Abstract
The aim of this study was to clarify the genetic backgrounds underlying the clinicopathological characteristics of urothelial carcinomas (UCs). Array comparative genomic hybridization analysis using a 244K oligonucleotide array was performed on 49 samples of UC tissue. Losses of 2q33.3–q37.3, 4p15.2–q13.1 and 5q13.3–q35.3 and gains of 7p11.2–q11.23 and 20q13.12–q13.2 were correlated with higher histological grade, and gain of 7p21.2–p21.12 was correlated with deeper invasion. Losses of 6q14.1–q27 and 17p13.3–q11.1 and gains of 19q13.12–q13.2 and 20q13.12–q13.33 were correlated with lymph vessel involvement. Loss of 16p12.2–p12.1 and gain of 3q26.32–q29 were correlated with vascular involvement. Losses of 5q14.1–q23.1, 6q14.1–q27, 8p22–p21.3, 11q13.5–q14.1 and 15q11.2–q22.2 and gains of 7p11.2–q11.22 and 19q13.12–q13.2 were correlated with the development of aggressive non-papillary UCs. Losses of 1p32.2–p31.3, 10q11.23–q21.1 and 15q21.3 were correlated with tumor recurrence. Unsupervised hierarchical clustering analysis based on copy number alterations clustered UCs into three subclasses: copy number alterations associated with genome-wide DNA hypomethylation, regional DNA hypermethylation on C-type CpG islands and genome-wide DNA hypo- and hypermethylation were accumulated in clusters A, B1 and B2, respectively. Tumor-related genes that may encode therapeutic targets and/or indicators useful for the diagnosis and prognostication of UCs should be explored in the above regions. Both genetic and epigenetic events appear to accumulate during urothelial carcinogenesis, reflecting the clinicopathological diversity of UCs.
Introduction
Urothelial carcinomas (UCs) are classified as superficial papillary carcinomas or non-papillary carcinomas according to their configuration (1). Papillary carcinomas usually remain non-invasive although patients need to undergo repeated urethrocystoscopic resection for recurrences. In contrast, non-papillary invasive carcinomas usually develop from widely spreading flat carcinomas in situ showing a higher histological grade, and their clinical outcome is poor. There is also an alternative pathway by which papillary carcinomas develop higher histological grades during repetitive recurrence and transform into non-papillary invasive carcinomas. Thus, UCs show marked clinicopathological diversity (2). In order to improve the efficiency of diagnosis and therapy, it is necessary to clarify the genetic backgrounds underlying the various clinicopathological characteristics of UCs.
Previous studies employing Southern blotting based on restriction enzyme length polymorphism, polymerase chain reaction (PCR)–loss of heterozygosity (LOH) analysis using microsatellite markers, comparative genomic hybridization (CGH) analysis and fluorescence in situ hybridization (FISH) have revealed chromosomal instability in UCs such as losses of 2q, 5q, 9q and 10q and gains of 5p, 7p, 8q, 11q and 20q (3–12). However, such approaches are not effective for defining the break points in detail. Although recently developed array-based technology has been applied to UCs (13–18), the resolution of the arrays employed was insufficient or correlations between copy number alterations and the clinicopathological parameters of UCs were not analyzed in detail. Therefore, the genetic backgrounds underlying urothelial carcinogenesis have not been fully clarified.
In addition, multistage carcinogenesis is known to comprise both genetic and epigenetic events (19–21). We have reported the accumulation of DNA methylation on C-type CpG islands (22) in a cancer-specific, but not age-dependent, manner and demonstrated protein overexpression of DNA methyltransferase 1, a major DNA methyltransferase, even in non-cancerous urothelia with no apparent histological changes obtained from patients with UCs (23,24), as a result of possible exposure to carcinogens in the urine at the precancerous stage. Accumulation of DNA methylation on C-type CpG islands associated with DNA methyltransferase 1 protein overexpression was more frequently evident in aggressive non-papillary UCs (23,24). DNA hypomethylation on pericentromeric satellite regions was significantly correlated with LOH on chromosome 9 in UCs (25). Moreover, we have identified optimal indicators for carcinogenetic risk estimation in histologically normal urothelia, and for prognostication in surgically resected specimens from patients with UCs (26) using the bacterial artificial chromosome array-based methylated CpG island amplification (BAMCA) method (27–29), which is suitable for overviewing the DNA methylation tendency of individual large regions among all chromosomes (30). Although these data indicated that not only genetic but also epigenetic alterations play significant roles in UC development, to our knowledge, the correlations between copy number alterations and DNA methylation profiles in UCs have not been examined in a genome-wide manner.
In the present study of 49 UCs, we analyzed copy number alterations by array CGH analysis using a high-resolution (244K) oligonucleotide array, DNA methylation alterations on a genome-wide scale using BAMCA and DNA methylation status on C-type CpG islands using bisulfite modification. We then examined the clinicopathological significance of copy number alterations and the correlations between alterations of copy number and those of DNA methylation.
Material and methods
Patients and tissue samples
Forty-nine samples (T1 to T49) of UCs of the urinary bladder, ureter and renal pelvis were obtained from specimens that had been surgically resected by radical cystectomy (16 patients) or nephroureterectomy (33 patients) at the National Cancer Center Hospital, Tokyo, Japan. The patients comprised 38 men and 11 women whose mean age was 68.59 ± 10.11 (mean ± standard deviation) years (range 49–85 years). Among the UCs, 19 and 30 were graded as low- and high-grade tumors, respectively, on the basis of the World Health Organization classification (31), and 34 and 15 were classed as superficial (pTis, pTa, pT1) and invasive (pT2 or more), respectively (31). Histological examination of UCs revealed lymph vessel involvement in 16 and vascular involvement in 9. On the basis of macroscopic examination, the UCs were divided into 28 papillary tumors and 21 non-papillary tumors. Five patients were positive for lymph node metastasis at the point of radical cystectomy or nephroureterectomy. Recurrence was diagnosed by urologists mainly on the basis of computed tomography, abdominal ultrasonography and urine cytological examinations. The mean observation period was 39.7 ± 31.8 months (mean ± standard deviation) and seven patients were positive for recurrence (lymph node metastasis, local recurrence and metastasis to the lung or bone in three, two and two patients, respectively). Clinicopathological parameters for each of the examined patients are summarized in supplementary Table S1, available at Carcinogenesis Online. This study was approved by the Ethics Committee of the National Cancer Center, Tokyo, Japan and was performed in accordance with the Declaration of Helsinki 1995. All patients gave their informed consent prior to their inclusion in this study.
Array CGH analysis
High-molecular-weight DNA from fresh-frozen tissue samples was extracted using phenol–chloroform, followed by dialysis. Array CGH was performed using a Human Genome CGH 244K Oligo Microarray Kit (Agilent Technologies, Santa Clara, CA). Labeling and hybridization were performed according to the manufacturer’s protocol (Protocol v4.0, June 2006). Briefly, 2 μg of DNA from the patient and from a sex-matched control were double digested with AluI and RsaI (Promega, Madison, WI) for 2 h at 37°C. The digested DNA was then labeled by random priming using an Agilent Genomic DNA Labeling Kit Plus. Patient DNA and control DNA were labeled with Cy5-dUTP and Cy3-dUTP, respectively, and the labeled DNAs were hybridized with human Cot I DNA at 65°C with rotation for 40 h. Arrays were analyzed using the Agilent DNA microarray scanner and the Agilent Feature Extraction software. Presentation of the results was obtained using the Agilent CGH Analytics software package.
