL, DeKosky ST. Post-mortem correlates of in vivo PiB-PET amyloid imaging in a typical case of Alzheimer’s disease. Brain. 2008;131:1630–1645. doi: 10.1093/brain/awn016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, Whitwell L, Ward C, Dale AM, Felmlee JP, Gunter JL, Hill DL, Killiany R, Schuff N, Fox-Bosetti S, Lin C, Studholme C, DeCarli CS, Krueger G, Ward HA, Metzger GJ, Scott KT, Mallozzi R, Blezek D, Levy J, Debbins JP, Fleisher AS, Albert M, Green R, Bartzokis G, Glover G, Mugler J, Weiner MW. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J.Magn Reson.Imaging. 2008a;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Lowe VJ, Senjem ML, Weigand SD, Kemp BJ, Shiung MM, Knopman DS, Boeve BF, Klunk WE, Mathis CA, Petersen RC. 11C PiB and structural MRI provide complementary information in imaging of Alzheimer’s disease and amnestic mild cognitive impairment. Brain. 2008b;131:665–680. doi: 10.1093/brain/awm336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Petersen RC, Xu YC, O'Brien PC, Smith GE, Ivnik RJ, Boeve BF, Waring SC, Tangalos EG, Kokmen E. Prediction of AD with MRI-based hippocampal volume in mild cognitive impairment. Neurology. 1999;52:1397–1403. doi: 10.1212/wnl.52.7.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klunk WE, Engler H, Nordberg A, Wang Y, Blomqvist G, Holt DP, Bergstrom M, Savitcheva I, Huang GF, Estrada S, Ausen B, Debnath ML, Barletta J, Price JC, Sandell J, Lopresti BJ, Wall A, Koivisto P, Antoni G, Mathis CA, Langstrom B. Imaging brain amyloid in Alzheimer’s disease with Pittsburgh Compound-B. Ann.Neurol. 2004;55:306–319. doi: 10.1002/ana.20009. [DOI] [PubMed] [Google Scholar]
- Kohannim O, Hua X, Hibar DP, Lee S, Chou YY, Toga AW, Jack CR, Jr, Weiner MW, Thompson PM. Boosting power for clinical trials using classifiers based on multiple biomarkers. Neurobiol.Aging. 2010;31:1429–1442. doi: 10.1016/j.neurobiolaging.2010.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau SM, Harvey D, Madison CM, Reiman EM, Foster NL, Aisen PS, Petersen RC, Shaw LM, Trojanowski JQ, Jack CR, Jr, Weiner MW, Jagust WJ. Comparing predictors of conversion and decline in mild cognitive impairment. Neurology. 2010;75:230–238. doi: 10.1212/WNL.0b013e3181e8e8b8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KK, Barnes J, Ridgway GR, Bartlett JW, Clarkson MJ, Macdonald K, Schuff N, Fox NC, Ourselin S. Automated cross-sectional and longitudinal hippocampal volume measurement in mild cognitive impairment and Alzheimer’s disease. Neuroimage. 2010;51:1345–1359. doi: 10.1016/j.neuroimage.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Sokal I, Quinn JF, Leverenz JB, Brodey M, Schellenberg GD, Kaye JA, Raskind MA, Zhang J, Peskind ER, Montine TJ. CSF tau/Abeta42 ratio for increased risk of mild cognitive impairment: a follow-up study. Neurology. 2007;69:631–639. doi: 10.1212/01.wnl.0000267428.62582.aa. [DOI] [PubMed] [Google Scholar]
- Lopez OL, Jagust WJ, DeKosky ST, Becker JT, Fitzpatrick A, Dulberg C, Breitner J, Lyketsos C, Jones B, Kawas C, Carlson M, Kuller LH. Prevalence and classification of mild cognitive impairment in the Cardiovascular Health Study Cognition Study: part 1. Arch Neurol. 2003;60:1385–1389. doi: 10.1001/archneur.60.10.1385. [DOI] [PubMed] [Google Scholar]
- Lorenzi M, Donohue M, Paternico D, Scarpazza C, Ostrowitzki S, Blin O, Irving E, Frisoni GB. Enrichment through biomarkers in clinical trials of Alzheimer’s drugs in patients with mild cognitive impairment. Neurobiol.Aging. 2010;31:1443–1451. doi: 10.1016/j.neurobiolaging.2010.04.036. 1451. [DOI] [PubMed] [Google Scholar]
- McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Hua X, Toga AW, Jack CR, Jr, Weiner MW, Thompson PM. Validation of a fully automated 3D hippocampal segmentation method using subjects with Alzheimer’s disease mild cognitive impairment, and elderly controls. Neuroimage. 2008;43:59–68. doi: 10.1016/j.neuroimage.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JC. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology. 1993;43:2412–2414. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- Petersen RC, Doody R, Kurz A, Mohs RC, Morris JC, Rabins PV, Ritchie K, Rossor M, Thal L, Winblad B. Current concepts in mild cognitive impairment. Arch.Neurol. 2001;58:1985–1992. doi: 10.1001/archneur.58.12.1985. [DOI] [PubMed] [Google Scholar]
- Petersen RC, Smith GE, Waring SC, Ivnik RJ, Tangalos EG, Kokmen E. Mild cognitive impairment: clinical characterization and outcome. Arch Neurol. 1999;56:303–308. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- Reiman EM, Langbaum JBS. Brain imaging in the evaluation of putative Alzheimer’s disease slowing, risk-reducing and prevention therapies. In: Jagust WJ, D’Esposito M, editors. Imaging the Aging Brain. New York: Oxford University Press; 2009. pp. 319–350. [Google Scholar]
