Abstract
Ecological speciation is the process by which barriers to gene flow between populations evolve due to adaptive divergence via natural selection. A relatively unexplored area in ecological speciation is the role of gene expression. Gene expression may be associated with ecologically important phenotypes not evident from morphology and play a role during colonization of new environments. Here we review two potential roles of gene expression in ecological speciation: (1) its indirect role in facilitating population persistence and (2) its direct role in contributing to genetically based reproductive isolation. We find indirect evidence that gene expression facilitates population persistence, but direct tests are lacking. We also find clear examples of gene expression having effects on phenotypic traits and adaptive genetic divergence, but links to the evolution of reproductive isolation itself remain indirect. Gene expression during adaptive divergence seems to often involve complex genetic architectures controlled by gene networks, regulatory regions, and “eQTL hotspots.” Nonetheless, we review how approaches for isolating the functional mutations contributing to adaptive divergence are proving to be successful. The study of gene expression has promise for increasing our understanding ecological speciation, particularly when integrative approaches are applied.
Keywords: adaptive divergence, eQTL, microarray, phenotypic plasticity, population persistence, qPCR, reproductive isolation, speciation, cis-regulatory mutation, gene expression
Ecological speciation
Natural selection is a central mechanism of evolutionary change within species. But to what extent is selection also responsible for the formation of new species (i.e., speciation)? Recent years have seen renewed efforts to address this question. Under one scenario, populations living in different ecological environments undergo adaptive genetic differentiation via divergent natural selection, and these same adaptive changes also result in the populations ceasing to exchange genes. Consistent with past work, we define this process of “ecological speciation” as one in which barriers to genetic exchange evolve between populations as a result of ecologically based divergent natural selection.1–5 Ecological speciation generally occurs because phenotypic traits under divergent selection, or those genetically correlated with them, incidentally affect reproductive isolation.6,7 Thus, ecological speciation can involve any type of reproductive barrier and can occur under any geographic arrangement of populations (allopatry, parapatry, and sympatry).1–3,5,8–12 Ecological speciation is distinguished from models of speciation which do not involve ecologically based divergent selection, such as speciation via genetic drift or the fixation of different incompatible mutations in populations experiencing similar selection.4,13
The process of ecological speciation makes explicit predictions. For example, it predicts that ecologically divergent pairs of populations will exhibit greater reproductive isolation than ecologically similar pairs of populations of similar age.11,14 Another prediction is that phenotypic traits involved in divergent adaptation will also cause reproductive isolation.15 For example, adaptive traits might directly reduce the fitness of immigrants and hybrids, due to a mismatch between immigrant and hybrid phenotypes and the ecological environment, generating “immigrant inviability” and extrinsic postmating isolation, respectively.16,17 Finally, ecological speciation predicts that neutral gene flow between populations will decrease as adaptive divergence increases.10,18 These predictions have now been supported numerous times using experiments or molecular data on levels of neutral gene flow (see Refs. 3–5 and 14). Additionally, some progress has been made in understanding the genetic basis of ecological speciation, but this stems primarily from Quantitative Trait Locus (QTL) and candidate gene studies.4,19,20 Here we focus on a largely unexplored issue: the role of gene expression in ecological speciation.
The role of gene expression warrants consideration because two events need to occur during the process of ecological speciation (following Ref. 21), and gene expression might strongly affect each of them. First, a key mechanism by which ecological divergence between populations occurs is via the colonization of new environments. In these cases, ecological speciation requires that newly founded populations persist in the colonized environments. Ernst Mayr7,8 especially espoused this “persistence view” of the role of ecology in speciation (for review see Refs. 21, 22). Second, populations in different environments need to evolve genetically based reproductive isolation. Gene expression might therefore promote ecological speciation in two ways: (1) indirectly by promoting population persistence or (2) more directly by affecting adaptive genetic divergence in traits causing reproductive isolation (Fig. 1; Ref. 23).
Figure 1.
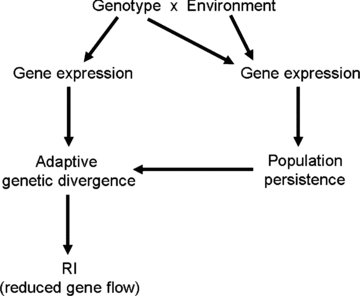
Conceptual diagram of the different ways the genetic and environmental components of gene expression might contribute to ecological speciation. Both components might contribute to population persistence, which is required for eventual speciation. The genetic components of gene expression could contribute to the adaptive genetic divergence, which drives ecological speciation. See text for details.
Here we review both putative roles for gene expression in ecological speciation. Because the study of gene expression and ecological speciation is in its infancy, our goals here are not only to review the existing literature and highlight what is already known, but also to provide a conceptual framework for thinking about the topic and point to especially promising avenues for further research. We begin by providing more detail on why studying gene expression might be fruitful for understanding speciation, followed by a discussion of how to measure gene expression. We then review the roles of gene expression in population persistence and in affecting adaptive genetic divergence.
What can the study of gene expression tell us about ecological speciation?
Gene expression is shaped by both genetic and environmental components, and can therefore be considered as a “molecular phenotype.”24 For example, the transcription rate of a gene can vary among genotypes such that it is a heritable phenotype.25–28 Gene expression might provide novel insights into speciation because gene expression profiles have the ability to uncover phenotypes, which would not readily be visible via traditional approaches. Our understanding of evolution has often been limited by our ability to define relevant phenotypes.29 For example, initial progress in understanding ecological speciation has necessarily focused on easily measured morphological, and to some extent, behavioral traits. In essence, gene expression might allow us to circumvent these limits by uncovering hidden phenotypes potentially of ecological relevance and phenotypes that are perhaps difficult or counter-intuitive to measure. This could be especially critical given that genome annotations to date currently stem mostly from model genetic organisms, and thus are lacking in ecological relevance.30 Identifying ecologically relevant expressed genes will thus likely increase the efficacy of genomics to address questions related to ecological speciation.31–33
For example, physiology has been grossly underrepresented in ecological speciation studies, presumably because of the difficulty associated with measuring these phenotypes.2,5 With current gene expression technologies, we can now examine many metabolic and mechanistic processes that were previously difficult to measure.34,35 This may be important because evolutionary changes in expression of physiological genes might sometimes precede morphological changes.7 Overall, the sensitivity achievable from modern gene expression technology (large numbers of genes in one assay, low transcript genes, and subtle gene expression differences) has allowed the study of specific organs and tissues and revealed that hidden phenotypes may stem from genes expressed in all of these tissues. For all of these reasons, gene expression studies have the potential for testing numerous hypotheses (i.e., numerous “traits” or genes), many of which an investigator would not necessarily think to test from previous research.36 The power of gene expression profiles as surrogate phenotypes is well established in fields such as genetic studies of disease research in humans29,37 but needs to be further implemented into ecological speciation studies.
Theoretically, this implementation seems possible. Johnson and Porter38 demonstrated that parallel directional selection on geographically isolated populations might lead to misregulation of gene expression that in turn may be associated with hybrid incompatibility. By modeling the evolution of a regulated pathway wherein hybrid incompatibility can arise as a consequence of misregulated gene expression, Johnson and Porter38 showed that parallel selection is expected to yield reproductive isolation regardless of the underlying mechanisms relating genotype to phenotype. In their analyses, population pairs experienced identical selection conditions and thus did not experience divergent selection. Nonetheless, these results suggest that the detection of gene misregulation may be a feasible starting point towards understanding the role of gene expression in ecological speciation, with the objective of measuring the level of hybrid incompatibility due to gene expression before finding the ultimate mutation responsible.39
In summary, gene expression studies may reveal the genes underlying adaptations that are difficult or impossible to measure in other ways, and these phenotypes may be of importance for initiating ecological divergence during speciation.40 Consequently, patterns of gene expression should be integrated into studies of ecological speciation, with a need for clearer predictions about how gene expression affects ecological speciation. Gene expression patterns may also provide insight into the underlying genetic architecture of ecological speciation and importantly, if it differs from other types of speciation.
How to study gene expression
Studies of gene expression measure the expression level of single genes, multiple genes, or the entire transcriptome (the latter defined as all the genes expressed in a cell, tissue or organism). The measure of expression is the abundance of transcribed messenger RNA (mRNA) molecules and is specific to the tissue, developmental stage, point in time, and taxon in which it is measured.24,41,42 Protein and mRNA abundances are highly correlated, that is why mRNA levels can be used as a proxy for differences in protein products.43 A variety of methods are now available to quantify gene expression and can be subdivided into two broad categories: (1) those for which (candidate) genes must be known in advance of quantification of expression and (2) those that quantify abundance for multiple genes and thus simultaneously identify genes of interest (Table 1). We treat each category in turn briefly here, and refer readers to previous reviews for greater detail.44,45
Table 1.