The results of array CGH were validated by FISH analysis. An LSI p16 (9p21) SpectrumOrange/CEP 9 SpectrumGreen Probe and an LSI p53 (17p13.1) SpectrumOrange Probe (Abbott/Vysis, Abbott Park, IL), corresponding to the CDKN2A and TP53 loci, respectively, were used. The FISH probes were hybridized to 5 μm thick sections of formalin-fixed, paraffin-embedded tissue specimens taken from a region immediately adjacent to that from which the corresponding fresh-frozen sample had been obtained within the same UC. Nuclei were stained with 4,5-diamidino-2-phenylindole.
BAC array-based methylated CpG island amplification
Because DNA methylation status is known to be organ specific (32), the reference DNA for analysis of the developmental stages of UCs should be obtained from the urothelium and not from other tissues or peripheral blood. Therefore, a mixture of normal urothelial DNA obtained from 11 male patients (C19 to C29) and 6 female patients (C30 to C35) without UCs was used as a reference for analyses of male and female test DNA samples, respectively. Of these 17 patients, 13 and 4 had undergone nephrectomy for renal cell carcinoma and nephrectomy for retroperitoneal sarcoma around the kidney, respectively. The mean age of the patients from whom normal urothelia had been obtained was 66.18 ± 10.49 (mean ± standard deviation) years (range 54–82 years). DNA methylation status was analyzed by BAMCA using a custom-made array (molecular cytogenetics Whole Genome Array-4500) harboring 4361 bacterial artificial chromosome (BAC) clones located throughout chromosomes 1–22, X and Y (33), as described previously (34,35). In 40 samples of UCs (T1 to T40), BAMCA had been performed and the results have already been published (26). For the present study, BAMCA was performed on nine additional samples of UCs (T41 to T49), and correlations between DNA methylation status and copy number alterations were examined in all 49 UCs.
Methylation-specific PCR and combined bisulfite restriction enzyme analysis
DNA methylation status on 5 C-type CpG islands was analyzed by methylation-specific polymerase chain reaction (MSP) and combined bisulfite restriction enzyme analysis (COBRA), as described previously (36). Briefly, bisulfite conversion was carried out using a CpGenome DNA Modification Kit (Chemicon International, Temecula, CA). DNA methylation status on CpG islands of the p16 and hMLH1 genes was determined by MSP using the primers described previously (36). The DNA methylation status of the methylated in tumor (MINT)-1, MINT-2 and MINT-12 clones was determined by COBRA using previously described primers and restriction enzymes (36). In 40 samples of UCs (T1 to T40), MSP and COBRA had been performed and the results have already been published (26). For the present study, MSP and COBRA were performed on nine additional samples of UCs (T41 to T49), and correlations between DNA methylation status and copy number alterations were examined in all 49 UCs.
Statistics
Correlations between copy number alterations and clinicopathological parameters of UCs were analyzed using the unpaired T-test. Based on Bonferroni correction for multiplicity of testing, differences at P < 0.00714 were considered significant. Unsupervised two-dimensional hierarchical clustering analysis of UCs was done using GeneSpring GX 10.0. Differences in the average number of array CGH probes showing copy number alterations, the average number of BAC clones showing DNA methylation alterations (hypo- and hypermethylation) and the average number of C-type CpG islands showing DNA methylation in UCs belonging to clusters A, B1 and B2 yielded by the unsupervised hierarchical clustering were analyzed using the Kruskal–Wallis test. Differences at P < 0.05 were considered significant.
Results
Validation of array CGH analysis by FISH
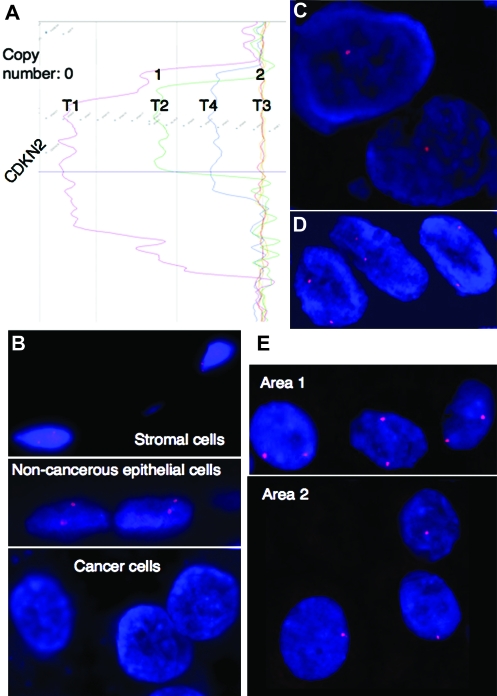
The array CGH analysis for copy number alterations was validated by FISH. Examples of array CGH profiles and FISH images of the four representative UCs (T1 to T4) are shown in Figure 1A–E, respectively. The signal ratios of the CDKN2A locus in T1, T2 and T3 corresponded to copy numbers of 0, 1 and 2, respectively, whereas the signal ratio in T4 did not correspond to any whole numbers (Figure 1A). The LSI p16 (9p21) SpectrumOrange/CEP 9 SpectrumGreen Probe corresponding to the CDKN2A gene revealed two signals in stromal cells and adjacent non-cancerous urothelial cells on the specimen of T1 (Figure 1B). The probe revealed zero, one and two signals in cancer cells in T1, T2 and T3, respectively (Figure 1B–D). FISH analysis revealed copy number heterogeneity within a UC: cancer cells showing two signals and those showing one signal were both observed in T4 (Figure 1E). These findings were able to explain the array CGH profile in T4 (Figure 1A). Similarly FISH analysis using the LSI p53 (17p13.1) SpectrumOrange Probe corresponding to the TP53 gene also validated the array CGH profiles (data not shown).
Fig. 1.
Validation of array CGH analysis by FISH. (A) Array CGH profiles of representative tissue specimens (T1 to T4). The signal ratios of the CDKN2 locus in T1, T2 and T3 corresponded to copy numbers of 0, 1 and 2, respectively, whereas the signal ratio in T4 did not correspond to any whole number. (B) Although the LSI p16 (9p21) SpectrumOrange/CEP 9 SpectrumGreen Probe corresponding to the CDKN2A gene revealed two signals in stromal cells and adjacent non-cancerous urothelial cells, it revealed no signal in cancer cells in T1. (C) FISH analysis using the same probe revealed one signal in cancer cells in T2. (D) FISH analysis using the same probe revealed two signals in cancer cells in T3. (E) FISH analysis using the same probe revealed copy number heterogeneity in T4: cancer cells in areas 1 and 2 showed two signals and one signal within a tumor, respectively. These findings can explain the array CGH profile of T4 in panel (A).
