Abstract
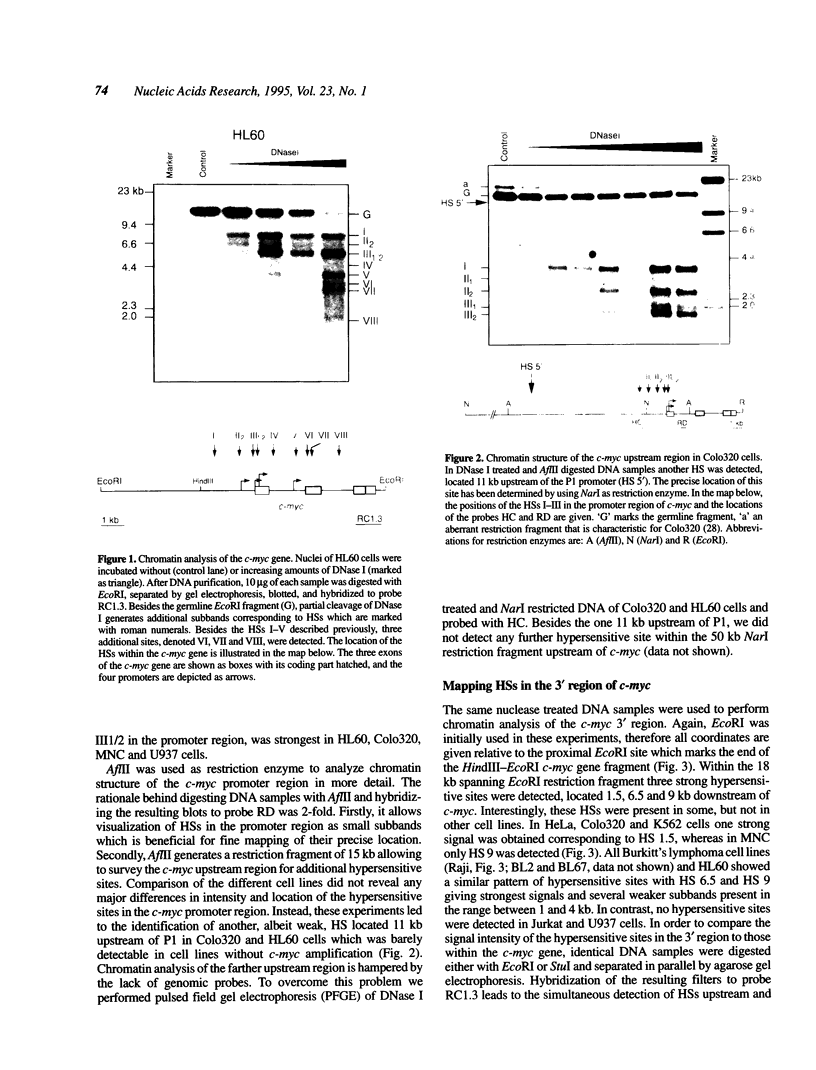
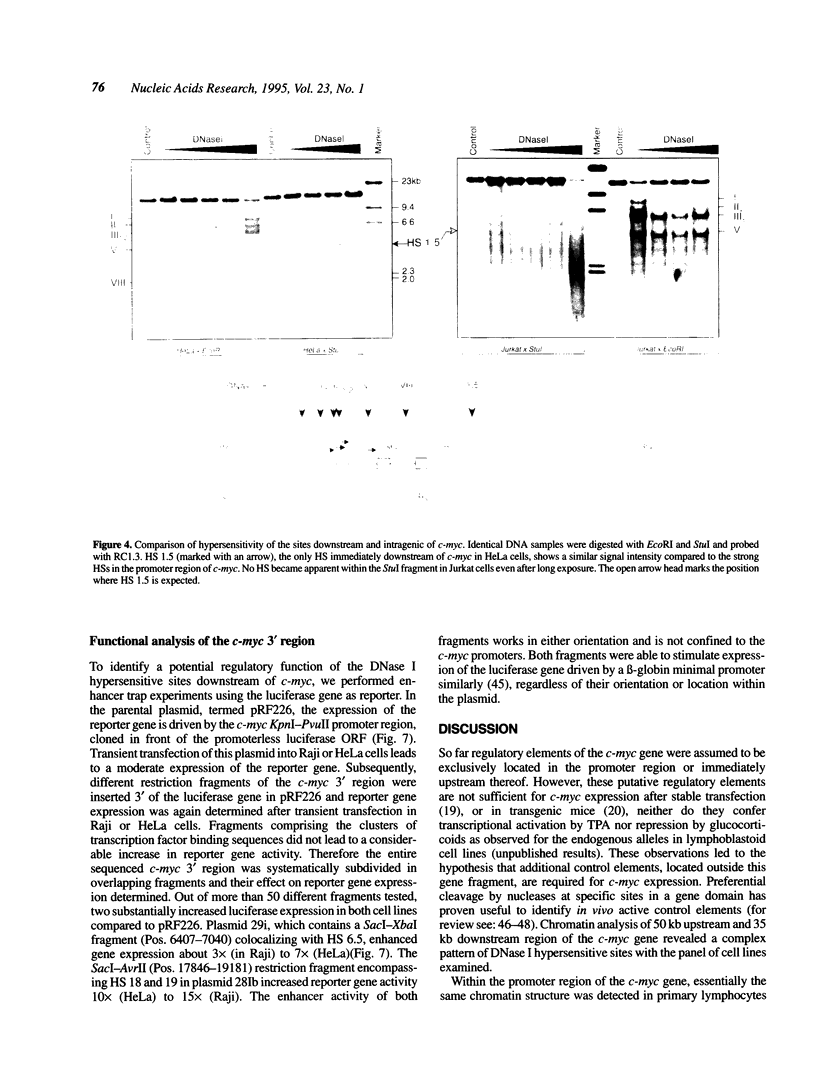
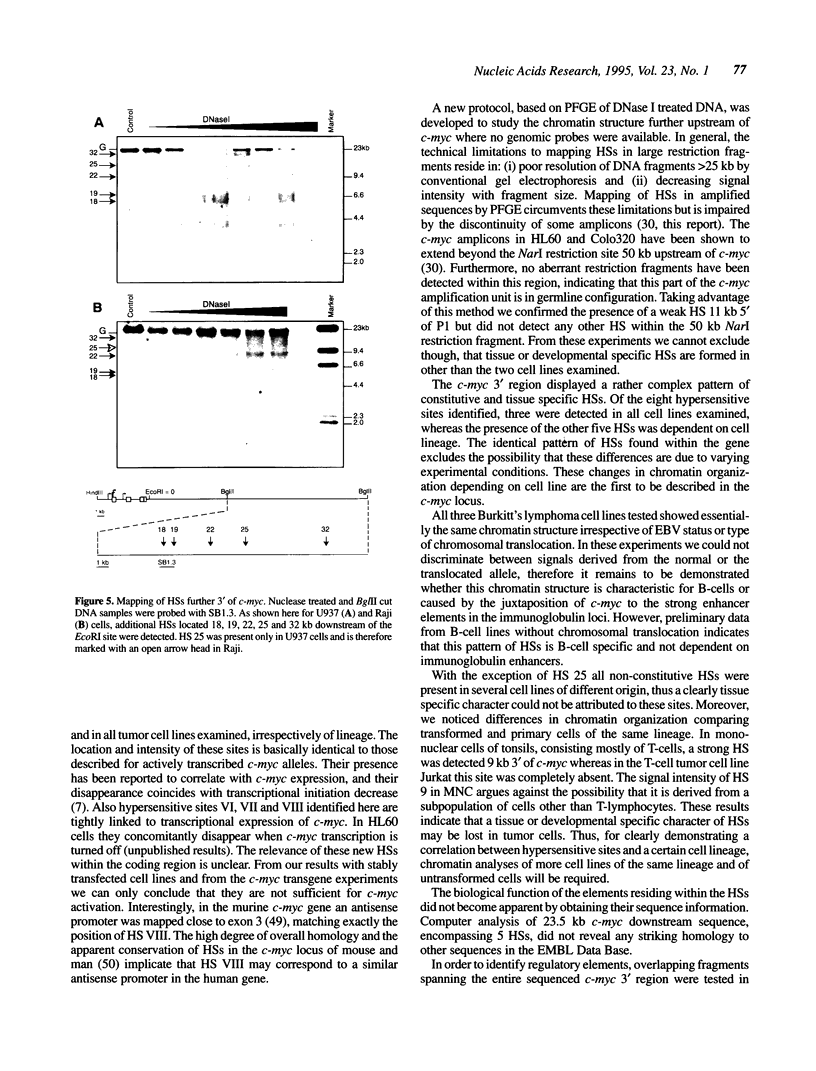
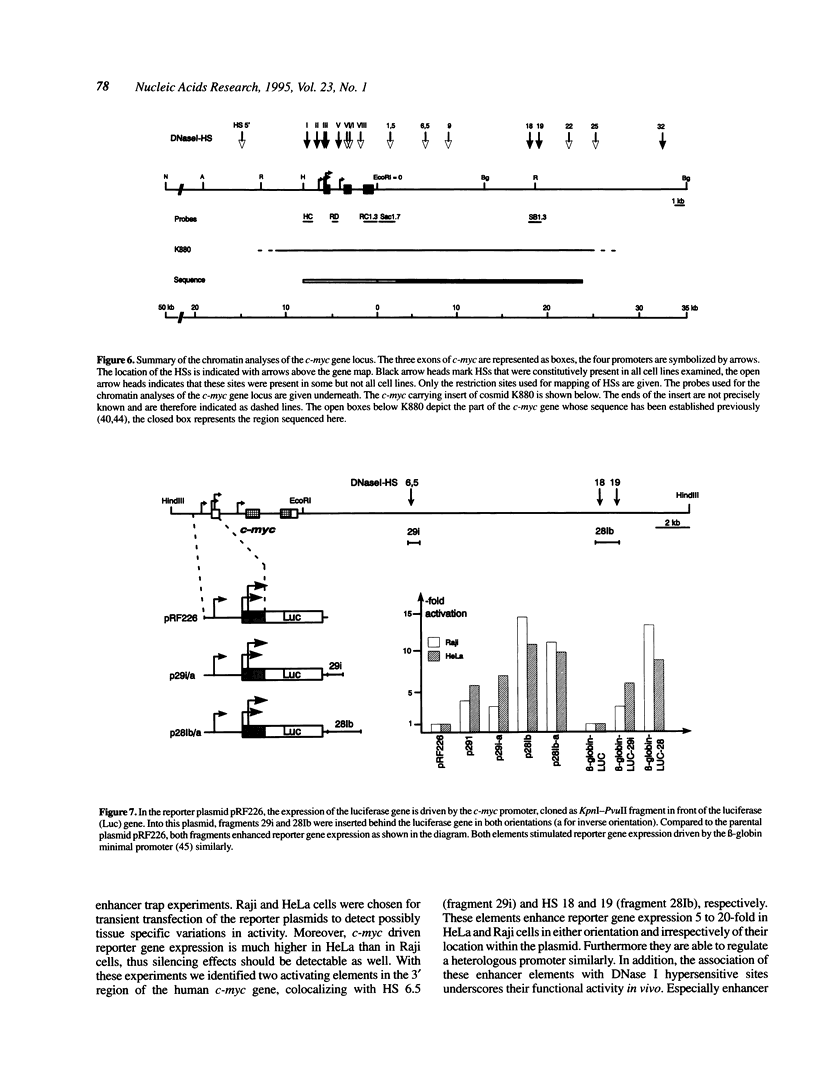
Expression of the proto-oncogene c-myc is tightly regulated in vivo. Transcription of c-myc is assumed to be controlled by a number of positive and negative cis-acting control elements located upstream or within exon 1 and intron 1. However, these regulatory elements are not sufficient for c-myc expression after stable transfection or in transgenic mice. Transcription of c-myc in vivo thus requires additional control elements located outside the tested HindIII-EcoRI gene fragment. In order to identify these putative additional control elements, we mapped DNase I hypersensitive sites around the human c-myc gene in nine different tumor cell lines and in primary lymphocytes. Within the coding and 5' region of the gene, an almost identical pattern of DNase I hypersensitive sites was detected in the various cells. In contrast, chromatin analysis of the c-myc 3' region revealed a complex pattern of constitutive and tissue-specific DNase I hypersensitive sites. In enhancer trap experiments we identified two cis-acting control elements, both co-localizing with DNase I hypersensitive sites, that stimulated c-myc transcription after transient transfection in Raji or HeLa cells. Both regulatory elements exerted their enhancer activity in either orientation and regardless of their location within the plasmids. Both elements also conferred activation on a heterologous promoter. The association of these enhancers with DNase I hypersensitive sites, indicating their functional activity in vivo, make them potential candidates for the postulated regulatory control element(s) required for c-myc expression in vivo.