Comparisons of gene expression methodologies. Technique denoted with (1) do not require a priori, whereas techniques denoted with (2) require that candidate genes are known ahead of time
| Technique | Approach | Pros | Cons | References |
|---|---|---|---|---|
| DDRT-PCR (1) Rarely used | A subset of differentially expressed genes | Inexpensive. Generates differentially expressed genes | Only good for genes in high abundance | 168 |
| RT-qPCR (2) | Individual candidate genes are compared | Precise, less expensive than some other methods. Great follow-up on transcriptome-wide techniques | Candidate genes are a prerequisite. Primer development can be difficult | 169 |
| Microarrays (1) | Up to tens of thousands of genes assayed at one time | Used to generate candidate genes. A large portion of the transcriptome easily screened | Only available for some taxa. Large development cost: expensive | 71, 170, 171 |
| SAGE (1) | Many genes assayed and sequenced at one time | Less development cost than microarrays. Does not require functional genome to be sequenced | Fairly expensive but getting cheaper | 172, 173 |
| Suppression subtractive hybridization (1) Rarely used | Identify genes differentially expressed | Inexpensive. Generates differentially expressed genes | Require sequencing to identify the physiological function of the differentially expressed genes | 54, 55 |
| Northern blot (2) | Probes for a single gene | The original gene expression tool | One gene at a time, limited utility in quantification | 46 |
| RNA-Seq via next generation sequencing (1) | Sequence all transcripts | the ultimate tool, price coming down | Expensive, computationally demanding | 80–82, 84, 174 |
We first outline methods that require candidate genes prior to analysis. These are cases where known genes are of a priori interest, for example, because of their function or association with ecological variables. The original gene expression technique is Northern blotting where RNA is extracted from a specific tissue and subjected to electrophoresis on a gel.46 The gel is then transferred to a nylon membrane that is washed with a labeled probe specific to the candidate gene of interest. If the gene was expressed and the transcript is present, the probe will hybridize and anneal to the membrane. Other samples may then be compared for the expression of this same gene. Northern blots allow the detection and only semiquantification of mRNA target sequences (the darker the band, the greater the expression).47 More sensitive techniques have since been developed. One such technique is retrotranscriptase quantitative polymerase chain reaction (RT-qPCR or qPCR).48,49 With qPCR, one converts mRNA to cDNA and then uses fluorescent probes specific to the cDNA in PCR to monitor the quantity of cDNA template. The PCR cycle associated with exponential growth of product is tightly associated with the quantity of the initial cDNA template, providing an estimate for the level of mRNA expression in the tissue. Depending on experimental design, qPCR can assess relative or absolute abundance of RNA. Since qPCR does not have the same technical problems as microarrays (see below), qPCR has emerged as a method to quantify and verify expression levels of candidate genes identified with large-scale transcriptomic studies.50,51
A second set of techniques does not require that candidate genes to be chosen prior to analysis. Within this set, two techniques are no longer in common use or have a limited history of use in ecological studies. The first are differential display techniques, which with real-time PCR (DDRT-PCR), can describe differences in gene expression between species.52 This strategy is based on the amplification of partial cDNA sequences from a pool of mRNA (of unknown genes being expressed) and is only useful when genes are abundantly expressed. Another method, suppression subtractive hybridization (SSH), employs PCR to differentially amplify cDNA.53 SSH has the advantage of identifying all the differentially expressed genes even at low abundance between nonmodel species.46 Few studies have used this method to compare gene expression profiles in divergent ecological conditions, presumably because of the availability of more sensitive and precise techniques.54,55
Currently, the most common technique for extensively assessing global gene expression profiles is microarrays, which are generally akin to a reverse Northern blot.56 Microarray experiments are performed by hybridizing “target” cDNA in solution from an experimental group or groups to the spots or “probes” that are fixed to the glass slide, often representing in the order of thousands of genes. Gene expression among groups for each spot are then compared according to their fluorescence intensities to detect up- or downregulated genes. Treatment sample is either competitively hybridized and compared to a common reference or another treatment (two-color experiment) or just the absolute intensity of a single treatment sample is measured (one-color experiment).57 The last decade has experienced an explosion of microarray studies in ecology and evolution, and we refer readers to past reviews for a more thorough treatment of the methodology (reviewed in Refs. 42, 49, 58–61).
Here, we focus on two-critical points; susceptibility of type I error and repeatability of the results. First, microarrays are a powerful tool but can be prone to type I errors stemming from the large number of comparisons involved and variation in experimental conditions (e.g., use of different tissues, treatments, ecological types, and species).41,62 Several methods exist for comparing samples on the arrays,63–65 along with many different data analysis programs, techniques for normalization, quality control, quantifying spot intensity, and correcting for multiple tests. Second, the repeatability of published microarray studies is arguably limited, especially when the sum of the expression data is unavailable.66,67 These discrepancies appear to be primarily due to incomplete data annotation or specification of data processing and analysis, rather than technical limitations. Standardized analytical procedures do not exist which can lead to contentious interpretations of the data. Although many journals now require that the data be submitted to acceptable public repositories upon conditional acceptance,68 more strict publication rules enforcing public data availability and explicit description of data processing and analysis will be needed to ensure repeatability.
Along these lines, it is important to note that microarrays are intended to be used with the species for which the chip has been developed,69 but some studies have demonstrated that microarrays can also be used in closely related species.49,70,71 Oligo microarrays have shorter fragments of cDNA spotted on the chip, so they are less ideal for cross-species work, since few numbers of polymorphism may affect hybridization greatly.72 However, cDNA arrays have longer DNA fragments increasing their potential usefulness to nontarget species. For example, the Genomics Research on Atlantic Salmon Project developed a salmonid microarray consisting of expressed sequence tags (ESTs) developed from both rainbow trout (Oncorhynchus mykiss) and Atlantic salmon (Salmo salar). This microarray has been successfully applied in many closely related salmonids, including other salmon (Salmoninae), whitefish (Coregoninae) and rainbow smelt (Osmerus mordax).73,74 Thus, cDNA microarrays can bridge differences between systems with plentiful genomic resources that are poorly understood ecologically, and systems with a well-known ecology but poorly developed genomic resources.41,75
An additional technique that assembles many short sequence tags (9–10 bp) excised from cDNA and inserts them in a 1 kbp vector for sequencing is serial analyses of gene expression (SAGE).76 Each tag in SAGE can be traced back to a single gene and the relative amount of that tag in the vector corresponds to the mRNA levels in the tissue collected. Thus, SAGE may be used to both identify expressed genes and quantify the relative amounts without any a priori ESTs or genomic resources. The latest iteration of this technique, SUPERSAGE assembles longer sequence tags (26 bp) from which primers for qPCR or potentially even oligo microarrays could be created.77
With the advent of next-generation sequence techniques, it is now possible to routinely sequence the entire transcriptome of each sample,78 even for nonmodel organisms.79 Like SAGE, this technique generates sequence data and transcript abundance.80,81 However, longer sequence reads have the power to discern alternative splice variants,82 alternative alleles,83 and single nucleotide polymorphisms (SNPs) within coding regions.84 This method will therefore be more precise in identifying and quantifying the transcription of closely related genes. As costs decrease and read lengths increase, this may ultimately replace all other transcriptomic methods.
In conclusion, a quickly expanding and improving suite of methods exist for the study of differential gene expression. The best method will depend on the research question, study organism, budget, and whether reasonable candidate genes are known a priori. One important gap in the literature is the lack of a comprehensive study comparing the accuracy and precision of these techniques. We now turn to more conceptually oriented questions of how gene expression might affect ecological speciation.
Gene expression and population persistence
“The creatures which can stand ‘the storm and the stress’ of the physical influences of the environment […] will live; while the others which cannot, will not.”—Baldwin85 (1896).
The first manner in which gene expression might affect speciation is via promoting population persistence. As exemplified by Baldwin's quotation above, once a population colonizes a new environment, it must persist if it is to speciate. Population establishment and persistence in a new environment may be facilitated by phenotypic plasticity (Fig. 2).86–90 Modulation of behavioral, morphological, or physiological traits via phenotypic plasticity could therefore occur before any adaptive genetic evolution occurs.23 Gene expression-mediated phenotypic plasticity may be described as reaction norms in gene expression with the molecular phenotype of gene expression-facilitating population persistence following colonization.30 Direct tests of this idea are lacking, but two lines of indirect evidence exist: (1) studies of plasticity in traits (morphology and behavior mostly) related to fitness and population persistence and (2) studies of gene expression responses during ecological shifts, particularly those resulting in exposure to ecological stress.
Figure 2.
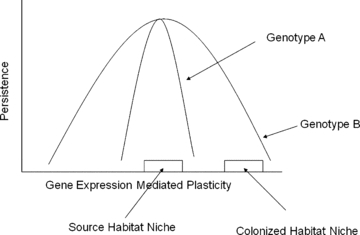
The effects of gene expression-mediated phenotypic plasticity (GMPP: y-axis) on colonization of new environments and subsequent population persistence. Genotype A has a lesser breadth of GMPP compared with genotype B. Both genotypes have high persistence in the source habitat, but genotype A has no potential to persist in a colonized habitat. Genotype B's GMPP allows persistence in the colonized habitat, allowing time for adaptive genetic divergence.
First, studies of plasticity in phenotypic traits related to fitness provide evidence for a role for plasticity in population persistence. However, such studies routinely lack evidence on how (or if) gene expression itself was involved. For example, Yeh and Price88 studied two populations of dark-eyed Junco birds, a native population in the mountains and a newly established population on the University of California San Diego (UCSD) campus. The UCSD population persisted for years despite significant environmental differences compared to the native habitat. Studies on the length of the breeding season, a classic trait dependent on temperature, revealed that the breeding season of the UCSD populations was twice as long as that of the ancestral populations, presumably due to more favorable climate (e.g., lack of snow) in the newly established population. Importantly, UCSD females displayed higher offspring production without a corresponding increase in mortality, suggesting that plasticity in breeding time was promoting colonization, population establishment, and persistence in the new environment. However, future studies are needed to examine if gene expression might be associated with the shifts in these life history traits.
Second, studies of gene expression response during ecological shifts support a role for gene expression in facilitating responses to ecological change (= stress). By ecological stress we mean simply a shift in ecology that affects the fitness of a population. In the last decade, such studies reporting evidence for a role of gene expression during ecological stress have increased (Table 2). Although these studies are critical toward understanding physiological stress response, detailed analysis of the visible phenotypes, and their explicit effects on population persistence, are needed. For instance, McCairns and Bernatchez91 examined adaptive divergence between freshwater and marine sticklebacks in a common garden experiment. Specifically, they measured fitness and survival to explore the role of gene expression at four candidate genes in response to osmoregulation. They found a significant correlation between gene expression and fitness and their results thus supported the hypothesis that ancestral plasticity for osmoregulation promoted adaptive divergence via heritable osmoregulation expression (sodium–potassium ATPase). These results are consistent with the hypothesis that gene expression modulation can promote adaptive divergence by allowing populations to persist in a changing environment, whereby fitness is maintained by plasticity.
Table 2.
