Abstract
The anti-skin carcinogenic effects of green tea catechins have been studied extensively in vitro and in vivo models but the precise epigenetic molecular mechanisms are still unclear. Accumulating data suggest that dietary phytochemicals may alter cancer risk by modifications of epigenetic processes in the cells. The present study was designed to investigate whether tea catechins, particularly (−)-epigallocatechin-3-gallate (EGCG), would modify epigenetic events to regulate DNA methylation-silenced tumor suppressor genes in skin cancer cells. DNA methylation, histone modifications and tumor suppressor gene expressions were studied in detail using human epidermoid carcinoma A431 cells as an in vitro model after EGCG treatment using cytostaining, western blotting, dot blot analysis, real-time polymerase chain reaction and enzymatic activity assays. Our study shows that EGCG treatment decreased global DNA methylation levels in A431 cells in a dose-dependent manner. EGCG decreased the levels of 5-methylcytosine, DNA methyltransferase (DNMT) activity, messenger RNA (mRNA) and protein levels of DNMT1, DNMT3a and DNMT3b. EGCG decreased histone deacetylase activity and increased levels of acetylated lysine 9 and 14 on histone H3 (H3-Lys 9 and 14) and acetylated lysine 5, 12 and 16 on histone H4 but decreased levels of methylated H3-Lys 9. Additionally, EGCG treatment resulted in re-expression of the mRNA and proteins of silenced tumor suppressor genes, p16INK4a and Cip1/p21. Together, our study provides new insight into the epigenetic mechanism of action of EGCG that may contribute to the chemoprevention of skin cancer and may have important implications for epigenetic therapy.
Introduction
Plant polyphenols offer promising new options for the development of more effective strategies for the prevention of cancer risk. (−)-Epigallocatechin-3-gallate (EGCG) is an active and major constituent of green tea or green tea polyphenols and has been shown to have anticarcinogenic properties (1–3). Epidemiological studies also suggested that regular consumption of green tea attenuates the risk of many cancers, including lung, colon, liver, pancreas and breast etc. (1–3). Topical treatment of EGCG or consumption of green tea polyphenols in drinking water of mice have been shown to inhibit skin cancer in rodent models, and anti-skin carcinogenic effects of EGCG have been linked with its anti-inflammatory, antioxidative and DNA repair mechanisms (1–4).
Epigenetic alterations in multiple genes are believed to play a crucial role in carcinogenesis including skin carcinogenesis. Epigenetic inactivation of genes by promoter hypermethylation has been recognized as an important mechanism by which tumor suppressor genes are shut down during development of tumors. Hypermethylation of CpG islands in the promoter region leads to silencing either by direct inhibition of transcription factor binding, or by attracting methylated DNA-binding proteins, recruiting other transcriptional repressors such as histone deacetylases (HDACs) and histone methyl transferases, resulting in transcriptionally inactive chromatin (5,6). DNA methylation at the 5′ position of cytosine is mediated by DNA methyltransferases (DNMTs). DNA methylation is the most characterized epigenetic mechanism that can be inherited without changing the DNA sequence (5,6). It also has been reported that approximately half of the tumor suppressor genes that are inactivated in sporadic cancers are more often inactivated by epigenetic than by genetic mechanisms. DNA hypermethylation is a major epigenetic mechanism in the silencing of the expression of tumor suppressor genes (7–9). DNA methylation is commonly associated with increased levels or altered functions of DNMTs and that initiate the methylation at position 5′ of cytosines of CpG dinucleotides (6,10). Additionally, histone modifications, particularly methylation and acetylation, may also be involved in transcriptional silencing of a number of genes in cancers. Recent investigations suggest that some dietary phytochemicals may prevent cancer by modifications of epigenetic processes (11–13).