Copy number alterations and their clinicopathological impact in UCs
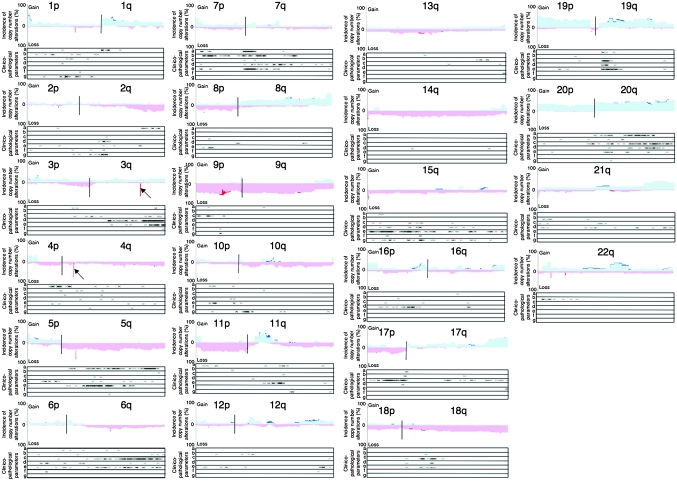
Figure 2 shows an overview of the copy number alterations on chromosomes 1–22 in all examined UCs. Chromosomal regions in which the incidence of copy number alterations in all examined UCs were ≥20% are summarized in Table I. If a UC showed copy number heterogeneity like that of T4 in Figure 1, the copy number observed in the major area within the tumor was described as the copy number of the UC in Figure 2 and Table I. On 3q26.1 and 4q13.2 (arrows in Figure 2), the incidence of homozygous deletion (copy number 0) on only 10 and 11 continuous oligonucleotide probes was high (59.2 and 67.3%, respectively, Table I). Although copy number polymorphism has been reported in the above region on 3q26.1, the UGT2B17 gene, which may be associated with smoking-related cancers (37), is the only gene reported to be located within the homozygously deleted region on 4q13.2.
Fig. 2.
Copy number alterations and their clinicopathological impacts in UCs. The incidence of copy number alterations on chromosomes 1–22 in UCs (T1 to T49) is shown. Gains (copy number: ≥3) and losses (copy number: 1 or 0) are shown in the upper and lower halves, respectively. Copy numbers of 0, 1, 3 and more are shown in dark red, light red, light blue and dark blue, respectively. The homozygously deleted regions on 3q26.1 and 4q13.2 are indicated by arrows. Locations of the array CGH probe on which copy number alterations were significantly correlated (unpaired T-test with Bonferroni correction, P < 0.00714) with histological grade (a), depth of invasion (b), lymph vessel involvement (c), vascular involvement (d), tumor configuration (papillary versus non-papillary, (e) lymph node metastasis (f) and recurrence (g) of UCs are indicated by ‘X’ under each of the histograms for chromosomes 1–22.
Table I.
Copy number alterations showing incidences of >20% in the examined UCs
| Chromosomal loci | Regionsa | CNb | LIc | HId | Chromosomal loci | Regionsa | CNb | LIc | HId |
| 1p36.33–1p36.31 | 000554287–006656854 | 3 | 22.4 | 65.3 | 10q21.1–10q21.3 | 057989564–069900014 | 1 | 20.4 | 26.5 |
| 1p36.21–1p36.12 | 016167535–020948798 | 3 | 20.4 | 22.4 | 10q22.1–10q23.1 | 071904528–073262876 | 3 | 20.4 | 24.5 |
| 1p31.1 | 072550248–072568068 | 0 | 20.4 | 20.4 | 10q23.1 | 083089747–087102060 | 1 | 20.4 | 24.5 |
| 1q12–1q31.1 | 142721264–187457065 | 3 | 20.4 | 40.8 | 10q23.2–10q24.31 | 089445919–102120260 | 1 | 20.4 | 26.5 |
| 1q31.1–1q44 | 190299165–247190770 | 3 | 20.4 | 30.6 | 10q25.1–10q26.13 | 105709333–124981616 | 1 | 20.4 | 26.5 |
| 2q32.1–2q32.2 | 185846809–190742545 | 1 | 20.4 | 20.4 | 10q26.3 | 133783634–135066163 | 3 | 20.4 | 22.4 |
| 2q32.3–2q37.3 | 192350477–242717069 | 1 | 20.4 | 32.7 | 11p15.5–11p15.4 | 000182372–003270486 | 3 | 28.6 | 32.7 |
| 3p25.3 | 009517826–011146712 | 3 | 22.4 | 26.5 | 11q21–11q22.1 | 096386431–097377420 | 1 | 20.4 | 20.4 |
| 3p25.2–3p25.1 | 012038778–015468042 | 3 | 20.4 | 32.7 | 11q22.1–11q23.3 | 097429396–116543936 | 1 | 20.4 | 26.5 |
| 3p21.31 | 048346498–050680352 | 3 | 20.4 | 24.5 | 11q23.3–11q24.2 | 124714078–125280002 | 1 | 20.4 | 20.4 |
| 3p14.1 | 065390626–065748270 | 1 | 20.4 | 20.4 | 11q24.2 | 124714078–125280002 | 1 | 20.4 | 20.4 |
| 3p14.1–3p13 | 068561452–072819381 | 1 | 20.4 | 24.5 | 11q24.2–11q25 | 126760808–132830348 | 1 | 20.4 | 22.4 |
| 3p12.3 | 076229787–076504425 | 1 | 20.4 | 20.4 | 11q25 | 133367262–133662626 | 1 | 20.4 | 20.4 |
| 3p12.3 | 077466215–078924208 | 1 | 20.4 | 20.4 | 12p13.33 | 001767600–002412124 | 3 | 20.4 | 20.4 |
| 3p12.3 | 079019795–079019854 | 1 | 20.4 | 20.4 | 12p13.31 | 006172174–007239121 | 3 | 20.4 | 22.4 |
| 3p12.3–3p12.2 | 081674283–083271792 | 1 | 20.4 | 24.5 | 12q13.13–12q13.2 | 050511140–053289666 | 3 | 22.4 | 34.7 |
| 3q21.3 | 128095046–130849578 | 3 | 20.4 | 22.4 | 12q24.23–12q24.31 | 119012815–122594513 | 3 | 20.4 | 20.4 |
| 3q26.1 | 163997228–164101835 | 0 | 51.0 | 59.2 | 13q13.3 | 036216743–038434639 | 1 | 20.4 | 22.4 |
| 3q27.1–3q27.2 | 184336189–186044074 | 3 | 20.4 | 20.4 | 13q21.1 | 053204015–057297991 | 1 | 20.4 | 22.4 |
| 3q29 | 194947608–198141010 | 3 | 20.4 | 20.4 | 13q34 | 112711763–114123908 | 3 | 20.4 | 22.4 |