Examples of studies showing that gene expression is affected by ecological stress. Under the assumption that gene expression allows populations to better persist in stressful environments, these studies indicate that differential gene expression can promote the colonization of, and subsequent persistence in, novel environments
| Organism | Study | Method | Environmental stressor | Proportion of genes affected by treatment | Major physiological function affected |
|---|---|---|---|---|---|
| Killfish Austrofundulus limnaeus | 171 | cDNA microarray | Daily and seasonal temperatures regimes | 11% | Molecular chaperones, cholesterol and fatty acids synthesis, membrane structure, solute carrier, carbohydrate metabolism, nitrogen metabolism, intermediary metabolism, cytoskeleton elements, protein turnover, complement and innate immunity, and cell growth and proliferation. |
| Bivalve Argopecten purpuratus | 175 | Suppression subtractive ESTs library + quantitative RT-PCR (candidate genes) | Copper tolerance | 8% | Cell differentiation, cellular communication, cytoskeleton, development and differentiation, energetic metabolism, protein regulation, respiratory chain, stress protein, translation and posttranslation processing, cellulose hydrolysis, and ribosomal protein. |
| Brazilian flounder Penalichtys orbignyanus | 176 | Semi-quantitative RT-PCR (candidate gene approach) | Hyperosmosis | 2 candidate genes up-regulated | Growth. |
| Coral fish Pomacentrus moluccensis | 177 | cDNA microarray | Prolonged heat and hypoxia | 2% (down-regulation mostly) | Cell adhesion, cell cycle and growth, cyskeleton, metabolism, protein processing, stress proteins, signal transduction, transcription, translation, and transport. |
| Arthropod Orchesella cincta | 27 | cDNA microarray | Cadmium | 14% (down-regulation mostly) | Translation, signal transduction, stress protein, redox state, general metabolism, chromatin remodeling, and proteolysis digestion. |
| Antarctic nematode Plectus murrayi | 178 | Suppression subtractive hybridization / ESTs library + quantitative RT-PCR | Desiccation resistance | 6% | Carbohydrate metabolism, amino-acid metabolism, lipid metabolism, xenobiotic metabolism, membrane transport, signal transduction, transcription, translation, replication, cell growth and death, and cell communication. |
| Thale cress Arabidopsis spp. | 179 | Oligo microarray | Salt, osmotic regulation, and temperature | 12 to 25% (up-regulation mostly) | Oxidative stress, membrane transport, phosphoregulation, transcription, circadian clock, fatty acid metabolism, stress protein, cytoskeleton, membrane protein, and carbohydrate metabolism. |
| Black cottonwood Populus trichocarpa | 180 | cDNA micorarray | Herbivory | 5% (up-regulation mostly) | Photosynthesis, general metabolism, transport, transcription, octadecanoid and ethylene signaling, detoxification and redox processes, and secondary metabolism. |
| Atlantic samon Salmo salar | 110 | cDNA microarray | Pathogens | 17% (up-regulation mostly) | Immunity-related genes, extracellular matrix component, electron and ion-transport chain, signal transduction, transcription, metal-binding protein, pyrimidine biosynthesis, protein degradation, localization and folding, DNA replication, and cell structure and adhesion. |
| Mycorrhizal involutus | 181 | cDNA microarray | Fungus Pillus Host specificity | 16% | Electron transport, lipid and fatty acid metabolism, transcription, sex determination, regulation of cell cycle, glycolysis, stress protein, protein biosynthesis, and aromatic compounds metabolism. |
Despite these advances, definitive tests demonstrating that gene expression facilitates population persistence are lacking. Such tests could be carried out using experimental evolution in the lab. For example, the genomics of Drosophila is increasingly well characterized, with some mutant lineages able or unable to cope with different stressors.92 Under controlled stress conditions, one could measure which genes are most strongly differentially expressed, while controlling for variation in ecologically relevant alternative alleles in different environments.34 The differentially expressed genes could then be knocked out in one “expression mutant” treatment (e.g., by the use of RNAi or destroying or inhibiting the promoter regions). Both mutant and control treatments would then be exposed to stress, simulating colonization of a new environment, and the population persistence of each compared. The prediction is that population persistence would be weaker for the mutant treatment. In principle, this experiment could even be conducted in the field.93 Similarly, gene expression studies of natural populations colonizing new environments may identify genes and pathways whose plasticity is essential to persistence in new environments.94
We note two additional points about the importance of gene expression for population persistence. First, populations and genes with much prestanding genetic variation may exhibit rapid evolution following the colonization of new environments95 and thus not require differential gene expression as strongly for population persistence. Second, purely environmentally induced gene expression (nonheritable molecular phenotypes) can still play an indirect role in speciation by facilitating population persistence and “buying the population time” for divergence in other, less plastic, evolvable traits (Fig. 1 in Ref. 23).
Gene expression and adaptive genetic divergence
The second manner in which gene expression might affect ecological speciation is by being associated with adaptive genetic divergence and reproductive isolation (Fig. 1). This forces a consideration of the link between divergent selection, adaptive genetic divergence, and reproductive isolation: loci under divergent selection and loci causing reproductive isolation are similar in exhibiting reduced introgression (and thus greater divergence) between populations relative to other loci.16,96–99 Indeed, an allele “a” that confers a poor fit of the phenotype to the environment can be selected against and contribute to speciation, whether the afflicted allele resides in one of the parental species (immigrant homozygote “aa”) or in a hybrid individual (heterozygote “Aa”). Recognizing that the adaptive genetic divergence, which results in selection against immigrants and hybrids, represents reproductive isolation itself helps clarify the relatedness of the two processes. Additionally, we stress that adaptive genetic divergence might incidentally cause the evolution of any form of reproductive isolation, including “nonecological” forms such as sexual isolation and intrinsic genetic incompatibilities in hybrids.5,100
Understanding the heritable component of gene expression will be fundamental toward understanding the genetics of ecological speciation. This is conceptually possible because the expression level for any given transcript is a phenotype that is influenced by both genetics and the environment. The genetic basis of gene transcription itself may exist prior to colonization of new environments or may actually evolve via genetic assimilation.101 We consider here two fundamental questions: (1) how substantial is the genetic component of gene expression and (2) can we elucidate whether or not this genetic component of gene expression is associated with adaptive divergence and reproductive isolation? Each question is addressed in a separate section. The main findings, as well as explicit directions for future research, are summarized in Table 3.
Table 3.
Summary of what is known about gene expression and ecological speciation, what is missing (= future directions), and how these gaps in our knowledge might be addressed
| Category/type of study | What is known | What is missing | How to address gaps in our understanding |
|---|---|---|---|
| Population persistence | |||
| 1. role of plasticity | Phenotypic plasticity can promote population persistence. | To what extent does this involve gene expression? | Add gene expression data to studies of phenotypic plasticity and population persistence. |
| 2. environmental stress | Gene expression may help populations deal with environmental stress. | Does gene expression during colonization of new environments actually promote population persistence? | Add information on population persistence to studies of gene expression in response to stress. |
| Heritability of expression divergence | |||
| 1. common garden and/or animal model | Expression divergence between populations can have a genetic basis, and can involve parallel evolution across independent populations. | How important is heritable gene expression divergence relative to other forms of genetic divergence (i.e., coding-region changes)? | Integrate studies of gene expression with studies examining functional mutations affecting trait divergence. |
| 2. eQTL | eQTL hotspots exist, exhibit signatures of divergent selection, and provide candidate gene regions for ecological speciation. Networks of gene interactions may be implicated in adaptive divergence. | What role do eQTL hotspots have in adaptive divergence and/or reproductive isolation? | Genome-wide studies will be integral to understanding how gene expression affects ecological speciation. |
| To what extent can we establish a mechanistic understanding of gene networks? | |||
| Can this inform us about the genetics of ecological speciation? | |||
| Does ecological speciation have a genetic architecture that is different from other types of speciation? If so, why? | |||
| Gene expression and reproductive isolation | |||
| Links between expression, adaptive divergence, and reproductive isolation | Gene expression divergence known to affect adaptive phenotypic divergence, and in some cases has been tied to adaptive genetic divergence. Underlying mutations rarely yet identified. | To what extent does expression divergence actually generate reproductive isolation, either ecologically based or other forms? | Quantify the extent to which expression divergence contributes to reproductive isolation of all forms. |
| To what extent can experimental studies of gene expression add to our understanding of the mechanisms of ecological speciation? | |||
| Can predictions be made about the likelihood of ecological speciation based on gene expression profiles? | |||
| Is divergence in gene expression associated with the causes of ecological speciation or the consequences? | |||
Genetic architecture of gene expression: heritability and eQTL mapping
Heritability of gene expression divergence
Gene transcription rates can vary among genotypes such that it is a heritable phenotype. Both the magnitude and rate of changes in gene transcription level in response to selection will depend on the heritability of gene expression.26,102 What proportion of the transcriptional variation in a population is attributable to genetic variation among individuals? Estimation of the heritability of gene expression is likely to be complicated because sources of transcriptional variation can vary tremendously among tissues within individuals, among individuals, and among populations.103 Although several studies have discovered gene expression differences between diverging populations, significant transcriptional differences need not reflect heritable genetic variation.104
A few studies have formally detected heritable gene transcription differences between populations. These studies quantified genetic differences using common garden experiments, which directly quantify levels of gene expression in the absence of environmental variation.105 For example, St-Cyr et al. quantified variation in gene expression for almost 4,000 genes in species pairs of lake whitefish from North American lakes under common garden conditions and found that 14% exhibited differences in transcription. These differences are therefore the heritable component of gene expression divergence. Remarkably, genes differentially expressed between species pairs in the common environment were similar to what had been previously identified in the wild. The collective results suggest a predominantly genetic control of differential transcription between these species pairs.106
In other studies, heritability of gene expression within a population has been estimated using parent–offspring or sibling regressions. For instance, studies of human gene expression have found that approximately 30% of genes have a significant heritable component.25,107 Estimating heritability for wild populations is also possible using restricted maximum likelihood (REML) “animal models” applied to multigenerational data from natural populations.108 When applied to pedigrees with multiple generations and low immigration rates, these models can reduce bias due to shared environment effects.109 Roberge et al.110 applied this approach to estimate heritability of gene expression in the Atlantic salmon genome, discovering that 16% of 6,500 gene transcripts had a heritable component of gene expression, on average explaining 40% of the variation in transcription profiles. These results compare to other median heritability estimates among genes with heritable transcription profiles ranging from 0.11 (in mice, Ref. 111) to 0.84 (in yeast, Ref. 112). Notably, studies estimating heritability using such approaches need to account for the fact that heritability within a population does not equate to heritable differences between populations. Overall, although there are no studies that have quantified the heritability of transcription profile differences underlying ecological speciation, these results suggest that the heritable component of gene expression exists, but is highly variable.
eQTL mapping
Analyses on the genetic architecture of transcriptome variation offers to further our understanding of the genetic basis of gene expression and adaptive divergence.113 By genetic architecture we mean quantifying the number, location, and effect sizes of genes contributing to adaptive divergence.114 In studies of genetic architecture, a QTL is defined as a region of the genome containing one or more genes that affect variation in a quantitative trait, identifiable by its linkage or association to polymorphic marker loci.115,116 Traditional, or “phenotypic” QTL (pQTL) uncover associations between genetic regions and traditional phenotypic traits such as morphology. Expression QTL (eQTL) map transcript abundance in the same manner as pQTL map “traditional” traits. eQTL mapping is emerging as a useful technique for localizing genomic regions contributing to gene expression divergence.115 eQTL studies are generally characterized by large numbers of phenotypes (e.g., the number of transcripts on a microarray), but the mapping is typically performed with fewer individuals, due to the still prohibitive cost of running the arrays. Although eQTL studies are still in their infancy, two general patterns have been observed: (1) the predominance of cis-localized eQTL and (2) the existence of genomic regions associated with the expression level of many transcripts (so-called eQTL “hotspots”). We consider each in turn.