The aim of the current study was to investigate whether EGCG would reactivate the expression of tumor suppressor genes, such as p16INK4a and Cip1/p21, in human skin carcinoma cells, if it is so then what is the underlying mechanism? For this purpose, we used a well-known human epidermoid carcinoma cell line A431 as an in vitro model. Our study demonstrates that treatment of skin cancer cells with EGCG reduced the levels of DNA methylation and DNMT activity, and that resulted in re-expression of messenger RNAs (mRNAs) and protein expressions of tumor suppressor genes (p16INK4a and Cip1/p21). EGCG treatment also inhibited HDAC activity and increases the levels of acetylated histones. However, these effects of EGCG were not observed in normal human epidermal keratinocytes (NHEK). These results suggest that EGCG can modulate the expression of anticancer genes via epigenetic processes involving DNA methylation and histone modifications. Furthermore, our findings suggest a novel epigenetic mechanism contributing to skin cancer chemoprevention by EGCG, a major and most active constituent of green tea polyphenols.
Material and methods
Cell lines and cell culture
Human skin cancer A431 and squamous cell carcinoma (SCC) 13 cells were purchased from the American Type Culture Collection (Manassas, VA) and NHEK were obtained from Cell Culture Core Facility of Skin Diseases Research Center at the University of Alabama at Birmingham, AL, USA. The cells were cultured as monolayers in Dulbecco’s modified Eagle’s medium supplemented with 10% heat inactivated fetal bovine serum, 100 μg/ml penicillin–streptomycin (Invitrogen, Carlsbad, CA), and maintained in a humidified atmosphere of 95% air and 5% CO2 at 37°C. The NHEK were cultured in keratinocyte growth medium supplemented with 5 ng/ml human recombinant epidermal growth factor and 0.05 mg/ml bovine pituitary extract (Gibco/Invitrogen, Carlsbad, CA) and maintained in an incubator under the conditions as described above. Cells were seeded at a density of 1 × 106 cells per petri dish and allowed to attach for 24 h before treatment with catechins or 5-aza-2′-deoxycytidine (5-aza-dc) for 3 or 6 days with media and treatments agents refreshed every 3 days. The subconfluent cells were treated with either various concentrations of EGCG or other catechins. Cells were treated with only a single dose of 5-aza-dc or trichostatin A (TSA).
Chemicals and antibodies
Green tea catechins, (−)-epicatechin (EC), (−)-gallocatechin (GC), (−)-epigallocatechin (EGC), (−)-epicatechin-gallate (ECG) and EGCG were obtained from Mitsui Norin Co. (Tokyo, Japan). Antibodies were purchased as follows: 5-methylcytosine (5-mC) from Calbiochem, EMD Biosciences (San Diego, CA), DNMT1, DNMT3a and DNMT3b from Imgenec Corporation (San Diego, CA), HDAC1 from Upstate Antibodies (New York, NY) and acetyl histone H4 and acetyl histone H3 from Abcam Antibodies (Cambridge, MA). RNA and DNA isolation kits were purchased from Invitrogen. The Methylamp™ Global DNA Methylation Quantification Kit and the EpiQuik DNMT Activity Assay Kit were purchased from Epigentek (New York, NY). Standardized real-time polymerase chain reaction (PCR) primers for DNMT1, DNMT3a, DNMT3b, p16INK4a and Cip1/p21 were obtained from SuperArray Biosciences (Fredrick, MD).
Global DNA methylation assay
The total genomic DNA was extracted from the cells, which were treated with EC, GC, EGC, ECG, EGCG or 5-aza-dc using the Qiagen ampR DNA Mini Kit (Qiagen Sciences, Maryland, MD) following the manufacturer’s instructions. The Global DNA methylation levels were determined using the Methylamp™ Global DNA Methylation Quantification Kit according to the manufacturer’s instructions. This analysis provides the levels of global DNA methylation, and is not specific to any particular gene. The methylated fraction of DNA is recognized by a 5-mC antibody. With this colorimetric kit, the amount of methylated DNA, which is proportional to the optical density intensity, is quantified through an enzyme-linked immunosorbent assay-like reaction.
