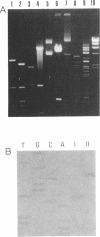
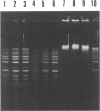
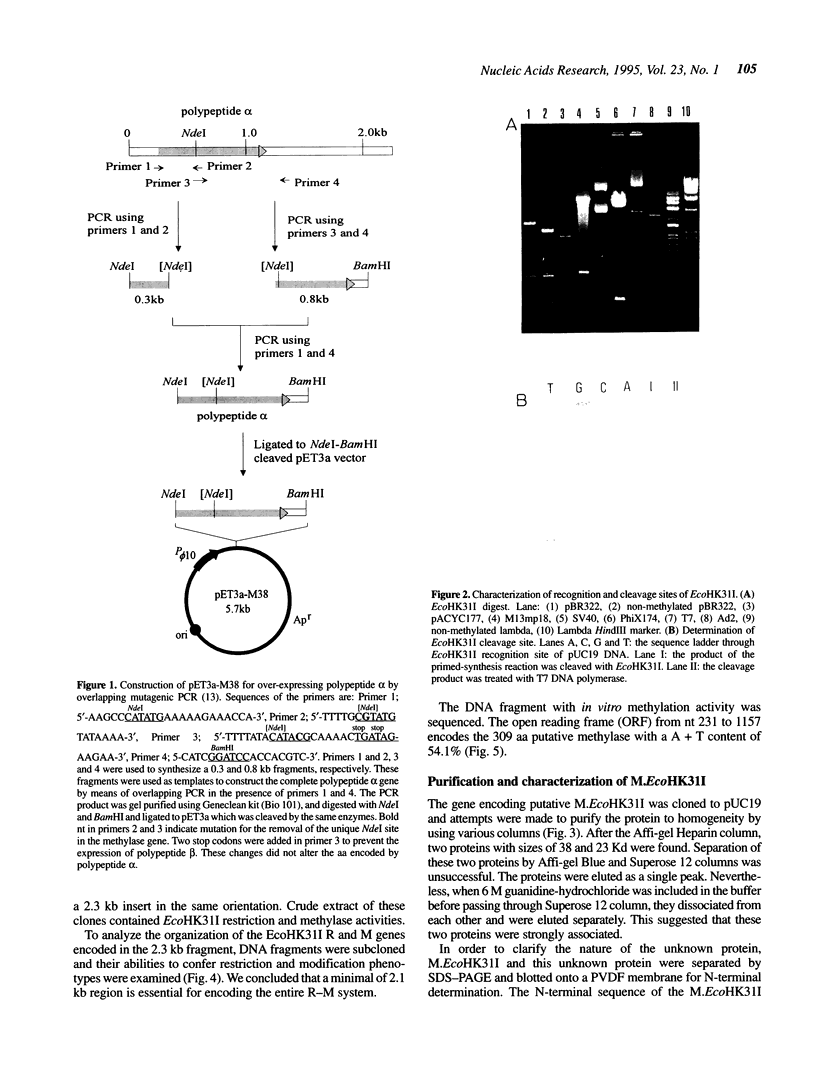
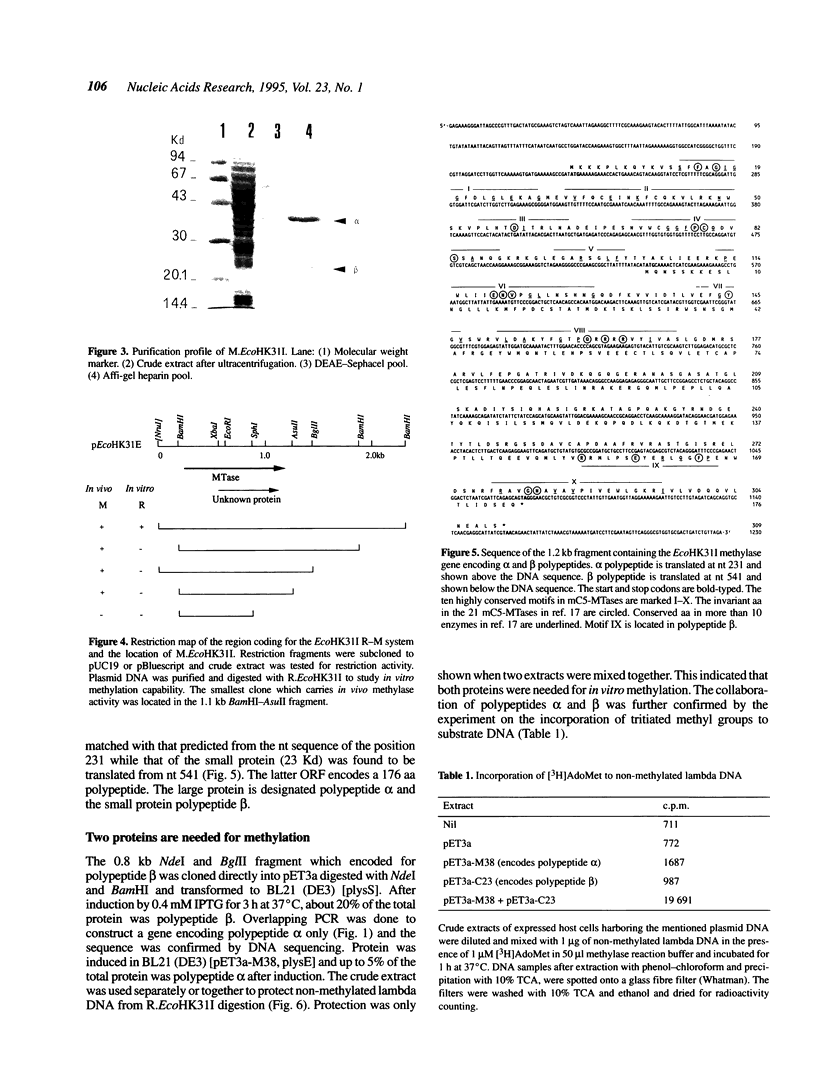
Abstract
The genes encoding EcoHK311 restriction-modification (R-M) system were isolated from a clinically-isolated Escherichia coli strain HK31. The entire R-M system of EcoHK311 is located in a 2.1 kb fragment. R.EcoHK311 is an isoschizomer of Eael which recognizes and cleaves Y decreases GGCCR. M.EcoHK31l consists of two polypeptides alpha and beta with sizes 309 and 176 aa, respectively. Polypeptide beta is encoded within aa, alternative reading frame of polypeptide alpha. All the conserved motifs in mC5-MTases can be found in polypeptide alpha except motif IX which is present in polypeptide beta. Polypeptides alpha and beta were separately synthesized in a T7 promoter controlled over-expression system and in vitro methylation occurred only when the two extracts were mixed and thus confirms that two polypeptides are required for methylation.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balganesh T. S., Reiners L., Lauster R., Noyer-Weidner M., Wilke K., Trautner T. A. Construction and use of chimeric SPR/phi 3T DNA methyltransferases in the definition of sequence recognizing enzyme regions. EMBO J. 1987 Nov;6(11):3543–3549. doi: 10.1002/j.1460-2075.1987.tb02681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N. L., Smith M. A general method for defining restriction enzyme cleavage and recognition sites. Methods Enzymol. 1980;65(1):391–404. doi: 10.1016/s0076-6879(80)65050-2. [DOI] [PubMed] [Google Scholar]
- Cheng X., Kumar S., Posfai J., Pflugrath J. W., Roberts R. J. Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-L-methionine. Cell. 1993 Jul 30;74(2):299–307. doi: 10.1016/0092-8674(93)90421-l. [DOI] [PubMed] [Google Scholar]