The segregation of eQTL has a local genomic context because there are two ways, denoted as cis or trans, that the level of transcript variation may map onto the genome,116–118 with each providing a different interpretation about genetic architecture. If the transcription profile maps within the gene region for the transcript in question, this association is referred to as cis or proximal eQTL. In contrast, if the transcription profile maps to another gene or genomic region it is referred to as a trans or distal eQTL.116 Cumulatively, the distribution of cis versus trans eQTL on the genome has shown that cis eQTL seem to have larger genetic effect sizes than trans eQTL and that there are more cis than trans eQTL in the genome,119 although the biological interpretation of this pattern remains obscure.117,120
Another emerging pattern is the existence of eQTL “hotspots”: genomic regions that are associated with the expression level of many transcripts.26,71 These hotspots may involve the distribution of eQTLs as well as transcriptional covariation between individuals in the mapping family.113,116 What do these hotspots tell us about the genetics of ecological speciation? First, they show that pQTL and eQTL can map to the same genomic regions. For example, recent studies mapped both eQTL and pQTL for morphological, life-history, and behavioral traits in dwarf and normal lake whitefish species pairs.19,71,121 Of 261 white muscle eQTL distributed over 24 linkage groups, 15 eQTL localized with overlapping pQTL.121,122 Strikingly, almost 90% of eQTL–pQTL colocalizations involved growth rate and condition factor, two traits central to the adaptive divergence of these species pairs.19 Of course, a caveat about overlapping eQTL and pQTLs is that the sizes of the QTL regions are often quite large, such that the apparent colocalization of these two types of QTL need not imply a functional relationship. Nonetheless, the genes within these regions harboring both eQTL and pQTL are arguably strong candidates for genes involved in ecological speciation.
Additionally, eQTL studies indicate that genomic regions involved in ecological speciation can be nonrandomly distributed across the genome. For example, in the same lake whitefish species pairs noted above, 50% of 249 eQTL identified in the brain were associated with only 12 hotspots distributed over eight linkage groups.71 A similar pattern was observed in muscle, where 41% of eQTL mapping to six hotspots across four linkage groups.121 These findings hint at the existence of localized “genomic islands” of expression divergence, as sometimes reported for islands of genetic differentiation in population genomic studies123,124 (but see Ref. 125).
Finally, eQTL have also informed us about the actual mechanisms of speciation, for example confirming that mapped genomic regions differentiated via divergent selection. The direction of additive eQTL reported by in the Whiteley et al.71 and Derome et al.121 were predominantly in one direction, suggesting a role for directional selection.2,126 eQTL hotspots have also been associated with molecular signatures of selection in natural populations. For example, in the whitefish species pairs, 10 loci were identified whose genetic divergence in nature exceeds neutral expectations. These are so-called outlier loci subject to divergent selection.124,127 Three of these outlier loci also corresponded to eQTL hotspots.122 Finally, eQTL hotspots may be an indication that coexpression involves a regulatory network such that speciation involves complex interactions between genes.119 Overall, eQTL studies can thus be used to infer the genomic distribution of expression profiles26 with eQTL distributions potentially informing the mechanisms of gene regulation,34 and providing insight into the process of speciation.
Other approaches to studying heritability of gene expression
The previously discussed approaches to studying the heritability of gene expression divergence may be thought of as top-down or a forward genetics approach: they start with the phenotype or entire transcriptome and work toward narrowing down to regions or genes implicated in adaptive divergence and ecological speciation. However, few QTL maps or genome scans exhibit sufficient resolution to find the exact functional genes or regulatory elements that contain the polymorphisms that are under selection.127,128 Moreover, mapping approaches may not be feasible in some organisms. Another approach which relies on sequence comparison of functional polymorphisms may be described as a gene expression approach to “reverse ecology”129: after differentially expressed functional genes are identified, sequences of the differentially expressed transcripts are compared by direct sequencing efforts that may uncover nonsynonymous mutations in the coding regions of the genes, or genetic polymorphisms in regulatory regions.130 Both of these steps may now be accomplished simultaneously with next generation sequencing.80 Thus, screening the transcriptome for gene expression differences, even in the absence of a QTL map or genome scan, could simultaneously start the search for functional polymorphisms.
Genetic component of gene expression: conclusions
Common garden and eQTL studies clearly demonstrate that gene expression divergence can have a heritable component and be associated with adaptive genetic divergence. Although progress has been made in identifying specific differentially expressed genomic regions contributing to adaptive divergence, identification of specific mutations, and characterization of interactions among genomic regions, remains a major challenge for future work.
Functional links between adaptive candidate gene expression, adaptive genetic divergence, and reproductive isolation
Even after genetic components of gene expression are identified, a major question remains: are these components associated with adaptive divergence and reproductive isolation? Several studies have demonstrated that reductions in hybrid fitness can be due to gene (mis)expression131 (for review Ref. 132), in some cases linking gene misexpression in hybrids to other factors previously identified as contributing to ecological speciation.39 Along these lines, a growing number of studies have now isolated and characterized specific candidate genes or patterns of gene expression associated with the adaptive divergence which drives ecological speciation (e.g., Refs. 71, 106, 133–135). Of these, none has demonstrated an actual association between gene expression and reproductive isolation (but see Ref. 39), usually because reproductive isolation itself was not explicitly considered, underlying mutations have not been identified, or mutations causing adaptive divergence lie in cis-regulatory rather coding regions of genes.136,137 To compensate, we work under the assumption that genes whose expression is associated with adaptive divergence might also impact the fitness of immigrant and hybrids and thus make a contribution to ecologically based reproductive isolation (“immigrant inviability” and extrinsic postmating isolation), albeit of unknown magnitude. Testing this assumption represents a major avenue for future research. The examples below thus illustrate both the promise and the difficulties associated with linking gene expression to ecological speciation.
Bmp4: beak shape and speciation in Darwin's finches
Darwin's finches arose via adaptive radiation on the Galapagos Islands.138 Beak morphology diverged adaptively among populations and species in response to divergent selection stemming from competition and use of seeds of differing size and hardness.139,140 Beak morphology might also contribute to reproductive isolation via song divergence141 or due to selection against immigrants and (intermediate) hybrids.142,143 Among species, higher levels of the bone morphogenetic protein 4 (Bmp4) expression are correlated with deeper beak shapes and over-expression of Bmp4 in chick embryos altered beak development in the predicted direction.144 These results provide compelling evidence that gene expression variation from Bmp4 affects morphological divergence among species of Darwin's finches (Fig. 3). Similar results occur for another gene, calmodulin (CaM145). However, due to a lack of common garden or mapping studies, there is as of yet no evidence that heritable differences in beak morphology are affected by Bmp4 or CaM. The mutations underlying beak size differences in Darwin's finches have not been identified. Thus, although there is good evidence that regulatory changes underlie morphological divergence among species of Darwin's finches, the ultimate link between gene expression and genetically based reproductive isolation (= speciation) is yet to be made.
Figure 3.
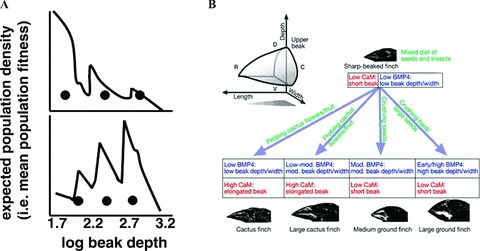
An example of the effects of gene expression in two genes (bone morphogenetic protein 4, bmp4, and calmodulin, CaM) on phenotypic traits of likely importance for ecological speciation in Geospiza, Darwin's finches. (A) Evidence for divergent selection on beak depth from reconstructions of adaptive landscapes. Lines depict the expected population density of a solitary granivorous finch species on two Galápagos islands (similar results were observed on 13 other islands). Dots depict mean log beak depths of actual populations for each curve. Distinct peaks in the adaptive landscape indicate divergent selection, as supported by the observation that actual beak depths differ among populations and tend to correspond to peaks in the landscape. Thus, selection against migrants between environments and intermediate hybrids would likely cause reproductive isolation. Modified from Schluter and Grant139 and reprinted with permission of the American Society for Naturalists. (B) Summary of the evidence that bmp- and CaM -dependent signaling regulates growth along different axes of bill morphology, facilitating the evolution of distinct beak morphologies in Darwin's finches. A beak of the sharp-beaked finch reflects a basal morphology for Geospiza. Abbreviations: C, caudal; D, dorsal; R, rostral; V, ventral. Modified from Abzhanov et al.145 and reprinted with permission of Nature.
Pitx1: pelvic reduction and speciation in threespine stickleback
Recently derived postglacial fish populations are among the most extensively studied systems of ecological speciation in nature (reviewed in Refs. 19, 146, 147). One such example is the threespine stickleback (Gasterosteus aculeatus) complex in which ecological divergence drove speciation between limnetic and benthic pairs within freshwater lakes, and between marine and freshwater populations.148,149 Ancestral marine and most derived freshwater stickleback have a robust pelvic apparatus, while at least 24 independent freshwater populations exhibit a greatly reduced or completely absent pelvic structure.137,150,151 Repeated parallel evolution is itself an indication that divergent selection drove evolution, with evidence pointing to predation and differences in ion concentration as the mechanisms of selection.152–155
Recent studies have examined the genetic basis of pelvic reduction. QTL studies repeatedly identified a single chromosomal region explaining more than two thirds the phenotypic variance in pelvic size.156–158 Yet, similar to Bmp4 in finches, the regulatory mutation contributing to differences in expression remained unknown until recently. Chan et al.137 reported that a small (501 bp) tissue specific enhancer (Pel) drives expression of the gene implicated in pelvic reduction (the Pitx1 gene137). Remarkably, small deletions functionally inactivated Pel in nine of 13 tested pelvic reduced populations. These regions exhibiting recurrent deletions, rather than the Pitx1 gene itself, appear to have been subject to positive selection.137 These results demonstrate that genetically based expression divergence contributed to adaptive divergence in pelvic morphology. However, direct links to between expression divergence and reproductive isolation remain to be established. The ability to conduct manipulative experiments in seminatural ponds (e.g., Refs. 95, 159) indicates that linking gene expression at Pitx1 to reproductive isolation (i.e., reduced fitness of immigrants and hybrids) is a distinct possibility.