M5-mC immunostaining
Cells were treated with various concentrations of EGCG (0, 5, 10 and 20 μg/ml) for 6 days and then harvested. A total of 1 × 105 to –2 × 105 cells were cytospun using a Cytospin 4 Equipment (Thermo Electron Corporation, Waltham, MA) at 1500 r.p.m. for 15 min and then processed for 5-mC cytostaining. Briefly, cells were permeabilized with 0.2% Triton X-100 in phosphate-buffered saline (PBS), washed with PBS for 10 min. The cells were then blocked with 3% preimmune goat serum in PBS for 30 min, followed by incubation with 3% H2O2 for 20 min to quench endogenous peroxidase. After washing the cells with PBS, cells were incubated with 5-mC-specific antibody (1:500, vol/vol; Calbiochem, Gibbstown, NJ) for 2 h, followed by sequential incubation of cells with biotinylated goat anti-mouse IgG1 and horseradish peroxidase-conjugated streptavidin and finally with diaminobenzidine substrate and counterstaining with methylene blue.
Dots blot analysis of DNA 5-mC
Cells were treated with EGCG for 6 days as described above. Genomic DNA was isolated using the DNA Isolation Kit (Qiagen Sciences) according to the manufacturer’s instructions, and dot blot analysis was performed as detailed previously (14). Briefly, genomic DNA (1 μg) was transferred onto a positively charged Hybond-enhanced chemiluminescence nitrocellulose membranes (Amersham Biosciences, Piscataway, NJ) using Bio-Dot Microfiltration Apparatus (Bio-Rad Laboratories, Hercules, CA), and fixed by baking the membrane for 30 min at 80°C. After blocking the non-specific-binding sites, the membrane was incubated with the antibody specific to 5-mC (1:500, vol/vol) followed by incubation with an horseradish peroxidase-conjugated secondary antibody. The membranes were then treated with enhanced chemiluminescence detection reagents and exposed to Kodak autoradiograph films. Equal DNA loading was verified by staining the membranes with 0.2% methylene blue. The intensity of each dot was measured by densitometry and normalized to total DNA.
DNMT activity assay
A431 or SCC 13 cells were treated for 3 or 6 days with various concentrations of EGCG or other catechins, such as EC, GC, EGC or ECG. After desired time point, cells were harvested and nuclear extracts were prepared from various treatment groups using Epiquik Nuclear Extraction Kit (Epigentek) following the manufacturer’s instructions. DNMT activity was determined using Epiquik DNMT Activity Assay Kit (Epigentek) according to the manufacturer’s protocol. Similarly, the effect of various catechins was also determined on DNMT activity in NHEK following identical protocol.
Quantitative mRNA analysis of DNMTs and tumor suppressor genes using real-time PCR
Total RNA was extracted from the cells of different treatment groups using Trizol Reagents Kit (Invitrogen) and complementary DNA was synthesized through the reverse transcription reaction (iScript complementary DNA Synthesis Kit; Bio-Rad Laboratories). Using SYBR Green/Fluorescein PCR Master Mix, complementary DNA was amplified using real-time PCR with a Bio-Rad MyiQ thermocycler and SYBR Green detection system (Bio-Rad Laboratories). Samples were run in duplicate to ensure amplification integrity. Manufacturer-supplied standardized primer pairs were used to measure the following: DNMT1, DNMT3a, DNMT3b, p16INK4a and Cip1/p21. The standard PCR conditions were: 95°C for 15 min and then 40 cycles at 95°C for 30 s, 55°C for 30 s and 72°C for 30 s, as recommended (SuperArray Bioscience Corporation, Frederick, MD). The mRNA expression levels of genes were normalized to the expression level of the housekeeping gene β-actin and relative to the average of all delta Ct-values in each sample using the cycle threshold (Ct) method.