| 3q29 | 198287059–1993214468 | 3 | 20.4 | 20.4 | 14q11.1–14q11.2 | 018149473–019665348 | 1 | 20.4 | 40.8 |
| 4p16.3–4p16.2 | 000041413–004080171 | 3 | 22.4 | 30.6 | 14q12 | 025347676–028443093 | 1 | 20.4 | 20.4 |
| 4p16.2–4p16.1 | 004863024–009205888 | 3 | 20.4 | 30.6 | 14q12–14q32.31 | 028746840–101366386 | 1 | 20.4 | 36.7 |
| 4p16.1 | 009410429–009800836 | 3 | 20.4 | 20.4 | 14q32.33 | 103609874–105504791 | 3 | 20.4 | 30.6 |
| 4q13.2 | 069057735–069165872 | 0 | 38.8 | 67.3 | 15q11.1–15q11.2 | 018683110–020387386 | 1 | 24.5 | 36.7 |
| 4q28.3 | 135481517–138478960 | 1 | 22.4 | 26.5 | 15q11.2 | 018683110–019435559 | 3 | 20.4 | 20.4 |
| 4q34.1 | 172444145–173706467 | 1 | 20.4 | 20.4 | 15q21.3 | 054364049–055292702 | 1 | 20.4 | 20.4 |
| 4q34.2–4q35.1 | 176641828–183811059 | 1 | 20.4 | 24.5 | 15q24.1–15q24.2 | 072125930–073833248 | 3 | 20.4 | 20.4 |
| 4q35.1 | 184728685–184971082 | 1 | 20.4 | 20.4 | 16p13.3 | 000028087–003208434 | 3 | 22.4 | 36.7 |
| 4q35.1–4q35.2 | 186466884–190719413 | 1 | 20.4 | 24.5 | 16p13.2 | 007398132–007610763 | 1 | 20.4 | 20.4 |
| 5p15.33 | 000075149–004092634 | 3 | 20.4 | 46.9 | 16p11.2 | 028394123–031439837 | 3 | 20.4 | 32.7 |
| 5p15.32–5p15.2 | 005757937–010857368 | 3 | 20.4 | 22.4 | 16q21 | 057623929–059070656 | 1 | 20.4 | 20.4 |
| 5p15.2–5p15.1 | 014200030–016428467 | 3 | 20.4 | 20.4 | 16q22.1 | 065296755–066623461 | 3 | 20.4 | 22.4 |
| 5q11.1–5q35.3 | 049595677–180644869 | 1 | 22.4 | 63.3 | 16q24.1–16q24.3 | 083207007–088690615 | 3 | 20.4 | 26.5 |
| 6p21.33–6p21.32 | 031497746–032281493 | 3 | 20.4 | 24.5 | 17p13.3–17p11.2 | 000029169–020234630 | 1 | 20.4 | 32.7 |
| 6p21.32–6p21.31 | 033247001–034187994 | 3 | 20.4 | 20.4 | 17q11.2 | 023533773–024473421 | 3 | 20.4 | 20.4 |
| 6p21.2–6p21.1 | 040188750–044391792 | 3 | 20.4 | 24.5 | 17q21.2–17q21.31 | 037792629–038922402 | 3 | 20.4 | 20.4 |
| 6q16.3–6q21 | 101107740–105665855 | 1 | 20.4 | 22.4 | 17q21.31 | 039360337–041458716 | 3 | 20.4 | 20.4 |
| 6q21–6q22.2 | 113047208–117916793 | 1 | 20.4 | 20.4 | 17q21.32–17q21.33 | 043930335–046303881 | 3 | 20.4 | 24.5 |
| 7p22.2 | 000140213–003449208 | 3 | 20.4 | 42.9 | 17q24.3–17q25.3 | 067481954–078653589 | 3 | 20.4 | 49.0 |
| 7p22.2–7p22.1 | 003871971–005993219 | 3 | 20.4 | 28.6 | 18p11.32–18p11.23 | 000004316–008103527 | 1 | 20.4 | 22.4 |
| 7p13 | 043944978–045169498 | 3 | 20.4 | 24.4 | 18p11.21 | 013020518–013601674 | 1 | 20.4 | 20.4 |
| 7q11.23 | 072356188–075985576 | 3 | 22.4 | 22.4 | 18q11.2 | 019251951–019903282 | 1 | 20.4 | 20.4 |
| 7q22.1 | 099307676–102120122 | 3 | 20.4 | 30.6 | 18q12.1–18q23 | 023887204–076111023 | 1 | 20.4 | 36.7 |
| 8p23.2–8p23.1 | 002209252–006655643 | 1 | 20.4 | 22.4 | 19p13.3–19p13.11 | 000064418–019716580 | 3 | 28.6 | 53.1 |
| 8p23.1 | 007040596–008140129 | 1 | 24.5 | 32.7 | 19q12–19q13.42 | 032981858–061360576 | 3 | 20.4 | 42.9 |
| 8p22 | 013056908–018810539 | 1 | 20.4 | 22.4 | 20p13–20q13.33 | 000008747–062379118 | 3 | 26.5 | 57.1 |
| 8p21.3–8p21.2 | 023372368–027249779 | 1 | 20.4 | 24.5 | 21p11.2 | 009896630–013600286 | 1 | 20.4 | 34.7 |
| 8p21.1–8p12 | 027678573–037057454 | 1 | 20.4 | 28.6 | 21q22.3 | 041606431–046914745 | 3 | 20.4 | 38.8 |
| 8q11.1–8q24.3 | 047062121–146264902 | 3 | 20.4 | 61.2 | 22q11.21 | 016646613–019038934 | 3 | 22.4 | 44.9 |
| 9p24.3–9p11.2 | 000153131–044199460 | 1 | 20.4 | 53.1 | 22q11.21 | 018989547–018989606 | 0 | 20.4 | 20.4 |
| 9p11.2–9q34.3 | 045419207–140241935 | 1 | 20.4 | 51.0 | 22q11.21 | 019835358–020440240 | 3 | 20.4 | 24.5 |
| 9p21.3 | 021698371–022372349 | 0 | 20.4 | 26.5 | 22q11.23 | 021944430–022991816 | 3 | 20.4 | 22.4 |
| 9p12–9p11.2 | 041970428–046018111 | 3 | 26.5 | 36.7 | 22q12.3–22q13.1 | 034773534–038422701 | 3 | 20.4 | 40.8 |
| 9p24.3 | 000153131–140241935 | 1 | 20.4 | 53.1 | 22q13.3–22q13.33 | 049000786–049565875 | 3 | 20.4 | 20.4 |
| 9q34.2–9q34.2 | 135191259–139424835 | 3 | 20.4 | 22.4 |
Based on NCBI36/hg18.
Copy number (If a UC shows copy number heterogeneity, the copy number observed in the major area within the tumor is considered to be the copy number of the UC).
Lowest incidence of copy number alterations in the chromosomal regions (%).
Highest incidence of copy number alterations in the chromosomal regions (%).