Other examples
There are many other examples of studies of gene expression and adaptation, but few make links to adaptive genetic divergence, and thus few pertain directly to ecological speciation. For instance, cichlid fish species have adapted to divergent light environments within lakes, via the effects of gene expression on the tuning of visual perceptual sensitivity.160 In this case, changes in gene expression contribute to sensory diversification in replicate radiations of cichlid fishes in the clear waters of Lake Malawi versus the turbid waters of Lake Victoria, and functional substitutions contributing to expression divergence were identified.161 These studies demonstrate important findings with respect to the molecular basis of ecologically driven sensory diversification, but again a direct demonstration that this contributed to reproductive isolation does not yet exist.
Mimetic wing coloration in Heliconius butterflies gives rise to wing patterns that show repeated convergence between species and have adaptive value in mimicry and mate choice, thus potentially associated with ecological speciation.162–165 Comparative gene expression between two species, H. erato and H. melpomeme, found that cinnabar expression correlated with the forewing band, providing good evidence that the expression of this gene gives rise to the red-banded phenotype in both species.162 Chamberlain et al.166 report similar associations between wing color and gene expression, but within polymorphic populations. Differences in the actual traits in these studies (wing color and pattern) are heritable, but once again functional mutations contributing to reproductive isolation are lacking.
On the other hand, recent genome-wide analyses of the transcriptome have demonstrated that complex patterns of gene misexpression may underlie reproductive isolation mechanisms in hybrids. Renault et al.39 contrasted gene expression divergence at key early developmental stages in species pairs of normal and dwarf whitefish (Coregonus clupeaformis) and their F1 hybrids to identify the main mode of action responsible for gene transcription and to discover key genes misexpressed in hybrids. Although only five of 5,000 transcripts differed in mean expression level between parentals and hybrids at the embryonic stage, 617 out of 5,300 transcripts differed significantly for 16-week-old juveniles. Remarkably, significant gene misexpression in backcross hybrids involved several genes, most notably the disruption of three key developmental genes involved in protein folding and mRNA translation. Overall, direct demonstrations of how gene expression causes reproductive isolation remains a major missing link in connecting the role of gene expression to ecological speciation. Once such demonstrations are made, it will be necessary to test whether, and how, expression divergence actually reduces gene flow between natural populations.
Conclusions and future directions
Gene expression is likely to be important for the two events required for ecological speciation: population persistence and the evolution of genetically based reproductive isolation. Studies of plasticity and population persistence have yet to address gene expression explicitly. When it comes to adaptive genetic divergence and reproductive isolation, gene expression divergence has been shown to be heritable and to contribute to adaptive genetic divergence, but links to the evolution of reproductive isolation remain indirect (see Table 3 for a summary of what is known, and what needs to be done next). Our review suggests that establishing this link will be challenging because the genetic architecture of ecological speciation can be controlled by gene networks and regulatory regions, rendering an understanding of the functional association between gene expression and adaptive divergence difficult. This implies that it may be difficult to make predictions about the likelihood of ecological speciation based on gene expression profiles until we have a better idea about the genetic architecture of ecological speciation and how it compares to other mechanisms of speciation.19,99 Nonetheless, isolating the mutations contributing to variation in adaptive traits, and then studying their effects on reproductive isolation, is a necessary task for understanding how gene expression affects ecological speciation.137 This is also important for establishing whether gene expression changes are associated with the causes of ecological speciation, or are the consequences. Such goals will likely be best achieved by integrating multiple molecular techniques with experimental studies of how different mutations (alleles) affect fitness and reproductive isolation.4,34,95,167
Acknowledgments
We thank Dolph Schluter for discussions about gene expression and speciation. We would like to thank three anonmymous reviewers for their constructive comments that greatly improved the manuscript. During part of the writing of this manuscript, S.A.P., H.C., and P.N. were hosted by the Institute for Advanced Study, Wissenschaftskolleg, Berlin. S.M.R. was funded by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (NSERC).
Conflicts of interest
The authors declare no conflicts of interest.
References
- 1.Funk DJ. Isolating a role for natural selection in speciation: host adaptation and sexual isolation in Neochlamisus bebbianae leaf beetles. Evolution. 1998;52:1744–1759. doi: 10.1111/j.1558-5646.1998.tb02254.x. [DOI] [PubMed] [Google Scholar]
- 2.Schluter D. The Ecology of Adaptive Radiation. Oxford: Oxford University Press; 2000. [Google Scholar]
- 3.Schluter D. Ecology and the origin of species. Trends Ecol. Evol. 2001;16:372–380. doi: 10.1016/s0169-5347(01)02198-x. [DOI] [PubMed] [Google Scholar]
- 4.Schluter D, Conte GL. Genetics and ecological speciation. Proc. Natl. Acad. Sci. USA. 2009;106:9955–9962. doi: 10.1073/pnas.0901264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rundle HD, Nosil P. Ecological speciation. Ecol. Lett. 2005;8:336–352. [Google Scholar]
- 6.Muller HJ. Isolating mechanisms, evolution, and temperature. Biol. Symp. 1942;6:71–125. [Google Scholar]
- 7.Mayr E. Animal Species and Evolution. Cambridge: Belknap Press; 1963. [Google Scholar]
- 8.Mayr E. Ecological factors in speciation. Evolution. 1947;1:263–288. [Google Scholar]
- 9.Lu G, Bernatchez L. Correlated trophic specialization and genetic divergence in sympatric lake whitefish ecotypes (Coregonus clupeaformis): support for the ecological speciation hypothesis. Evolution. 1999;53:1491–1505. doi: 10.1111/j.1558-5646.1999.tb05413.x. [DOI] [PubMed] [Google Scholar]
- 10.Ogden R, Thorpe RS. Molecular evidence for ecological speciation in tropical habitats. Proc. Natl. Acad. Sci. USA. 2002;99:13612–13615. doi: 10.1073/pnas.212248499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Funk DJ, Nosil P, Etges WJ. Ecological divergence exhibits consistently positive associations with reproductive isolation across disparate taxa. Proc. Natl. Acad. Sci. USA. 2006;103:3209–3213. doi: 10.1073/pnas.0508653103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vines TH, Schluter D. Strong assortative mating between allopatric sticklebacks as a by-product of adaptation to different environments. Proc. R. Soc. B. 2006;273:911–916. doi: 10.1098/rspb.2005.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schluter D. Evidence for ecological speciation and its alternative. Science. 2009;323:737–741. doi: 10.1126/science.1160006. [DOI] [PubMed] [Google Scholar]
- 14.Funk DJ, Filchak KE, Feder JL. Herbivorous insects: model systems for the comparative study of speciation ecology. Genetica. 2002;116:251–267. [PubMed] [Google Scholar]
- 15.Jiggins CD, et al. Reproductive isolation caused by colour pattern mimicry. Nature. 2001;411:302–305. doi: 10.1038/35077075. [DOI] [PubMed] [Google Scholar]
- 16.Nosil P, Vines TH, Funk DJ. Perspective: reproductive isolation caused by natural selection against immigrants from divergent habitats. Evolution. 2005;59:705–719. [PubMed] [Google Scholar]
- 17.Rundle HD, Whitlock MC. A genetic interpretation of ecologically dependent isolation. Evolution. 2001;55:198–201. doi: 10.1111/j.0014-3820.2001.tb01284.x. [DOI] [PubMed] [Google Scholar]
- 18.Nosil P. Speciation with gene flow could be common. Mol. Ecol. 2008;17:2103–2106. doi: 10.1111/j.1365-294X.2008.03715.x. [DOI] [PubMed] [Google Scholar]
- 19.Rogers SM, Bernatchez L. The genetic architecture of ecological speciation and the association with signatures of selection in natural lake whitefish (Coregonas sp. Salmonidae) species pairs. Mol. Biol. Evol. 2007;24:1423–1438. doi: 10.1093/molbev/msm066. [DOI] [PubMed] [Google Scholar]
- 20.Via S, West J. The genetic mosaic suggests a new role for hitchhiking in ecological speciation. Mol. Ecol. 2008;17:4334–4345. doi: 10.1111/j.1365-294X.2008.03921.x. [DOI] [PubMed] [Google Scholar]
- 21.Schluter D. Ecological causes of speciation. In: Howard DJ, Berlocher SH, editors. Endless Forms: Species and Speciation. Oxford: Oxford University Press; 1998. pp. 114–129. [Google Scholar]
- 22.Levin DA. Ecological speciation: the role of disturbance. Syst. Bot. 2004;29:225–233. [Google Scholar]
- 23.Price TD, Qvarnstrom A, Irwin DE. The role of phenotypic plasticity in driving genetic evolution. Proc. R. Soc. B. 2003;270:1433–1440. doi: 10.1098/rspb.2003.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ranz JM, Machado CA. Uncovering evolutionary patterns of gene expression using microarrays. Trends Ecol. Evol. 2006;21:29–37. doi: 10.1016/j.tree.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Schadt EE, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 26.Gibson G, Weir B. The quantitative genetics of transcription. Trends Genet. 2005;21:616–623. doi: 10.1016/j.tig.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 27.Roelofs D, et al. Adaptive differences in gene expression associated with heavy metal tolerance in the soil arthropod Orchesella cincta. Mol. Ecol. 2009;18:3227–3239. doi: 10.1111/j.1365-294X.2009.04261.x. [DOI] [PubMed] [Google Scholar]