Full text
PDF

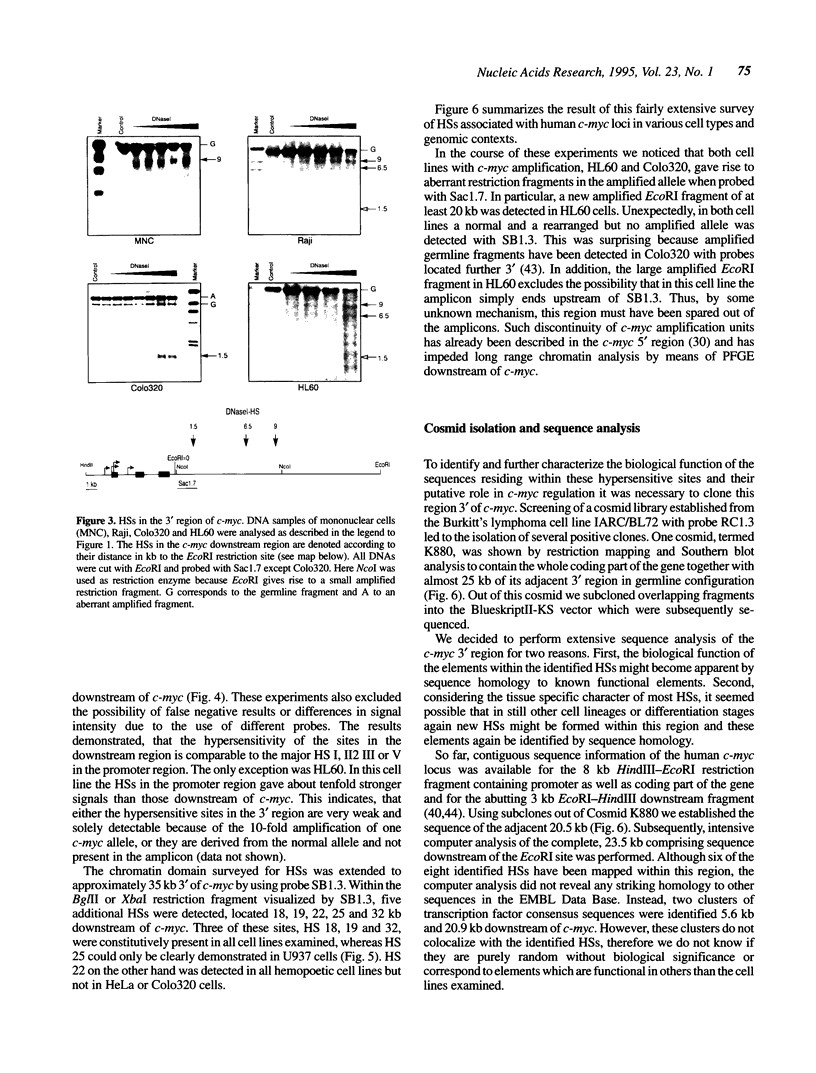


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams C. C., Workman J. L. Nucleosome displacement in transcription. Cell. 1993 Feb 12;72(3):305–308. doi: 10.1016/0092-8674(93)90109-4. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986 Jun 12;321(6071):702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Cole M. D. The myc oncogene: its role in transformation and differentiation. Annu Rev Genet. 1986;20:361–384. doi: 10.1146/annurev.ge.20.120186.002045. [DOI] [PubMed] [Google Scholar]
- Collins S. J., Ruscetti F. W., Gallagher R. E., Gallo R. C. Terminal differentiation of human promyelocytic leukemia cells induced by dimethyl sulfoxide and other polar compounds. Proc Natl Acad Sci U S A. 1978 May;75(5):2458–2462. doi: 10.1073/pnas.75.5.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross S. H., Little P. F. A cosmid vector for systematic chromosome walking. Gene. 1986;49(1):9–22. doi: 10.1016/0378-1119(86)90381-1. [DOI] [PubMed] [Google Scholar]
- Dyson P. J., Littlewood T. D., Forster A., Rabbitts T. H. Chromatin structure of transcriptionally active and inactive human c-myc alleles. EMBO J. 1985 Nov;4(11):2885–2891. doi: 10.1002/j.1460-2075.1985.tb04018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson P. J., Rabbitts T. H. Chromatin structure around the c-myc gene in Burkitt lymphomas with upstream and downstream translocation points. Proc Natl Acad Sci U S A. 1985 Apr;82(7):1984–1988. doi: 10.1073/pnas.82.7.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick D., Berger R., Polack A., Bornkamm G. W. Transcription of c-myc in human mononuclear cells is regulated by an elongation block. Oncogene. 1987;2(1):61–65. [PubMed] [Google Scholar]
- Eick D., Bornkamm G. W. Transcriptional arrest within the first exon is a fast control mechanism in c-myc gene expression. Nucleic Acids Res. 1986 Nov 11;14(21):8331–8346. doi: 10.1093/nar/14.21.8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin S. C. Chromatin structure and gene activity. Curr Opin Cell Biol. 1990 Jun;2(3):437–445. doi: 10.1016/0955-0674(90)90125-x. [DOI] [PubMed] [Google Scholar]
- Elgin S. C. The formation and function of DNase I hypersensitive sites in the process of gene activation. J Biol Chem. 1988 Dec 25;263(36):19259–19262. [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Gazin C., Dupont de Dinechin S., Hampe A., Masson J. M., Martin P., Stehelin D., Galibert F. Nucleotide sequence of the human c-myc locus: provocative open reading frame within the first exon. EMBO J. 1984 Feb;3(2):383–387. doi: 10.1002/j.1460-2075.1984.tb01816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson J. S., Croker B. P. Chromatin structure changes suggest a compensatory response to c-myc gene amplification in malignant fibrous histiocytoma. J Cell Biochem. 1992 Jun;49(2):148–156. doi: 10.1002/jcb.240490207. [DOI] [PubMed] [Google Scholar]