HDAC activity assay
Effect of tea catechins on HDAC activity in cancer cells was assessed using the HDAC Colorimetric Activity Assay Kit (Active Motif, Carlsbad, CA) according to the manufacturer’s instructions. Briefly, A431 cells were treated with either various tea catechins or various concentrations of EGCG or TSA for 3 or 6 days, then harvested and nuclear protein fractions were prepared using Epiquik Nuclear Extraction Kit (Epigentek) following the manufacturer’s instructions. Forty micrograms of nuclear proteins from each treatment group were incubated with a colorimetric HDAC substrate supplied with the kit for 3 h after which the reaction was developed with assay development buffer supplied with kit. Nuclear extract prepared from the A431 cells treated with 300 nM of TSA served as a positive control.
Cell lysates and western blotting
Analysis of protein levels was performed using western blotting. Cell lysates from different treatment groups were prepared as described previously (14,15). Proteins (25–40 μg protein) were electrophoresed on premade 10% Tris–glycine gels (Invitrogen) and then transferred onto nitrocellulose membranes. After blocking in freshly prepared PBS containing 3% non-fat dry milk at room temperature for 30 min, the membranes were incubated with antibodies against total histone H3 (1:2000), acetylated H3-Lys 9, H3-Lys 14, acetylated H4-Lys 5, H4-Lys 12 and H4-Lys 16 (Millipore, Billerica, MA) at 4°C overnight followed by a anti-rabbit peroxidase-conjugated secondary antibody at 1:1000 dilution (Santa Cruz Biotechnology, Santa Cruz, CA). Protein bands were visualized on X-ray film using an enhanced chemiluminescence system (Amersham Life Science, Piscataway, NJ). To verify equal protein loading, membranes were washed and reprobed with anti-H1 antibody (Millipore). Similarly, the expression levels of DNMT1, DNMT3a and DNMT3b, Cip1/p21 and p16INK4a were also determined in cell lysates using western blot analysis, and equal protein loading was verified using anti-β actin antibody. Experiments were repeated three times, and thus three western blots were run in each experiment, and representative blot is shown in each case.
Statistical analysis
The data for global DNA methylation levels, DNMT activity and HDAC activity are expressed as percentages with the basal levels in skin cancer cells taken as 100%. Student’s t-test was used to determine the statistical difference between treatment groups. The results of the real-time PCR are expressed as the means ± standard deviation. The data were considered significant if P < 0.05.
Results
Effect of green tea catechins and 5-aza-dc on global DNA methylation levels in skin cancer cells
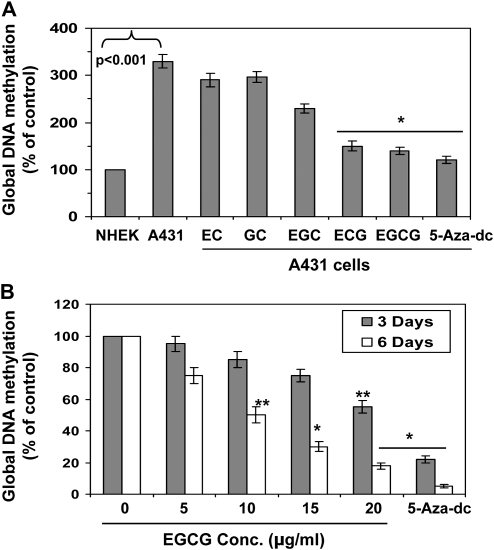
First to examine whether green tea polyphenolic components, such as EC, GC, EGC, ECG and EGCG, have epigenetic effects on DNA methylation levels, and if it is, then which component has superior effects than others. For this purpose, A431 cells were treated with equimolar concentration (25 μM) of EC, GC, EGC, ECG and EGCG for 6 days. NHEK were used as a control. At the termination of the experiments, cells were harvested, DNA isolated and subjected to the analysis of global DNA methylation levels using Global DNA Methylation kit and data were presented in terms of percent of control (NHEK) cells. As shown in Figure 1A, the level of global DNA methylation in A431 cells was >3-fold higher compared with NHEK. The inhibitory effect on DNA methylation (demethylation) of EC and GC was less compared with other catechin derivatives found in green tea. The inhibitory effect of ECG and EGCG was significantly higher than other catechins in decreasing the levels of DNA methylation in A431 cells when cells were treated for 6 days. The treatment of A431 cells with 5-aza-dc (5 μM) also significantly decreased the level of global DNA methylation and used as a positive control.