Chromosomal loci on which copy number alterations were significantly correlated with clinicopathological parameters of UCs are shown in Figure 2. The clinicopathological impacts of the copy number alterations are also summarized in Figure 3. For example, loss of 1p32.2–p31.3 was correlated with UC recurrence. Loss of 2q33.3–q37.3 was correlated with higher histological grade. Gain of 3q26.32–q29 was correlated with vascular involvement. Loss of 4p15.2–q13.1 was correlated with higher histological grade. Losses of 5q13.3–q35.3 and 5q14.1–q23.1 were correlated with higher histological grade and tumor configuration (development of non-papillary tumors), respectively. Loss of 6q14.1–q27 was correlated with both lymph vessel involvement and tumor configuration. Gains of 7p21.2–p21.12, 7p11.2–q11.22 and 7p11.2–q11.23 were correlated with deeper invasion, tumor configuration and higher histological grade, respectively. Loss of 8p22–p21.3 was correlated with tumor configuration. Loss of 10q11.23–q21.1 was correlated with UC recurrence. Loss of 11q13.5–q14.1 was correlated with tumor configuration. Losses of 15q11.2–q22.2 and 15q21.3 were correlated with tumor configuration and recurrence, respectively. Loss of 16p12.2–p12.1 was correlated with vascular involvement of UCs. Loss of 17p13.3–q11.1 was correlated with lymph vessel involvement. Gain of 19q13.12–q13.2 was correlated with lymph vessel involvement and tumor configuration. Gains of 20q13.12–q13.2 and 20q13.12–q13.33 were correlated with higher histological grade and lymph vessel involvement, respectively. On the other hand, although the incidences of 8q gain and 9p, 11p and 14q loss were generally high in UCs, such copy number alterations were not evidently correlated with any clinicopathological parameters.
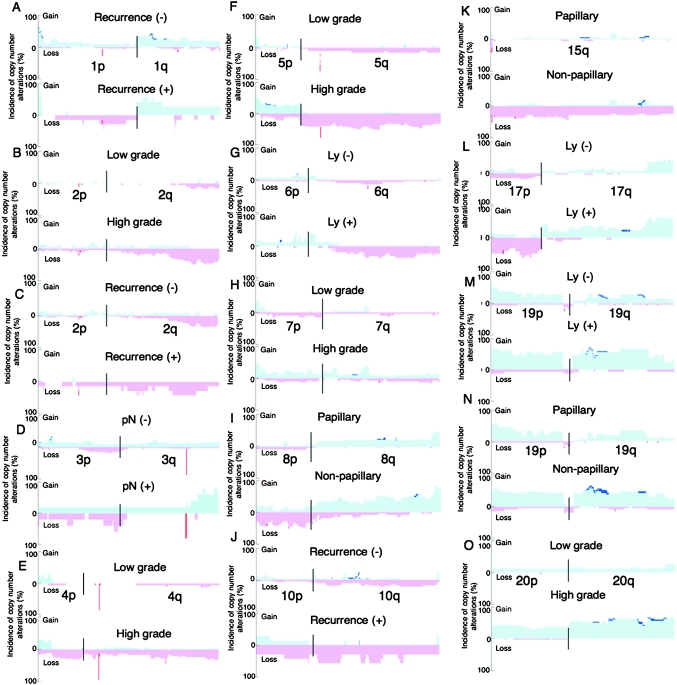
Fig. 3.
Correlations between copy number alterations on representative chromosomes and clinicopathological parameters of UCs. The 49 UCs (T1 to T49) were divided into recurrence-negative (n = 42) and -positive (n = 7) cases (A, C and J), histologically low-grade (n = 19) and high-grade (n = 30) tumors (B, E, F, H and O), lymph node metastasis (pN)-negative (n = 44) and -positive (n = 5) tumors (D), lymph vessel involvement (Ly)-negative (n = 33) and -positive (n = 16) tumors (G, L and M), and papillary (n = 28) and non-papillary (n = 21) tumors (I, K and N). −, negative; +, positive. The incidence of copy number alterations on chromosomes 1 (A), 2 (B and C), 3 (D), 4 (E), 5 (F), 6 (G), 7 (H), 8 (I), 10 (J), 15 (K), 17 (L), 19 (M and N) and 20 (O) in each of the UC groups is shown. Gains (copy number: ≥3) and losses (copy number: 1 or 0) are indicated in the upper and lower halves, respectively. Copy numbers of 0, 1, 3 and more are shown in dark red, light red, light blue and dark blue, respectively.
Unsupervised hierarchical clustering of UCs based on array CGH data
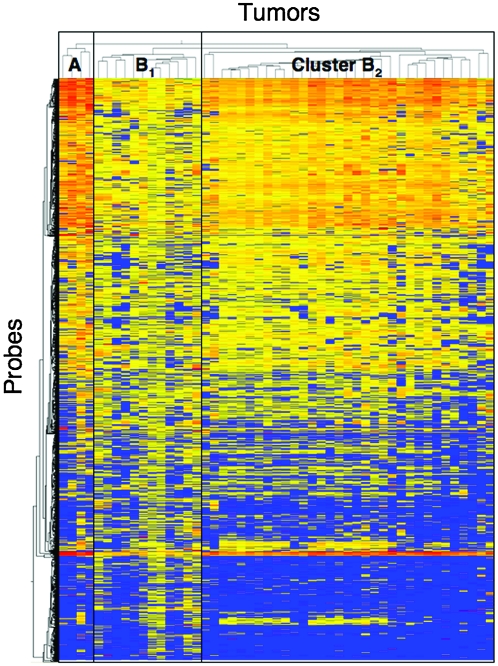
Using two-dimensional unsupervised hierarchical clustering analysis based on copy numbers and all array CGH probes, the 49 UCs were clustered into three subclasses, clusters A, B1 and B2 (Figure 4), which contained 4, 12 and 33 tumors, respectively. The average number of probes on which loss (copy number 1 or 0) or gain (≥3) was detected was significantly higher in cluster A (99 499 ± 29 879) than in cluster B1 (63 324 ± 40 064) and cluster B2 (46853 ± 35000, P = 0.0271). As shown in Table II, the average number of probes on which gain (≥3) was detected was significantly higher in cluster A than in clusters B1 and B2 (P = 0.0153), whereas the difference in the average number of probes on which loss (1 or 0) was detected among clusters A, B1 and B2 did not reach statistical significance. The average number of probes on which a copy number of >3 was detected was significantly higher in cluster A than in clusters B1 and B2 (P = 0.0053). These data indicated that copy number alterations, especially chromosomal gain, were accumulated in cluster A in comparison with clusters B1 and B2.
Fig. 4.
Unsupervised two-dimensional hierarchical clustering analysis based on array CGH analysis of UCs (T1 to T49). Forty-nine patients with UCs were hierarchically clustered into three subclasses, clusters A (n = 4), B1 (n = 12) and B2 (n = 33), based on copy numbers. Copy numbers of 0 or 1 (loss), 2 (no change) and ≥3 (gain) on each probe are shown in blue, yellow and red, respectively. The cluster trees for tumors and probes are shown at the top and to the left of the panel, respectively.
Table II.
Correlation between genetic clustering of UCs and copy number alterations and DNA methylation status
| Copy number and DNA methylation status | Genetic clustering of UCs | ||
| Cluster A | Cluster B1 | Cluster B2 | |
| Average numbers of array CGH probes showing copy number alterations | |||
| Loss (1 or 0) | 24 525 ± 15404 | 40 826 ± 31644 | 26 448± 28 462 |
| Gain (≥3) | 74 974± 38013* | 22 498 ± 15484 | 20 405 ± 17 369 |
| Gain (>3) | 1897 ± 1001† | 236 ± 315 | 209 ± 361 |
| Average numbers of BAC clones showing DNA methylation alterations | |||
| DNA hypomethylation | 312 ± 44‡ | 189 ± 95 | 236± 91 |
| DNA hypermethylation | 334± 85 | 254± 112 | 287± 77 |
| Average numbers of C-type CpG islands showing DNA methylation | 0.75 ± 0.96 | 1.33 ± 0.98§ | 0.58 ± 0.79 |
P = 0.0153 to clusters B1 and B2.