- Gross D. S., Garrard W. T. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- Guilhot S., Petridou B., Syed-Hussain S., Galibert F. Nucleotide sequence 3' to the human c-myc oncogene; presence of a long inverted repeat. Gene. 1988 Dec 10;72(1-2):105–108. doi: 10.1016/0378-1119(88)90131-x. [DOI] [PubMed] [Google Scholar]
- Hay N., Bishop J. M., Levens D. Regulatory elements that modulate expression of human c-myc. Genes Dev. 1987 Sep;1(7):659–671. doi: 10.1101/gad.1.7.659. [DOI] [PubMed] [Google Scholar]
- Jain V. K., Magrath I. T. A chemiluminescent assay for quantitation of beta-galactosidase in the femtogram range: application to quantitation of beta-galactosidase in lacZ-transfected cells. Anal Biochem. 1991 Nov 15;199(1):119–124. doi: 10.1016/0003-2697(91)90278-2. [DOI] [PubMed] [Google Scholar]
- Joos S., Haluska F. G., Falk M. H., Henglein B., Hameister H., Croce C. M., Bornkamm G. W. Mapping chromosomal breakpoints of Burkitt's t(8;14) translocations far upstream of c-myc. Cancer Res. 1992 Dec 1;52(23):6547–6552. [PubMed] [Google Scholar]
- Jücker M., Roebroek A. J., Mautner J., Koch K., Eick D., Diehl V., Van de Ven W. J., Tesch H. Expression of truncated transcripts of the proto-oncogene c-fps/fes in human lymphoma and lymphoid leukemia cell lines. Oncogene. 1992 May;7(5):943–952. [PubMed] [Google Scholar]
- Kumar S., Leffak M. Conserved chromatin structure in c-myc 5'flanking DNA after viral transduction. J Mol Biol. 1991 Nov 5;222(1):45–57. doi: 10.1016/0022-2836(91)90736-p. [DOI] [PubMed] [Google Scholar]
- Kumar S., Leffak M. DNA topology of the ordered chromatin domain 5' to the human c-myc gene. Nucleic Acids Res. 1989 Apr 11;17(7):2819–2833. doi: 10.1093/nar/17.7.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavenu A., Pournin S., Babinet C., Morello D. The cis-acting elements known to regulate c-myc expression ex vivo are not sufficient for correct transcription in vivo. Oncogene. 1994 Feb;9(2):527–536. [PubMed] [Google Scholar]
- Lenoir G. M., Preud'homme J. L., Bernheim A., Berger R. Correlation between immunoglobulin light chain expression and variant translocation in Burkitt's lymphoma. Nature. 1982 Jul 29;298(5873):474–476. doi: 10.1038/298474a0. [DOI] [PubMed] [Google Scholar]
- Lenoir G. M., Vuillaume M., Bonnardel C. The use of lymphomatous and lymphoblastoid cell lines in the study of Burkitt's lymphoma. IARC Sci Publ. 1985;(60):309–318. [PubMed] [Google Scholar]