- Düsterhöft A., Erdmann D., Kröger M. Stepwise cloning and molecular characterization of the HgiDI restriction-modification system from Herpetosiphon giganteus Hpa2. Nucleic Acids Res. 1991 Mar 11;19(5):1049–1056. doi: 10.1093/nar/19.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R., Krummel B., Saiki R. K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988 Aug 11;16(15):7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreman C., de Waard A. Agmenellum quadruplicatum M.AquI, a novel modification methylase. J Bacteriol. 1990 Jan;172(1):266–272. doi: 10.1128/jb.172.1.266-272.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimasauskas S., Kumar S., Roberts R. J., Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994 Jan 28;76(2):357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- Klimasauskas S., Nelson J. L., Roberts R. J. The sequence specificity domain of cytosine-C5 methylases. Nucleic Acids Res. 1991 Nov 25;19(22):6183–6190. doi: 10.1093/nar/19.22.6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimasauskas S., Timinskas A., Menkevicius S., Butkienè D., Butkus V., Janulaitis A. Sequence motifs characteristic of DNA[cytosine-N4]methyltransferases: similarity to adenine and cytosine-C5 DNA-methylases. Nucleic Acids Res. 1989 Dec 11;17(23):9823–9832. doi: 10.1093/nar/17.23.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Cheng X., Klimasauskas S., Mi S., Posfai J., Roberts R. J., Wilson G. G. The DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 1994 Jan 11;22(1):1–10. doi: 10.1093/nar/22.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauster R., Trautner T. A., Noyer-Weidner M. Cytosine-specific type II DNA methyltransferases. A conserved enzyme core with variable target-recognizing domains. J Mol Biol. 1989 Mar 20;206(2):305–312. doi: 10.1016/0022-2836(89)90480-4. [DOI] [PubMed] [Google Scholar]
- McClelland M., Nelson M. The effect of site-specific DNA methylation on restriction endonucleases and DNA modification methyltransferases--a review. Gene. 1988 Dec 25;74(1):291–304. doi: 10.1016/0378-1119(88)90305-8. [DOI] [PubMed] [Google Scholar]