- 28.Whitehead A, Crawford DL. Variation within and among species in gene expression: raw material for evolution. Mol. Ecol. 2006;15:1197–1211. doi: 10.1111/j.1365-294X.2006.02868.x. [DOI] [PubMed] [Google Scholar]
- 29.Nevins JR, Potti A. Mining gene expression profiles: expression signatures as cancer phenotypes. Nat. Rev. Genet. 2007;8:601–609. doi: 10.1038/nrg2137. [DOI] [PubMed] [Google Scholar]
- 30.Aubin-Horth N, Renn SCP. Genomic reaction norms: using integrative biology to understand molecular mechanisms of phenotypic plasticity. Mol. Ecol. 2009;18:3763–3780. doi: 10.1111/j.1365-294X.2009.04313.x. [DOI] [PubMed] [Google Scholar]
- 31.Landry CR, Aubin-Horth N. Ecological annotation of genes and genomes through ecological genomics. Mol. Ecol. 2007;16:4419–4421. doi: 10.1111/j.1365-294X.2007.03504.x. [DOI] [PubMed] [Google Scholar]
- 32.Pena-Castillo L, Hughes TR. Why are there still over 1000 uncharacterized yeast genes? Genetics. 2007;176:7–14. doi: 10.1534/genetics.107.074468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carroll LS, Potts WK. Advances in the Study of Behavior. Vol. 36. Elsevier Academic Press. San Diego; 2006. Functional genomics requires ecology; pp. 173–215. [Google Scholar]
- 34.Dalziel AC, Rogers SM, Schulte PM. Linking genotypes to phenotypes and fitness: how mechanistic biology can inform molecular ecology. Mol. Ecol. 2009;18:4997–5017. doi: 10.1111/j.1365-294X.2009.04427.x. [DOI] [PubMed] [Google Scholar]
- 35.Scott CP, Williams DA, Crawford DL. The effect of genetic and environmental variation on metabolic gene expression. Mol. Ecol. 2009;18:2832–2843. doi: 10.1111/j.1365-294X.2009.04235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noor MAF, Feder JL. Speciation genetics: evolving approaches. Nat. Rev. Genet. 2006;7:851–861. doi: 10.1038/nrg1968. [DOI] [PubMed] [Google Scholar]
- 37.Liotta L, Petricoin E. Molecular profiling of human cancer. Nat. Rev. Genet. 2000;1:48–56. doi: 10.1038/35049567. [DOI] [PubMed] [Google Scholar]
- 38.Johnson NA, Porter AH. Rapid speciation via parallel, directional selection on regulatory genetic pathways. J. Theor. Biol. 2000;205:527–542. doi: 10.1006/jtbi.2000.2070. [DOI] [PubMed] [Google Scholar]
- 39.Renaut S, Nolte AW, Bernatchez L. Gene expression divergence and hybrid misexpression between lake whitefish species pairs (Coregonus spp. Salmonidae) Mol. Biol. Evol. 2009;26:925–936. doi: 10.1093/molbev/msp017. [DOI] [PubMed] [Google Scholar]
- 40.Feder ME, Mitchell-Olds T. Evolutionary and ecological functional genomics. Nat. Rev. Genet. 2003;4:651–657. doi: 10.1038/nrg1128. [DOI] [PubMed] [Google Scholar]
- 41.Thomas MA, Klaper R. Genomics for the ecological toolbox. Trends Ecol. Evol. 2004;19:439–445. doi: 10.1016/j.tree.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 42.Goetz FW, MacKenzie S. Functional genomics with microarrays in fish biology and fisheries. Fish Fish. 2008;9:378–395. [Google Scholar]
- 43.Gracey AY. Interpreting physiological responses to environmental change through gene expression profiling. J. Exp. Biol. 2007;210:1584–1592. doi: 10.1242/jeb.004333. [DOI] [PubMed] [Google Scholar]
- 44.Yuan JS, et al. Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7:12. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karlen Y, et al. Statistical significance of quantitative PCR. BMC Bioinformatics. 2007;8:131. doi: 10.1186/1471-2105-8-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berger SL, Kimmel AR. Guide to Molecular Cloning Techniques. New York: Academic Press; 1987. [Google Scholar]
- 47.Schlamp K, et al. BlotBase: a northern blot database. Gene. 2008;427:47–50. doi: 10.1016/j.gene.2008.08.026. [DOI] [PubMed] [Google Scholar]
- 48.Rasmussen R, et al. Quantitative PCR by continuous fluorescence monitoring of a double strand DNA specific binding dye. Biochemica. 1998;2:8–11. [Google Scholar]
- 49.Van Straalen NM, Roelofs D. An Introduction to Ecological Genomics. New York: Oxford University Press; 2006. [Google Scholar]
- 50.Etienne W, et al. Comparison of mRNA gene expression by RT-PCR and DNA microarray. BioTechniques. 2004;36:618–626. doi: 10.2144/04364ST02. [DOI] [PubMed] [Google Scholar]
- 51.Rajeevan MS, et al. Use of real-time quantitative PCR to validate the results of cDNA array and differential display PCR technologies. Methods. 2001;25:443–451. doi: 10.1006/meth.2001.1266. [DOI] [PubMed] [Google Scholar]
- 52.Liang P, Pardee AB. Differential display of eukaryotic messenger-RNA by means of polymerase chain-reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 53.Diatchenko L, et al. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shaw KL, Danley PD. Behavioral genomics and the study of speciation at a porous species boundary. German Zool. Soc. 2003;106:261–273. doi: 10.1078/0944-2006-00129. [DOI] [PubMed] [Google Scholar]
- 55.Jones H, Ostrowski M, Scanlan DJ. A suppression subtractive hybridization approach reveals niche-specific genes that may be involved in predator avoidance in marine Synechococcus isolates. Appl. Environ. Microbiol. 2006;72:2730–2737. doi: 10.1128/AEM.72.4.2730-2737.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schena M, et al. Quantitative monitoring of gene-expression patterns with a complementary-DNA mircroarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 57.Patterson TA, et al. Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nat. Biotechnol. 2006;24:1140–1150. doi: 10.1038/nbt1242. [DOI] [PubMed] [Google Scholar]
- 58.Bowtell D, Sambrook J. DNA Micorarrays: A Molecular Cloning Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]
- 59.Shimkets RA. Gene Expression Profiling. Methods and Protocols. Totowa: Humana Press; 2004. [Google Scholar]
- 60.Childs G, et al. Printing spotted glass microarrays. In: Bowtell D, Sambrook J, editors. DNA Microarrays: A Molecular Cloning Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2003. pp. 61–100. [Google Scholar]
- 61.Nielsen JL, Pavey SA. Perspectives: gene expression in fisheries management. Curr. Zool. 2010;56:157–174. [Google Scholar]
- 62.Yang HU, Churchill G. Estimating p-values in small microarray experiments. Bioinformatics. 2007;23:38–43. doi: 10.1093/bioinformatics/btl548. [DOI] [PubMed] [Google Scholar]
- 63.Ball CA, et al. An introduction to microarry bioinformatics. In: Bowtell D, Sambrook J, editors. DNA Microarrays: A Molecular Cloning Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2003. pp. 509–601. [Google Scholar]
- 64.Naidoo S, Denby KJ, Berger DK. Microarray experiments: considerations for experimental design. S. Afr. J. Sci. 2005;101:347–354. [Google Scholar]
- 65.Oleksiak MF, Churchill GA, Crawford DL. Variation in gene expression within and among natural populations. Nat. Genet. 2002;32:261–266. doi: 10.1038/ng983. [DOI] [PubMed] [Google Scholar]
- 66.Allison DB, et al. Microarray data analysis: from disarray to consolidation and consensus (vol 7, p. 55, 2006) Nat. Rev. Genet. 2006;7:406–406. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- 67.Ioannidis JPA, et al. Repeatability of published microarray gene expression analyses. Nat. Genet. 2009;41:149–155. doi: 10.1038/ng.295. [DOI] [PubMed] [Google Scholar]
- 68.Ball CA, et al. Submission of microarray data to public repositories. PLoS Biol. 2004;2:1276–1277. doi: 10.1371/journal.pbio.0020317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bar-Or C, Czosnek H, Koltai H. Cross-species microarray hybridizations: a developing tool for studying species diversity. Trends Genet. 2007;23:200–207. doi: 10.1016/j.tig.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 70.Kammenga JE, et al. Microarray challenges in ecology. Trends Ecol. Evol. 2007;22:273–279. doi: 10.1016/j.tree.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 71.Whiteley AR, et al. The phenomics and expression quantitative trait locus mapping of brain transcriptomes regulating adaptive divergence in lake whitefish species pairs (Coregonus sp.) Genetics. 2008;180:147–164. doi: 10.1534/genetics.108.089938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.von Schalburg KR, et al. Expansion of the genomics research on Atlantic salmon Salmo salar L. project (GRASP) microarray tools. J. Fish Biol. 2008;72:2051–2070. [Google Scholar]
- 73.von Schalburg KR, et al. Fish and chips: various methodologies demonstrate utility of a 16,006-gene salmonid microarray. BMC Genomics. 2005;6:126. doi: 10.1186/1471-2164-6-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Koop BF, et al. A salmonid EST genomic study: genes, duplications, phylogeny and microarrays. BMC Genomics. 2008;9:595. doi: 10.1186/1471-2164-9-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McKay JK, Stinchcombe JR. Ecological genomics of model eukaryotes. Evolution. 2008;62:2953–2957. doi: 10.1111/j.1558-5646.2008.00536.x. [DOI] [PubMed] [Google Scholar]
- 76.Velculescu VE, et al. Serial analysis of gene-expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 77.Matsumura H, et al. SuperSAGE array: the direct use of 26-base-pair transcript tags in oligonucleotide arrays. Nat. Methods. 2006;3:469–474. doi: 10.1038/nmeth882. [DOI] [PubMed] [Google Scholar]
- 78.Wall PK, et al. Comparison of next generation sequencing technologies for transcriptome characterization. BMC Genomics. 2009;10:347. doi: 10.1186/1471-2164-10-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vera JC, et al. Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. Mol. Ecol. 2008;17:1636–1647. doi: 10.1111/j.1365-294X.2008.03666.x. [DOI] [PubMed] [Google Scholar]
- 80.Wolf JBW, et al. Nucleotide divergence vs. gene expression differentiation: comparative transcriptome sequencing in natural isolates from the carrion crow and its hybrid zone with the hooded crow. Mol. Ecol. 2010;19:162–175. doi: 10.1111/j.1365-294X.2009.04471.x. [DOI] [PubMed] [Google Scholar]
- 81.Goetz F, et al. A genetic basis for the phenotypic differentiation between siscowet and lean lake trout (Salvelinus namaycush) Mol. Ecol. 2010;19:176–196. doi: 10.1111/j.1365-294X.2009.04481.x. [DOI] [PubMed] [Google Scholar]