Fig. 1.
Effect of green tea catechins (25 μM) on global DNA methylation in human A431 skin cancer cells. (A) Effect of EC, GC, EGC, ECG, EGCG and 5-aza-dc (5 μM) on global DNA methylation levels in A431 cells. Cells were treated with different catechins for 6 days and the levels of global DNA methylation was determined using Global DNA Methylation kit. NHEK served as a control. (B) Dose-dependent effect of EGCG on the Global DNA methylation levels in A431 cells. Cells were treated with EGCG for 3 or 6 days. Data are presented in terms of percent of control (non-EGCG-treated) group, which was assigned a value of 100%, and as means ± standard deviations, n = 3. Significant difference versus non-EGCG-treated controls; *P < 0.001, **P < 0.01.
We were also interested to examine the time-dependent effect of green tea catechin on suppressing the levels of DNA methylation in skin cancer cells. As EGCG was more effective in reducing the levels of DNA methylation level than others, we selected EGCG for further detailed studies. EGCG was selected based on the following reasons: (i) EGCG has been shown to be most active component compared with other catechins and (ii) EGCG is also a major component (>50%) among other catechins or polyphenols found in green tea (14). For this purpose, A431 cells were treated with various concentration of EGCG for 3 and 6 days. At the termination of the experiment, cells were harvested and DNA was isolated for the analysis of global DNA methylation level. As shown in Figure 1B, treatment of cells with EGCG was resulted in reduction in the levels of DNA methylation in a concentration-dependent manner. However, the levels of global DNA methylation in A431 cells were significantly reduced (P < 0.01 and P < 0.001) after 6 days of treatment with EGCG than the treatment of cells with EGCG after 3 days. 5-Aza-dc is a potent DNA demethylation agent and used as a positive control. These data suggest that DNA methylation process can be reversed but the process appears slow and 6 days time point is best effective time period for demethylation of DNA by EGCG.
EGCG treatment induces 5-mC demethylation in DNA of A431 cells
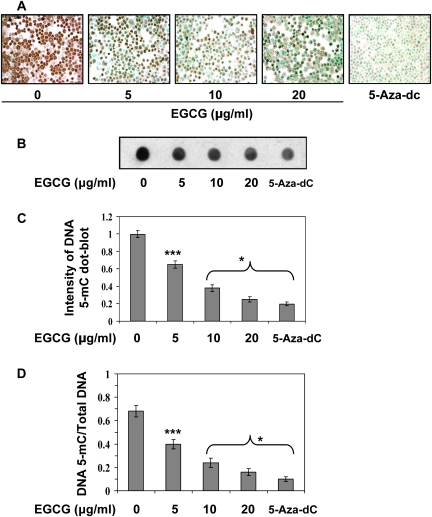
Further to examine the dose-dependent effect of EGCG on DNA demethylation, A431 cells were treated with EGCG for 6 days, and cells were harvested for immunocytostaining of DNA methylation using antibody specific to 5-mC. Cells were also treated with 5-aza-dc as a positive control. As shown by 5-mC-positive cells, EGCG treatment resulted in reduction of 5-mC-positive cells in a concentration-dependent manner compared with non-EGCG-treated A431 cells (Figure 2A). Treatment of EGCG resulted in reduction of the number of 5-mC-positive cells in a dose-dependent manner (20–65%, P < 0.001). To further verify the effect of EGCG on DNA demethylation, total genomic DNA was isolated from A431 cells and analysed by dot blot assay using an anti-5-mC antibody. As shown in Figure 2B and C, treatment of A431 cells with EGCG or 5-aza-dc significantly decreased methylation of total DNA. These results suggest that EGCG may have a broad effect on DNA demethylation. Furthermore, the intensity of individual dot blots was measured by densitometry and levels of 5-mC were normalized by total DNA (Figure 2D). This analysis indicates a significant decrease (P < 0.05 and P < 0.001) in DNA methylation in terms of total amount of DNA by EGCG treatment of A431 cells.