- Mi S., Roberts R. J. How M.MspI and M.HpaII decide which base to methylate. Nucleic Acids Res. 1992 Sep 25;20(18):4811–4816. doi: 10.1093/nar/20.18.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pósfai G., Kim S. C., Szilák L., Kovács A., Venetianer P. Complementation by detached parts of GGCC-specific DNA methyltransferases. Nucleic Acids Res. 1991 Sep 25;19(18):4843–4847. doi: 10.1093/nar/19.18.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pósfai J., Bhagwat A. S., Pósfai G., Roberts R. J. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 1989 Apr 11;17(7):2421–2435. doi: 10.1093/nar/17.7.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Sugisaki H., Yamamoto K., Takanami M. The HgaI restriction-modification system contains two cytosine methylase genes responsible for modification of different DNA strands. J Biol Chem. 1991 Jul 25;266(21):13952–13957. [PubMed] [Google Scholar]
- Trautner T. A., Balganesh T. S., Pawlek B. Chimeric multispecific DNA methyltransferases with novel combinations of target recognition. Nucleic Acids Res. 1988 Jul 25;16(14A):6649–6658. doi: 10.1093/nar/16.14.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead P. R., Brown N. L. EaeI: a restriction endonuclease from Enterobacter aerogenes. FEBS Lett. 1983 May 2;155(1):97–101. doi: 10.1016/0014-5793(83)80217-8. [DOI] [PubMed] [Google Scholar]
- Wilson G. G. Organization of restriction-modification systems. Nucleic Acids Res. 1991 May 25;19(10):2539–2566. doi: 10.1093/nar/19.10.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Tao T., Wilson G. G., Blumenthal R. M. The M.AluI DNA-(cytosine C5)-methyltransferase has an unusually large, partially dispensable, variable region. Nucleic Acids Res. 1993 Feb 25;21(4):905–911. doi: 10.1093/nar/21.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]