- Reiman EM, Caselli RJ, Chen K, Alexander GE, Bandy D, Frost J. Declining brain activity in cognitively normal apolipoprotein E ε4 heterozygotes: A foundation for using positron emission tomography to efficiently test treatments to prevent Alzheimer’s disease. Proc.Natl.Acad.Sci.U.S.A. 2001;98:3334–3339. doi: 10.1073/pnas.061509598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, Chen K, Alexander GE, Caselli RJ, Bandy D, Osborne D, Saunders AM, Hardy J. Functional brain abnormalities in young adults at genetic risk for late-onset Alzheimer’s dementia. Proc.Natl.Acad.Sci.U.S.A. 2004;101:284–289. doi: 10.1073/pnas.2635903100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, Chen K, Alexander GE, Caselli RJ, Bandy D, Osborne D, Saunders AM, Hardy J. Correlations between apolipoprotein E epsilon4 gene dose and brain-imaging measurements of regional hypometabolism. Proc.Natl.Acad.Sci.U.S.A. 2005;102:8299–8302. doi: 10.1073/pnas.0500579102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey A. L’examen psychologique dans les cas d’encephalopathie tramatique. Archives de Psychologie. 1941;28:215–285. [Google Scholar]
- Risacher SL, Saykin AJ, West JD, Shen L, Firpi HA, McDonald BC. Baseline MRI predictors of conversion from MCI to probable AD in the ADNI cohort. Curr.Alzheimer Res. 2009;6:347–361. doi: 10.2174/156720509788929273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen WG, Mohs RC, Davis KL. A new rating scale for Alzheimer’s disease. Am J Psychiatry. 1984;141:1356–1364. doi: 10.1176/ajp.141.11.1356. [DOI] [PubMed] [Google Scholar]
- Sakamoto S, Matsuda H, Asada T, Ohnishi T, Nakano S, Kanetaka H, Takasaki M. Apolipoprotein E genotype and early Alzheimer’s disease: a longitudinal SPECT study. J.Neuroimaging. 2003;13:113–123. [PubMed] [Google Scholar]
- Schott JM, Bartlett JW, Barnes J, Leung KK, Ourselin S, Fox NC. Reduced sample sizes for atrophy outcomes in Alzheimer’s disease trials: baseline adjustment. Neurobiol.Aging. 2010;31:1452–1462. doi: 10.1016/j.neurobiolaging.2010.04.011. 1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuff N, Woerner N, Boreta L, Kornfield T, Shaw LM, Trojanowski JQ, Thompson PM, Jack CR, Jr, Weiner MW. MRI of hippocampal volume loss in early Alzheimer’s disease in relation to ApoE genotype and biomarkers. Brain. 2009a;132:1067–1077. doi: 10.1093/brain/awp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuff N, Woerner N, Boreta L, Kornfield T, Shaw LM, Trojanowski JQ, Thompson PM, Jack CR, Jr, Weiner MW. MRI of hippocampal volume loss in early Alzheimer’s disease in relation to ApoE genotype and biomarkers. Brain. 2009b;132:1067–1077. doi: 10.1093/brain/awp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw LM, Vanderstichele H, Knapik-Czajka M, Clark CM, Aisen PS, Petersen RC, Blennow K, Soares H, Simon A, Lewczuk P, Dean R, Siemers E, Potter W, Lee VM, Trojanowski JQ. Cerebrospinal fluid biomarker signature in Alzheimer’s disease neuroimaging initiative subjects. Ann.Neurol. 2009a;65:403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw LM, Vanderstichele H, Knapik-Czajka M, Clark CM, Aisen PS, Petersen RC, Blennow K, Soares H, Simon A, Lewczuk P, Dean R, Siemers E, Potter W, Lee VM, Trojanowski JQ. Cerebrospinal fluid biomarker signature in Alzheimer’s disease neuroimaging initiative subjects. Ann Neurol. 2009b;64:403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Pol LA, Hensel A, van der Flier WM, Visser PJ, Pijnenburg YA, Barkhof F, Gertz HJ, Scheltens P. Hippocampal atrophy on MRI in frontotemporal lobar degeneration and Alzheimer’s disease. J Neurol Neurosurg.Psychiatry. 2006;77:439–442. doi: 10.1136/jnnp.2005.075341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell JL, Przybelski SA, Weigand SD, Knopman DS, Boeve BF, Petersen RC, Jack CR., Jr 3D maps from multiple MRI illustrate changing atrophy patterns as subjects progress from mild cognitive impairment to Alzheimer’s disease. Brain. 2007;130:1777–1786. doi: 10.1093/brain/awm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DF, Rosenberg PB, Zhou Y, Kumar A, Raymont V, Ravert HT, Dannals RF, Nandi A, Brasic JR, Ye W, Hilton J, Lyketsos C, Kung HF, Joshi AD, Skovronsky DM, Pontecorvo MJ. In vivo imaging of amyloid deposition in Alzheimer disease using the radioligand 18F-AV-45 (florbetapir [corrected] F 18) J.Nucl.Med. 2010;51:913–920. doi: 10.2967/jnumed.109.069088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakushev I, Landvogt C, Buchholz HG, Fellgiebel A, Hammers A, Scheurich A, Schmidtmann I, Gerhard A, Schreckenberger M, Bartenstein P. Choice of reference area in studies of Alzheimer’s disease using positron emission tomography with fluorodeoxyglucose-F18. Psychiatry Res. 2008;164:143–153. doi: 10.1016/j.pscychresns.2007.11.004. [DOI] [PubMed] [Google Scholar]