P = 0.0053 to clusters B1 and B2.
P = 0.0487 to clusters B1 and B2.
P = 0.0412 to clusters A and B2.
Correlation between genetic clustering of UCs and DNA methylation status revealed by BAMCA, MSP and COBRA
As shown in Table II, the average number of BAC clones showing DNA hypomethylation was significantly higher in cluster A than in clusters B1 and B2 (P = 0.0487), whereas there were no significant differences in the average number of BAC clones showing DNA hypermethylation among the three clusters. The incidence of DNA methylation on CpG islands of the p16 and hMLH1 genes and the MINT-1, MINT-2 and MINT-12 clones was 11 of 49 (detected/analyzed, 22.4%), 1 of 49 (2.0%), 9 of 49 (18.4%), 1 of 49 (2.0%) and 11 of 49 (22.4%), respectively. As shown in Table II, the average number of methylated C-type CpG islands was significantly higher in cluster B1 than in clusters A and B2 (P = 0.0412). Taken together, the data suggested that copy number alterations associated with overall DNA hypomethylation and regional DNA hypermethylation on C-type CpG islands were accumulated in clusters A and B1, respectively, when defined on the basis of copy number alterations.
Discussion
We and other groups have demonstrated copy number alterations in UCs for each chromosome or chromosome arm by Southern blotting, PCR–LOH and CGH analyses (3,4,6,7,9–11,25). Several array CGH analyses of UCs have also been performed using tiling BAC arrays (15,16,18). However, such analyses were unable to define the break points in detail. We here examined copy number alterations in UCs using a high-resolution (244K) oligonucleotide array capable of defining break points more precisely.
Copy numbers not corresponding to whole numbers were detected in the array CGH profiles of some UCs. In such cases, FISH analysis revealed copy number heterogeneity even within a single UC (e.g. cancer cells showing both two signals and one signal can be seen in T4 in Figure 1E). In UCs, heterogeneity of cellular atypia is frequently observed in histological specimens: a small area showing higher grade cellular atypia develops within a low-grade UC or cancer cells gain higher grade cellular atypia before they start to disrupt the basal membrane and invade into subepithelial tissues. It is feasible that copy number heterogeneity corresponds to such histological heterogeneity during the multistep malignant progression of UCs.
Our meticulous examination revealed the clinicopathological impacts of copy number alterations at various chromosomal loci (Figures 2 and 3 !supportNestedAnchors]>]>). Losses (copy number 1 or 0) of 2q33.3–q37.3, 4p15.2–q13.1 and 5q13.3–q35.3 and gains (copy number ≥3) of 7p11.2–q11.23 and 20q13.12–q13.2 were significantly correlated with higher histological grade of UCs. Gain of 7p21.2–p21.12 was significantly correlated with deeper invasion. Losses of 6q14.1–q27 and 17p13.3–q11.1 and gains of 19q13.12–q13.2 and 20q13.12–q13.33 were significantly correlated with lymph vessel involvement. Loss of 16p12.2–p12.1 and gain of 3q26.32–q29 were significantly correlated with vascular involvement. Losses of 5q14.1–q23.1, 6q14.1–q27, 8p22–p21.3, 11q13.5–q14.1 and 15q11.2–q22.2 and gains of 7p11.2–q11.22 and 19q13.12–q13.2 were significantly correlated with tumor configuration (development of a non-papillary tumor). Possibly affected genes, which are located at such chromosomal loci and for which correlations with growth, motility and invasiveness of tumor cells and tumorigenesis have already been reported, are listed in supplementary Table S2, available at Carcinogenesis Online. Significant correlations between copy number alterations on such loci and clinicopathological parameters reflecting the malignant potential of UCs may be at least partly attributable to silencing or activation of the listed genes. Moreover, such chromosomal loci are important targets for exploration of unidentified tumor-related genes that participate in the malignant progression of UCs; the products of such genes may become target molecules for therapy of UCs. In addition, losses of 1p32.2–p31.3, 10q11.23–q21.1 and 15q21.3 were significantly correlated with recurrence of UCs: copy numbers at such chromosomal loci may become indicators for prognostication of patients with UCs (estimation of recurrence risk using surgically resected specimens).
On the other hand, although the incidence of gain of the entire arm of chromosome 8q and losses of the entire arm of chromosomes 9q, 11p and 14q were not significantly correlated with any of the examined clinicopathological parameters reflecting the malignant potential of UCs, the incidence of such copy number alterations was generally high. Such copy number alterations may occur in the earlier stage of development of both papillary and non-papillary UCs. Therefore, gatekeeper genes for urothelial carcinogenesis may exist on 8q, 9q, 11p and 14q. Combinations of the copy numbers of 8q, 9q, 11p and 14q could become applicable as indicators for the early diagnosis of UCs based on examination of urinary sediments and tissue specimens.
Moreover, the incidence of homozygous deletion on only 11 continuous oligonucleotide probes on 4q13.2 was high (67.3%) and the UGT2B17 gene is located within this homozygously deleted lesion. Copy number polymorphism of the UGT2B17 gene is reportedly associated with smoking-related cancer development (37), and a significant association between UCs and smoking has been demonstrated epidemiologically (38). Since there are many family genes, the exact copy numbers of the UGT2B17 gene were evaluated by quantitative PCR using specific primer sets (supplementary Table S3 is available at Carcinogenesis Online). Levels of expression of messenger RNA (mRNA) for the UGT2B17 gene normalized relative to the expression of glyceraldehyde-3-phosphate dehydrogenase mRNA were also examined by quantitative reverse transcription (RT)–PCR analysis (supplementary Table S3 is available at Carcinogenesis Online) in 37 of the 49 UCs for which RNA samples were available. Quantitative RT–PCR data for the UGT2B17 gene in 28 UCs showing homozygous deletion (copy number 0) was 3.53 ± 6.40, being significantly lower than that in 9 UCs not showing it (61.61 ± 98.32, P = 0.008176). Since the homozygous deletion actually resulted in gene silencing, the correlation between the copy number of the UGT2B17 gene and susceptibility to UCs should be further examined.