- Levine R. A., McCormack J. E., Buckler A., Sonenshein G. E. Transcriptional and posttranscriptional control of c-myc gene expression in WEHI 231 cells. Mol Cell Biol. 1986 Nov;6(11):4112–4116. doi: 10.1128/mcb.6.11.4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipp M., Schilling R., Wiest S., Laux G., Bornkamm G. W. Target sequences for cis-acting regulation within the dual promoter of the human c-myc gene. Mol Cell Biol. 1987 Apr;7(4):1393–1400. doi: 10.1128/mcb.7.4.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little P. F., Cross S. H. A cosmid vector that facilitates restriction enzyme mapping. Proc Natl Acad Sci U S A. 1985 May;82(10):3159–3163. doi: 10.1073/pnas.82.10.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozzio C. B., Lozzio B. B. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood. 1975 Mar;45(3):321–334. [PubMed] [Google Scholar]
- MacGregor G. R., Caskey C. T. Construction of plasmids that express E. coli beta-galactosidase in mammalian cells. Nucleic Acids Res. 1989 Mar 25;17(6):2365–2365. doi: 10.1093/nar/17.6.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mango S. E., Schuler G. D., Steele M. E., Cole M. D. Germ line c-myc is not down-regulated by loss or exclusion of activating factors in myc-induced macrophage tumors. Mol Cell Biol. 1989 Aug;9(8):3482–3490. doi: 10.1128/mcb.9.8.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcu K. B., Bossone S. A., Patel A. J. myc function and regulation. Annu Rev Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- Mengle-Gaw L., Rabbitts T. H. A human chromosome 8 region with abnormalities in B cell, HTLV-I+ T cell and c-myc amplified tumours. EMBO J. 1987 Jul;6(7):1959–1965. doi: 10.1002/j.1460-2075.1987.tb02458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möllers B., Klages S., Wedel A., Cross M., Spooncer E., Dexter T. M., Renkawitz R. The mouse M-lysozyme gene domain: identification of myeloid and differentiation specific DNasel hypersensitive sites and of a 3'-cis acting regulatory element. Nucleic Acids Res. 1992 Apr 25;20(8):1917–1924. doi: 10.1093/nar/20.8.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K. Sequences involved in accurate and efficient transcription of human c-myc genes microinjected into frog oocytes. Mol Cell Biol. 1986 Nov;6(11):4093–4098. doi: 10.1128/mcb.6.11.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PULVERTAFT J. V. CYTOLOGY OF BURKITT'S TUMOUR (AFRICAN LYMPHOMA). Lancet. 1964 Feb 1;1(7327):238–240. doi: 10.1016/s0140-6736(64)92345-1. [DOI] [PubMed] [Google Scholar]
- Palumbo A. P., Boccadoro M., Battaglio S., Corradini P., Tsichlis P. N., Huebner K., Pileri A., Croce C. M. Human homologue of Moloney leukemia virus integration-4 locus (MLVI-4), located 20 kilobases 3' of the myc gene, is rearranged in multiple myelomas. Cancer Res. 1990 Oct 15;50(20):6478–6482. [PubMed] [Google Scholar]
- Polack A., Strobl L., Feederle R., Schweizer M., Koch E., Eick D., Wiegand H., Bornkamm G. W. The intron enhancer of the immunoglobulin kappa gene activates c-myc but does not induce the Burkitt-specific promoter shift. Oncogene. 1991 Nov;6(11):2033–2040. [PubMed] [Google Scholar]