- 82.Ferguson L, et al. Characterization of a hotspot for mimicry: assembly of a butterfly wing transcriptome to genomic sequence at the HmYb/Sb locus. Mol. Ecol. 2010;19:240–254. doi: 10.1111/j.1365-294X.2009.04475.x. [DOI] [PubMed] [Google Scholar]
- 83.Fontanillas P, et al. Key considerations for measuring allelic expression on a genomic scale using high-throughput sequencing. Mol. Ecol. 2010;19:212–227. doi: 10.1111/j.1365-294X.2010.04472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Renaut S, Nolte AW, Bernatchez L. Mining transcriptome sequences towards identifying adaptive single nucleotide polymorphisms in lake whitefish species pairs (Coregonus spp. Salmonidae) Mol. Ecol. 2010;19:115–131. doi: 10.1111/j.1365-294X.2009.04477.x. [DOI] [PubMed] [Google Scholar]
- 85.Baldwin JM. A new factor in evolution. Am. Nat. 1896;30:441–451. [Google Scholar]
- 86.Via S, Lande R. Genotype-environment interaction and the evolution of phenotypic plasticity. Evolution. 1985;39:505–522. doi: 10.1111/j.1558-5646.1985.tb00391.x. [DOI] [PubMed] [Google Scholar]
- 87.Robinson BW, Dukas R. The influence of phenotypic modifications on evolution: the Baldwin effect and modern perspectives. Oikos. 1999;85:582–589. [Google Scholar]
- 88.Yeh PJ, Price TD. Adaptive phenotypic plasticity and the successful colonization of a novel environment. Am. Nat. 2004;164:531–542. doi: 10.1086/423825. [DOI] [PubMed] [Google Scholar]
- 89.Reusch TBH, Wood TE. Molecular ecology of global change. Mol. Ecol. 2007;16:3973–3992. doi: 10.1111/j.1365-294X.2007.03454.x. [DOI] [PubMed] [Google Scholar]
- 90.Svanback R, Pineda-Krch M, Doebeli M. Fluctuating population dynamics promotes the evolution of phenotypic plasticity. Am. Nat. 2009;174:176–189. doi: 10.1086/600112. [DOI] [PubMed] [Google Scholar]
- 91.McCairns RJS, Bernatchez L. Adaptive divergence between freshwater and marine sticklebacks: insights into the role of phenotypic plasticity from an integrated analysis of candidate gene expression. Evolution. 2010;64:1029–1047. doi: 10.1111/j.1558-5646.2009.00886.x. [DOI] [PubMed] [Google Scholar]
- 92.Sorensen JG, Loeschcke V, Kristensen TN. Lessons from the use of genetically modified Drosophila melanogaster in ecological studies: Hsf mutant lines show highly trait-specific performance in field and laboratory thermal assays. Funct. Ecol. 2009;23:240–247. [Google Scholar]
- 93.Knight CA, et al. Expression profiling and local adaptation of Boechera holboellii populations for water use efficiency across a naturally occurring water stress gradient. Mol. Ecol. 2006;15:1229–1237. doi: 10.1111/j.1365-294X.2006.02818.x. [DOI] [PubMed] [Google Scholar]
- 94.Juenger TE, et al. Natural genetic variation in whole-genome expression in Arabidopsis thaliana: the impact of physiological QTL introgression. Mol. Ecol. 2006;15:1351–1365. doi: 10.1111/j.1365-294X.2006.02774.x. [DOI] [PubMed] [Google Scholar]
- 95.Barrett RDH, Schluter D. Adaptation from standing genetic variation. Trends Ecol. Evol. 2008;23:38–44. doi: 10.1016/j.tree.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 96.Barton NH, Hewitt GM. Adaptation, speciation and hybrid zones. Nature. 1989;341:497–503. doi: 10.1038/341497a0. [DOI] [PubMed] [Google Scholar]
- 97.Mallet J. A species definition for the Modern Synthesis. Trends Ecol. Evol. 1995;10:294–299. doi: 10.1016/0169-5347(95)90031-4. [DOI] [PubMed] [Google Scholar]
- 98.Mallet J. Hybridization as an invasion of the genome. Trends Ecol. Evol. 2005;20:229–237. doi: 10.1016/j.tree.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 99.Wu C. The genic view of the process of speciation. J. Evol. Biol. 2001;14:851–865. [Google Scholar]
- 100.Rogers SM, Bernatchez L. The genetic basis of intrinsic and extrinsic postzygotic reproductive isolation jointly promoting speciation in the lake whitefish species complex (Coregonus clupeaformis) J. Evol. Biol. 2006;19:1979–1994. doi: 10.1111/j.1420-9101.2006.01150.x. [DOI] [PubMed] [Google Scholar]
- 101.Gibson G. The environmental contribution to gene expression profiles. Nat. Rev. Genet. 2008;9:575–581. doi: 10.1038/nrg2383. [DOI] [PubMed] [Google Scholar]
- 102.Falconer DS, Mackay TFC. Quantitative Genetics. Essex: Benjamin Cummings; 1996. [Google Scholar]
- 103.Whitehead A, Crawford DL. Neutral and adaptive variation in gene expression. Proc. Natl. Acad. Sci. USA. 2006;103:5425–5430. doi: 10.1073/pnas.0507648103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Roff DA. A centennial celebration for quantitative genetics. Evolution. 2007;61:1017–1032. doi: 10.1111/j.1558-5646.2007.00100.x. [DOI] [PubMed] [Google Scholar]
- 105.Lai Z, et al. Natural variation in gene expression between wild and weedy populations of Helianthus annuus. Genetics. 2008;179:1881–1890. doi: 10.1534/genetics.108.091041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Derome N, Bernatchez L. The transcriptomics of ecological convergence between 2 limnetic coregonine fishes (Salmonidae) Mol. Biol. Evol. 2006;23:2370–2378. doi: 10.1093/molbev/msl110. [DOI] [PubMed] [Google Scholar]
- 107.Monks SA, et al. Genetic inheritance of gene expression in human cell lines. Am. J. Hum. Genet. 2004;75:1094–1105. doi: 10.1086/426461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kruuk LEB. Estimating genetic parameters in natural populations using the ‘animal model’. Phil. Trans. R. Soc. B. 2004;359:873–890. doi: 10.1098/rstb.2003.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kruuk LEB, Hadfield JD. How to separate genetic and environmental causes of similarity between relatives. J. Evol. Biol. 2007;20:1890–1903. doi: 10.1111/j.1420-9101.2007.01377.x. [DOI] [PubMed] [Google Scholar]
- 110.Roberge C, Guderley H, Bernatchez L. Genomewide identification of genes under directional selection: gene transcription QST scan in diverging Atlantic salmon subpopulations. Genetics. 2007;177:1011–1022. doi: 10.1534/genetics.107.073759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chesler EJ, et al. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat. Genet. 2005;37:233–242. doi: 10.1038/ng1518. [DOI] [PubMed] [Google Scholar]
- 112.Brem RB, et al. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 113.Rockman MV, Kruglyak L. Genetics of global gene expression. Nat. Rev. Genet. 2006;7:862–872. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- 114.Rieseberg LH. Genetic mapping as a tool for studying speciation. In: Soltis DE, Soltis PS, Doyle JJ, editors. Molecular systematics of plants. 2nd ed. New York: Chapman and Hall Inc.; 1998. pp. 459–487. [Google Scholar]
- 115.Gilad Y, Rifkin SA, Pritchard JK. Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet. 2008;24:408–415. doi: 10.1016/j.tig.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mackay TFC, Stone EA, Ayroles JF. The genetics of quantitative traits: challenges and prospects. Nat. Rev. Genet. 2009;10:565–577. doi: 10.1038/nrg2612. [DOI] [PubMed] [Google Scholar]
- 117.Hubner N, et al. Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat. Genet. 2005;37:243–253. doi: 10.1038/ng1522. [DOI] [PubMed] [Google Scholar]
- 118.Kirst M, et al. Genetic architecture of transcript-level variation in differentiating xylem of a eucalyptus hybrid. Genetics. 2005;169:2295–2303. doi: 10.1534/genetics.104.039198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wentzell AM, et al. Linking metabolic QTLs with network and cis-eQTLs controlling biosynthetic pathways. PLoS Genet. 2007;3:1687–1701. doi: 10.1371/journal.pgen.0030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Landry CR, et al. Compensatory cis–trans evolution and the dysregulation of gene expression in interspecific hybrids of Drosophila. Genetics. 2005;171:1813–1822. doi: 10.1534/genetics.105.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Derome N, et al. Pervasive sex-linked effects on transcription regulation as revealed by expression quantitative trait loci mapping in lake whitefish species pairs (Coregonus sp., salmonidae) Genetics. 2008;179:1903–1917. doi: 10.1534/genetics.107.086306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bernatchez L, et al. On the origin of species: insights from the ecological genomics of whitefish. Phil. Trans. R. Soc. B. 2010;365:1783–1800. doi: 10.1098/rstb.2009.0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Turner TL, Hahn MW, Nuzhdin SV. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 2005;3:1572–1578. doi: 10.1371/journal.pbio.0030285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nosil P, Funk DJ, Ortiz-Barrientos D. Divergent selection and heterogeneous genomic divergence. Mol. Ecol. 2009;18:375–402. doi: 10.1111/j.1365-294X.2008.03946.x. [DOI] [PubMed] [Google Scholar]
- 125.Michel AP, et al. Widespread genomic divergence during sympatric speciation. Proc. Natl. Acad. Sci. USA. 2010;107:9724–9729. doi: 10.1073/pnas.1000939107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Orr HA. Testing natural selection vs. genetic drift in phenotypic evolution using quantitative trait locus data. Genetics. 1998;149:2099–2104. doi: 10.1093/genetics/149.4.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Beaumont C, et al. A genome scan with AFLP((TM)) markers to detect fearfulness-related QTLs in Japanese quail. Anim. Genet. 2005;36:401–407. doi: 10.1111/j.1365-2052.2005.01336.x. [DOI] [PubMed] [Google Scholar]
- 128.Nielsen R. Molecular signatures of natural selection. Annu. Rev. Genet. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- 129.Li YF, et al. “Reverse ecology” and the power of population genomics. Evolution. 2008;62:2984–2994. doi: 10.1111/j.1558-5646.2008.00486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cassone BJ, et al. Differential gene expression in incipient species of Anopheles gambiae. Mol. Ecol. 2008;17:2491–2504. doi: 10.1111/j.1365-294X.2008.03774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Landry CR, Hartl DL, Ranz JM. Genome clashes in hybrids: insights from gene expression. Heredity. 2007;99:483–493. doi: 10.1038/sj.hdy.6801045. [DOI] [PubMed] [Google Scholar]
- 132.Ortiz-Barrientos D, Counterman BA, Noor MAF. Gene expression divergence and the origin of hybrid dysfunctions. Genetica. 2007;129:71–81. doi: 10.1007/s10709-006-0034-1. [DOI] [PubMed] [Google Scholar]