UCs were grouped into three subclasses, clusters A, B1 and B2, based on copy number alterations. In cluster A, copy number alterations, especially chromosomal gains, revealed by array CGH analysis, and DNA hypomethylation revealed by BAMCA were both accumulated in a genome-wide manner. DNA hypomethylation may result in chromosomal instability through changes in chromatin configuration and enhancement of chromosomal recombination (39). Although such correlation between DNA hypomethylation and chromosomal instability has been observed in experimental models (40) and human immunodeficiency, centromeric instability and facial anomalies syndrome (41) and cancers (25,42), details of the DNA methylation status around each of the chromosome breakpoints are still unclear. UCs in cluster A may be ideal for examination of DNA methylation status around breakpoints to further clarify the molecular mechanisms responsible for chromosomal instability resulting from DNA methylation alterations. Cluster B1 showed accumulation of regional DNA hypermethylation on C-type CpG islands. In addition, chromosomal losses tended to be accumulated in cluster B1 in comparison with clusters A and B2, although such differences did not reach statistical significance. The cancer phenotype associated with accumulation of DNA methylation on C-type CpG islands is defined as the CpG island methylator phenotype, and such accumulation is generally associated with frequent silencing of tumor-related genes due to DNA hypermethylation only and/or a two-hit mechanism involving DNA hypermethylation and LOH in human cancers of various organs (22). Silencing of tumor-related genes due to DNA hypermethylation and chromosomal losses may be critical for the development of UCs belonging to cluster B1. In cluster B2, the number of BAC clones showing both DNA hypo- and hypermethylation by BAMCA was rather high, and the number of probes showing loss or gain by array CGH was rather low, in comparison with cluster B1, although such differences did not reach statistical significance. In addition to copy number alterations, genome-wide DNA methylation alterations may also participate in the development of UCs belonging to cluster B2.
The number of CpG sites in CpG islands and repetitive sequences in 5′ regions, introns, exons and non-coding regions on BAC clones showing DNA hypomethylation in UCs are summarized in supplementary Table S4, available at Carcinogenesis. DNA hypomethylation was observed in BAC clones including both CpG islands and repetitive sequences, possibly resulting in activation of tumor-related genes and/or parasitic elements and loss of chromosomal integrity.
Silencing of representative genes on affected chromosomal loci was confirmed using quantitative RT–PCR analysis (supplementary Table S3 is available at Carcinogenesis Online). Although DNA methylation of the p16 gene was detected using MSP, quantitative examination using pyrosequencing (supplementary Table S3 is available at Carcinogenesis Online) revealed generally low DNA methylation levels (1.82 ± 0.65%) in all UCs. Therefore, correlations between copy numbers based on array CGH analysis and mRNA expression levels based on quantitative RT–PCR analysis were examined. The p16 gene was silenced in 11 UCs showing homozygous deletion (copy number, 0; quantitative RT–PCR data, 1.24 ± 1.20), whereas the mRNA expression level in 27 UCs not showing it was 104.1 ± 205.11 (P = 0.00000357). On the other hand, the DNA methylation level of the CXCL12 gene was 12.59 ± 18.43% for the UCs as a whole. The CXCL12 gene was silenced in 2 UCs with DNA methylation levels of ≥50% (mRNA expression level: 1.81 ± 1.00) but not in 34 UCs with DNA methylation levels of <50% (mRNA expression level: 24.45 ± 34.04). The level of expression of mRNA for the ERBB4 gene in 18 UCs showing a DNA methylation level of ≥5% and/or chromosomal loss (copy number 0 or 1) was 59.1 ± 101.2 and tended to be lower than that in 20 UCs with a DNA methylation level of <5% and a copy number of 2 (128.4 ± 259.3), suggesting the possibility of inactivation due to a combination of DNA hypermethylation and chromosomal loss, although such differences did not reach statistically significant levels. Taken together, the data suggest that genetic and epigenetic alterations (copy number alterations and DNA methylation alterations) are not mutually exclusive during urothelial carcinogenesis. Reflecting the clinicopathological diversity and histological heterogeneity of UCs, genetic and epigenetic events appear to accumulate in a complex manner during the developmental stage of individual tumors.
Supplementary material
Supplementary Tables S1–S4 can be found at http://carcin.oxfordjournals.org/
Funding
Grant-in-Aid for the Third Term Comprehensive 10 Years Strategy for Cancer Control (H19-3-002) from the Ministry of Health, Labor and Welfare of Japan; Grant-in-Aid for Cancer Research (21-2-2) from the Ministry of Health, Labor and Welfare of Japan; Grant (P06012) from the New Energy and Industrial Technology Development Organization (NEDO); Program for Promotion of Fundamental Studies in Health Sciences (10–42) of the National Institute of Biomedical Innovation (NiBio).
Acknowledgments
N.N. is an awardee of a research resident fellowship from the Foundation for Promotion of Cancer Research in Japan.
Conflicts of Interest Statement: None declared.
Glossary
Abbreviations
- BAC
bacterial artificial chromosome
- BAMCA
bacterial artificial chromosome array-based methylated CpG island amplification
- CGH
comparative genomic hybridization
- COBRA
combined bisulfite restriction enzyme analysis
- FISH
fluorescence in situ hybridization
- LOH
loss of heterozygosity
- mRNA
messenger RNA
- MINT
methylated in tumor
- MSP
methylation-specific polymerase chain reaction
- RT
reverse transcription
- PCR
polymerase chain reaction
- UC
urothelial carcinoma
References
- 1.Kakizoe T, et al. Relationship between papillary and nodular transitional cell carcinoma in the human urinary bladder. Cancer Res. 1988;48:2299–2303. [PubMed] [Google Scholar]
- 2.Kakizoe T. Development and progression of urothelial carcinoma. Cancer Sci. 2006;97:821–828. doi: 10.1111/j.1349-7006.2006.00264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knowles M, et al. Allelotype of human bladder cancer. Cancer Res. 1994;54:531–538. [PubMed] [Google Scholar]
- 4.Orlow I, et al. Deletion of the p16 and p15 genes in human bladder tumors. J. Natl Cancer Inst. 1995;87:1524–1529. doi: 10.1093/jnci/87.20.1524. [DOI] [PubMed] [Google Scholar]
- 5.Sauter G, et al. c-myc copy number gains in bladder cancer detected by fluorescence in situ hybridization. Am. J. Pathol. 1995;146:1131–1139. [PMC free article] [PubMed] [Google Scholar]
- 6.Simoneau AR, et al. Evidence for two tumor suppressor loci associated with proximal chromosome 9p to q and distal chromosome 9q in bladder cancer and the initial screening for GAS1 and PTC mutations. Cancer Res. 1996;56:5039–5043. [PubMed] [Google Scholar]
- 7.Richter J, et al. Marked genetic differences between stage pTa and stage pT1 papillary bladder cancer detected by comparative genomic hybridization. Cancer Res. 1997;57:2860–2864. [PubMed] [Google Scholar]
- 8.Wagner U, et al. Chromosome 8p deletions are associated with invasive tumor growth in urinary bladder cancer. Am. J. Pathol. 1997;151:753–759. [PMC free article] [PubMed] [Google Scholar]
- 9.Hovey RM, et al. Genetic alterations in primary bladder cancers and their metastases. Cancer Res. 1998;58:3555–3560. [PubMed] [Google Scholar]