- Quinn L. A., Moore G. E., Morgan R. T., Woods L. K. Cell lines from human colon carcinoma with unusual cell products, double minutes, and homogeneously staining regions. Cancer Res. 1979 Dec;39(12):4914–4924. [PubMed] [Google Scholar]
- Ray R., Thomas S., Miller D. M. Mouse fibroblasts transformed with the human c-myc gene express a high level of mRNA but a low level of c-myc protein and are non-tumorigenic in nude mice. Oncogene. 1989 May;4(5):593–600. [PubMed] [Google Scholar]
- Siebenlist U., Bressler P., Kelly K. Two distinct mechanisms of transcriptional control operate on c-myc during differentiation of HL60 cells. Mol Cell Biol. 1988 Feb;8(2):867–874. doi: 10.1128/mcb.8.2.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Hennighausen L., Battey J., Leder P. Chromatin structure and protein binding in the putative regulatory region of the c-myc gene in Burkitt lymphoma. Cell. 1984 Jun;37(2):381–391. doi: 10.1016/0092-8674(84)90368-4. [DOI] [PubMed] [Google Scholar]
- Spencer C. A., Groudine M. Control of c-myc regulation in normal and neoplastic cells. Adv Cancer Res. 1991;56:1–48. doi: 10.1016/s0065-230x(08)60476-5. [DOI] [PubMed] [Google Scholar]
- Spicer D. B., Sonenshein G. E. An antisense promoter of the murine c-myc gene is localized within intron 2. Mol Cell Biol. 1992 Mar;12(3):1324–1329. doi: 10.1128/mcb.12.3.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobl L. J., Eick D. Hold back of RNA polymerase II at the transcription start site mediates down-regulation of c-myc in vivo. EMBO J. 1992 Sep;11(9):3307–3314. doi: 10.1002/j.1460-2075.1992.tb05409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L. K., Showe L. C., Croce C. M. Analysis of the 3' flanking region of the human c-myc gene in lymphomas with the t(8;22) and t(2;8) chromosomal translocations. Nucleic Acids Res. 1986 May 27;14(10):4037–4050. doi: 10.1093/nar/14.10.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundström C., Nilsson K. Establishment and characterization of a human histiocytic lymphoma cell line (U-937). Int J Cancer. 1976 May 15;17(5):565–577. doi: 10.1002/ijc.2910170504. [DOI] [PubMed] [Google Scholar]
- Tsichlis P. N., Lee J. S., Bear S. E., Lazo P. A., Patriotis C., Gustafson E., Shinton S., Jenkins N. A., Copeland N. G., Huebner K. Activation of multiple genes by provirus integration in the Mlvi-4 locus in T-cell lymphomas induced by Moloney murine leukemia virus. J Virol. 1990 May;64(5):2236–2244. doi: 10.1128/jvi.64.5.2236-2244.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westin G., Gerster T., Müller M. M., Schaffner G., Schaffner W. OVEC, a versatile system to study transcription in mammalian cells and cell-free extracts. Nucleic Acids Res. 1987 Sep 11;15(17):6787–6798. doi: 10.1093/nar/15.17.6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wet J. R., Wood K. V., DeLuca M., Helinski D. R., Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987 Feb;7(2):725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]