- 133.Lexer C, Lai Z, Rieseberg LH. Candidate gene polymorphisms associated with salt tolerance in wild sunflower hybrids: implications for the origin of Helianthus paradoxus, a diploid hybrid species. New Phytol. 2004;161:225–233. doi: 10.1046/j.1469-8137.2003.00925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Colosimo PF, et al. Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science. 2005;307:1928–1933. doi: 10.1126/science.1107239. [DOI] [PubMed] [Google Scholar]
- 135.Steiner CC, Weber JN, Hoekstra HE. Adaptive variation in beach mice produced by two interacting pigmentation genes. PLoS Biol. 2007;5:1880–1889. doi: 10.1371/journal.pbio.0050219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Stern DL, Orgogozo V. The loci of evolution: how predictable is genetic evolution? Evolution. 2008;62:2155–2177. doi: 10.1111/j.1558-5646.2008.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chan YF, et al. From trait to base pairs: parallel evolution of pelvic reduction in three-spined sticklebacks occurs by repeated deletion of a tissue-specific pelvic enhancer at Pitx1. Mech. Dev. 2009;126:S14–S15. [Google Scholar]
- 138.Grant PR. Ecology and Evolution of Darwin's Finches. Princeton: Princeton University Press; 1986. [Google Scholar]
- 139.Schluter D, Grant PR. Determinants of morphological patterns in communities of Darwin's finches. American Naturalist. 1984;123:175–196. [Google Scholar]
- 140.Grant PR, Grant BR. Evolution of character displacement in Darwin's finches. Science. 2006;313:224–226. doi: 10.1126/science.1128374. [DOI] [PubMed] [Google Scholar]
- 141.Podos J. Correlated evolution of morphology and vocal signal structure in Darwin's finches. Nature. 2001;409:185–188. doi: 10.1038/35051570. [DOI] [PubMed] [Google Scholar]
- 142.Grant PR, Grant BR. Pedigrees, assortative mating and speciation in Darwin's finches. Proc. R. Soc. B. 2008;275:661–668. doi: 10.1098/rspb.2007.0898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hendry AP, et al. Disruptive selection in a bimodal population of Darwin's finches. Proc. R. Soc. B. 2009;276:753–759. doi: 10.1098/rspb.2008.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Abzhanov A, et al. Bmp4 and morphological variation of beaks in Darwin's finches. Science. 2004;305:1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- 145.Abzhanov A, et al. The calmodulin pathway and evolution of elongated beak morphology in Darwin's finches. Nature. 2006;442:563–567. doi: 10.1038/nature04843. [DOI] [PubMed] [Google Scholar]
- 146.McKinnon JS, Rundle HD. Speciation in nature: the threespine stickleback model systems. Trends Ecol. Evol. 2002;17:480–488. [Google Scholar]
- 147.Hendry AP. Ecological speciation! Or the lack thereof? Can. J. Fish. Aquat. Sci. 2009;66:1383–1398. [Google Scholar]
- 148.Schluter D, McPhail JD. Ecological character displacement and speciation in sticklebacks. Am. Nat. 1992;140:85–108. doi: 10.1086/285404. [DOI] [PubMed] [Google Scholar]
- 149.McKinnon JS, et al. Evidence for ecology's role in speciation. Nature. 2004;429:294–298. doi: 10.1038/nature02556. [DOI] [PubMed] [Google Scholar]
- 150.Bell MA. Interacting evolutionary constraints in pelvic reduction of threespine sticklebacks, Gasterosteus aculeatus (Pisces, Gasterosteidae) Biol. J. Linn. Soc. 1987;31:347–382. [Google Scholar]
- 151.Gow JL, et al. Ecological predictions lead to the discovery of a benthic-limnetic sympatric species pair of threespine stickleback in Little Quarry Lake, British Columbia. Can. J. Zool. 2008;86:564–571. [Google Scholar]
- 152.Reimchen TE. Spine deficiency and polymorphism in a population of Gasterosteus aculeatus—an adaptation to predators. Can. J. Zool. 1980;58:1232–1244. [Google Scholar]
- 153.Bell MA, et al. Evolution of pelvic reduction in threespine stickleback fish—a test of competing hypotheses. Evolution. 1993;47:906–914. doi: 10.1111/j.1558-5646.1993.tb01243.x. [DOI] [PubMed] [Google Scholar]
- 154.Reimchen TE, Nosil P. Temporal variation in divergent selection on spine number in threespine stickleback. Evolution. 2002;56:2472–2483. doi: 10.1111/j.0014-3820.2002.tb00172.x. [DOI] [PubMed] [Google Scholar]
- 155.Vamosi SM. Predation sharpens the adaptive peaks: survival trade-offs in sympatric sticklebacks. Ann. Zool. Fenn. 2002;39:237–248. [Google Scholar]
- 156.Shapiro MD, et al. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature. 2004;428:717–723. doi: 10.1038/nature02415. [DOI] [PubMed] [Google Scholar]
- 157.Cresko WA, et al. Parallel genetic basis for repeated evolution of armor loss in Alaskan threespine stickleback populations. Proc. Natl. Acad. Sci. USA. 2004;101:6050–6055. doi: 10.1073/pnas.0308479101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Coyle SM, Huntingford FA, Peichel CL. Parallel evolution of Pitx1 underlies pelvic reduction in Scottish threespine stickleback (Gasterosteus aculeatus) J. Hered. 2007;98:581–586. doi: 10.1093/jhered/esm066. [DOI] [PubMed] [Google Scholar]
- 159.Schluter D. Experimental evidence that competition promotes divergence in adaptive radiation. Science. 1994;266:798–801. doi: 10.1126/science.266.5186.798. [DOI] [PubMed] [Google Scholar]
- 160.Carleton KL, Kocher TD. Cone opsin genes of African cichlid fishes: tuning spectral sensitivity by differential gene expression. Mol. Biol. Evol. 2001;18:1540–1550. doi: 10.1093/oxfordjournals.molbev.a003940. [DOI] [PubMed] [Google Scholar]
- 161.Hofmann HA. Evolution of cichlid mating systems: how social behavior sculpts brains and genomes. Integr. Comp. Biol. 2009;49:E78–E78. [Google Scholar]
- 162.Ferguson LC, Jiggins CD. Shared and divergent expression domains on mimetic Heliconius wings. Evol. Dev. 2009;11:498–512. doi: 10.1111/j.1525-142X.2009.00358.x. [DOI] [PubMed] [Google Scholar]
- 163.Mallet J, Gilbert LE. Why are there so many mimicry rings—correlations between habitat, behavior and mimicry in heliconious butterflies. Biol. J. Linn. Soc. 1995;55:159–180. [Google Scholar]
- 164.Kapan DD. Three-butterfly system provides a field test of mullerian mimicry. Nature. 2001;409:338–340. doi: 10.1038/35053066. [DOI] [PubMed] [Google Scholar]
- 165.Jiggins CD. Ecological speciation in mimetic butterflies. Bioscience. 2008;58:541–548. [Google Scholar]
- 166.Chamberlain NL, et al. Polymorphic butterfly reveals the missing link in ecological speciation. Science. 2009;326:847–850. doi: 10.1126/science.1179141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Storz JF, Wheat CW. Integrating evolutionary and functional approaches to infer adaptation at specific loci. Evolution. doi: 10.1111/j.1558-5646.2010.01044.x. In press. doi: 10.1111/j.1558-5646.2010.01044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Reiland J, Noor MAF. Little qualitative RNA misexpression in sterile male F1 hybrids of Drosophila pseudoobscura and D. persimilis. BMC Evol. Biol. 2002;2:16. doi: 10.1186/1471-2148-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kobayashi N, et al. Magp4 gene may contribute to the diversification of cichlid morphs and their speciation. Gene. 2006;373:126–133. doi: 10.1016/j.gene.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 170.Derome N, Duchesne P, Bernatchez L. Parallelism in gene transcription among sympatric lake whitefish (Coregonus clupeaformis Mitchill) ecotypes. Mol. Ecol. 2006;15:1239–1249. doi: 10.1111/j.1365-294X.2005.02968.x. [DOI] [PubMed] [Google Scholar]
- 171.Podrabsky JE, Somero GN. Changes in gene expression associated with acclimation to constant temperatures and fluctuating daily temperatures in an annual killifish Austrofundulus limnaeus. J. Exp. Biol. 2004;207:2237–2254. doi: 10.1242/jeb.01016. [DOI] [PubMed] [Google Scholar]
- 172.Bernier JC, et al. Differential gene expression between fall- and spring-run Chinook salmon assessed by long serial analysis of gene expression. Trans. Am. Fish. Soc. 2008;137:1378–1388. [Google Scholar]
- 173.Molina C, et al. SuperSAGE: the drought stress-responsive transcriptome of chickpea roots. BMC Genomics. 2008;9:553. doi: 10.1186/1471-2164-9-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Elmer KZ, et al. Rapid evolution and selection inferred from the transcriptomes of sympatric crater lake cichlid fishes. Mol. Ecol. 2010;19:197–211. doi: 10.1111/j.1365-294X.2009.04488.x. [DOI] [PubMed] [Google Scholar]
- 175.Zapata M, et al. Transcriptomic response of Argopecten purpuratus post-larvae to copper exposure under experimental conditions. Gene. 2009;442:37–46. doi: 10.1016/j.gene.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 176.Meier KM, et al. Increased growth hormone (GH), growth hormone receptor (GHR), and insulin-like growth factor I (IGF-I) gene transcription after hyperosmotic stress in the Brazilian flounder Paralichthys orbignyanus. Fish Physiol. Biochem. 2009;35:501–509. doi: 10.1007/s10695-008-9287-1. [DOI] [PubMed] [Google Scholar]
- 177.Kassahn KS, et al. From transcriptome to biological function: environmental stress in an ectothermic vertebrate, the coral reef fish Pomacentrus moluccensis. BMC Genomics. 2007;8:358. doi: 10.1186/1471-2164-8-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Adhikari BN, Wall DH, Adams BJ. Desiccation survival in an Antarctic nematode: molecular analysis using expressed sequenced tags. BMC Genomics. 2009;10:69. doi: 10.1186/1471-2164-10-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kreps JA, et al. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002;130:2129–2141. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ralph S, et al. Genomics of hybrid poplar (Populus trichocarpa x deltoides) interacting with forest tent caterpillars (Malacosoma disstria): normalized and full-length cDNA libraries, expressed sequence tags, and a cDNA microarray for the study of insect-induced defences in poplar. Mol. Ecol. 2006;15:1275–1297. doi: 10.1111/j.1365-294X.2006.02824.x. [DOI] [PubMed] [Google Scholar]
- 181.Le Quere A, et al. Screening for rapidly evolving genes in the ectomycorrhizal fungus Paxillus involutus using cDNA microarrays. Mol. Ecol. 2006;15:535–550. doi: 10.1111/j.1365-294X.2005.02796.x. [DOI] [PubMed] [Google Scholar]