- 10.Richter J, et al. Patterns of chromosomal imbalances in advanced urinary bladder cancer detected by comparative genomic hybridization. Am. J. Pathol. 1998;153:1615–1621. doi: 10.1016/S0002-9440(10)65750-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simon R, et al. Chromosomal aberrations associated with invasion in papillary superficial bladder cancer. J. Pathol. 1998;185:345–351. doi: 10.1002/(SICI)1096-9896(199808)185:4<345::AID-PATH109>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 12.Olesen SH, et al. Mitotic checkpoint genes hBUB1, hBUB1B, hBUB3 and TTK in human bladder cancer, screening for mutations and loss of heterozygosity. Carcinogenesis. 2001;22:813–815. doi: 10.1093/carcin/22.5.813. [DOI] [PubMed] [Google Scholar]
- 13.Hoque MO, et al. Genome-wide genetic characterization of bladder cancer: a comparison of high-density single-nucleotide polymorphism arrays and PCR-based microsatellite analysis. Cancer Res. 2003;63:2216–2222. [PubMed] [Google Scholar]
- 14.Veltman JA, et al. Array-based comparative genomic hybridization for genome-wide screening of DNA copy number in bladder tumors. Cancer Res. 2003;63:2872–2880. [PubMed] [Google Scholar]
- 15.Blaveri E, et al. Bladder cancer stage and outcome by array-based comparative genomic hybridization. Clin. Cancer Res. 2005;11:7012–7022. doi: 10.1158/1078-0432.CCR-05-0177. [DOI] [PubMed] [Google Scholar]
- 16.Heidenblad M, et al. Tiling resolution array CGH and high density expression profiling of urothelial carcinomas delineate genomic amplicons and candidate target genes specific for advanced tumors. BMC Med. Genomics. 2008;1:3. doi: 10.1186/1755-8794-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zieger K, et al. Chromosomal imbalance in the progression of high-risk non-muscle invasive bladder cancer. BMC Cancer. 2009;9:149. doi: 10.1186/1471-2407-9-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindgren D, et al. Combined gene expression and genomic profiling define two intrinsic molecular subtypes of urothelial carcinoma and gene signatures for molecular grading and outcome. Cancer Res. 2010;70:3463–3472. doi: 10.1158/0008-5472.CAN-09-4213. [DOI] [PubMed] [Google Scholar]
- 19.Kanai Y, et al. Alterations of DNA methylation associated with abnormalities of DNA methyltransferases in human cancers during transition from a precancerous to a malignant state. Carcinogenesis. 2007;28:2434–2442. doi: 10.1093/carcin/bgm206. [DOI] [PubMed] [Google Scholar]
- 20.Kanai Y. Alterations of DNA methylation and clinicopathological diversity of human cancers. Pathol. Int. 2008;58:544–558. doi: 10.1111/j.1440-1827.2008.02270.x. [DOI] [PubMed] [Google Scholar]
- 21.Kanai Y. Genome-wide DNA methylation profiles in precancerous conditions and cancers. Cancer Sci. 2010;101:36–45. doi: 10.1111/j.1349-7006.2009.01383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toyota M, et al. CpG island methylator phenotype in colorectal cancer. Proc. Natl Acad. Sci. USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakagawa T, et al. Increased DNA methyltransferase 1 protein expression in human transitional cell carcinoma of the bladder. J. Urol. 2003;170:2463–2466. doi: 10.1097/01.ju.0000095919.50869.c9. [DOI] [PubMed] [Google Scholar]
- 24.Nakagawa T, et al. DNA hypermethylation on multiple CpG islands associated with increased DNA methyltransferase DNMT1 protein expression during multistage urothelial carcinogenesis. J. Urol. 2005;173:1767–1771. doi: 10.1097/01.ju.0000154632.11824.4d. [DOI] [PubMed] [Google Scholar]
- 25.Nakagawa T, et al. DNA hypomethylation on pericentromeric satellite regions significantly correlates with loss of heterozygosity on chromosome 9 in urothelial carcinomas. J. Urol. 2005;173:243–246. doi: 10.1097/01.ju.0000141577.98902.49. [DOI] [PubMed] [Google Scholar]
- 26.Nishiyama N, et al. Genome-wide DNA methylation profiles in urothelial carcinomas and urothelia at the precancerous stage. Cancer Sci. 2010;101:231–240. doi: 10.1111/j.1349-7006.2009.01330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Misawa A, et al. Methylation-associated silencing of the nuclear receptor 1I2 gene in advanced-type neuroblastomas, identified by bacterial artificial chromosome array-based methylated CpG island amplification. Cancer Res. 2005;65:10233–10242. doi: 10.1158/0008-5472.CAN-05-1073. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka K, et al. Frequent methylation-associated silencing of a candidate tumor-suppressor, CRABP1, in esophageal squamous-cell carcinoma. Oncogene. 2007;26:6456–6468. doi: 10.1038/sj.onc.1210459. [DOI] [PubMed] [Google Scholar]
- 29.Sugino Y, et al. Epigenetic silencing of prostaglandin E receptor 2 (PTGER2) is associated with progression of neuroblastomas. Oncogene. 2007;26:7401–7413. doi: 10.1038/sj.onc.1210550. [DOI] [PubMed] [Google Scholar]
- 30.Arai E, et al. DNA methylation profiles in precancerous tissue and cancers: carcinogenetic risk estimation and prognostication based on DNA methylation status. Epigenomics. 2010;2:467–481. doi: 10.2217/epi.10.16. [DOI] [PubMed] [Google Scholar]
- 31.Eble JN, et al. Tumours of the Urinary System and Male Genital Organs. World Health Organization Classification of Tumours. Pathology and Genetics. Lyon: IARC Press; 2004. [Google Scholar]
- 32.Illingworth R, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inazawa J, et al. Comparative genomic hybridization (CGH)-arrays pave the way for identification of novel cancer-related genes. Cancer Sci. 2004;95:559–563. doi: 10.1111/j.1349-7006.2004.tb02486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arai E, et al. Genome-wide DNA methylation profiles in both precancerous conditions and clear cell renal cell carcinomas are correlated with malignant potential and patient outcome. Carcinogenesis. 2009;30:214–221. doi: 10.1093/carcin/bgn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arai E, et al. Genome-wide DNA methylation profiles in liver tissue at the precancerous stage and in hepatocellular carcinoma. Int. J. Cancer. 2009;125:2854–2862. doi: 10.1002/ijc.24708. [DOI] [PubMed] [Google Scholar]
- 36.Arai E, et al. Regional DNA hypermethylation and DNA methyltransferase (DNMT) 1 protein overexpression in both renal tumors and corresponding nontumorous renal tissues. Int. J. Cancer. 2006;119:288–296. doi: 10.1002/ijc.21807. [DOI] [PubMed] [Google Scholar]
- 37.Lazarus P, et al. Genotype-phenotype correlation between the polymorphic UGT2B17 gene deletion and NNAL glucuronidation activities in human liver microsomes. Pharmacogenet. Genomics. 2005;15:769–778. doi: 10.1097/01.fpc.0000175596.52443.ef. [DOI] [PubMed] [Google Scholar]
- 38.Alberg AJ, et al. Cigarette smoking and bladder cancer: a new twist in an old saga? J. Natl Cancer Inst. 2009;101:1525–1526. doi: 10.1093/jnci/djp385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones PA, et al. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Howard G, et al. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene. 2008;27:404–408. doi: 10.1038/sj.onc.1210631. [DOI] [PubMed] [Google Scholar]
- 41.Hansen RS, et al. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc. Natl Acad. Sci. USA. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saito Y, et al. Overexpression of a splice variant of DNA methyltransferase 3b, DNMT3b4, associated with DNA hypomethylation on pericentromeric satellite regions during human hepatocarcinogenesis. Proc. Natl Acad. Sci. USA. 2002;99:10060–10065. doi: 10.1073/pnas.152121799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.