Fig. 2.
Treatment of A431 cells with various concentrations of EGCG for 6 days decreased the levels of 5-mC dose dependently. Treatment of cells with 5-aza-dc was used as a positive control. (A) Cytostaining of 5-mC-postive cells using a 5-mC-specific antibody. (B) Dot blot analysis of 5-mC in DNA extracted from various treatment groups of cells. (C) Dot blot data are presented in terms of relative density of dot blots as means ± standard deviations in different treatment groups (n = 3). (D) Effects of EGCG and 5-aza-dc on genomic DNA methylation in A431 cells. DNA methylation at 5-cytosine was detected using an anti-5-mC antibody and samples loading was determined by staining total DNA with methylene blue. The intensity of individual dots was measured by densitometry and levels of 5-mC were normalized by total DNA. Data are mean ± standard deviation (n = 3). Significant difference versus non-EGCG-treated control; *P < 0.001, ***P < 0.05.
EGCG and 5-aza-dc induce DNA 5-mC demethylation and reduce global DNA methylation level in human SCC 13 cells
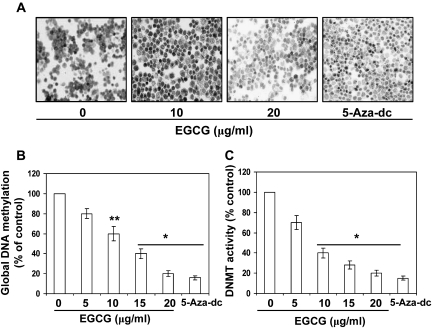
Just to verify whether EGCG and 5-aza-dc have similar epigenetic effects on other skin cancer cell lines, we examined the effect of EGCG and 5-aza-dc on another human SCC 13 cell line. For this purpose, SCC 13 cells were treated with EGCG or 5-aza-dc for 6 days similar to A431 cells. Cells were harvested and immunocytostaining was performed to detect 5-mC-positive cells. As shown in Figure 3A, EGCG treatment resulted in reduction of 5-mC-positive cells in SCC 13 cells compared with non-EGCG-treated SCC 13 cells. Treatment of cells with EGCG was resulted in reduction in the levels of global DNA methylation in a concentration-dependent manner (Figure 3B). Cells treated with 5-aza-dc served as a positive control.
Fig. 3.
Treatment of SCC 13 cells with EGCG for 6 days decreased the levels of 5-mC. Treatment of cells with 5-aza-dc was used as a positive control. Cytostaining of 5-mC-postive cells using a 5-mC-specific antibody. (B) Dose-dependent effect of EGCG on the Global DNA methylation levels in SCC 13 cells. Cells were treated with EGCG for 6 days. Data are presented in terms of percent of control (non-EGCG-treated) group, which was assigned a value of 100%, and as means ± standard deviation, n = 3. (C) Treatment of SCC 13 cells with EGCG or 5-aza-dc for 6 days inhibits DNMT activity. Total DNMT activity in nuclear extracts was determined using the DNMT Activity Assay Kit. Data are presented in terms of percentage versus the results using non-EGCG-treated controls, which was assigned a value of 100%, and as means ± standard deviations; n = 3. Significant difference versus non-EGCG-treated controls; *P < 0.001, **P < 0.01.
EGCG and 5-aza-dc decrease DNMT activity, mRNA and protein expression of DNMTs in A431 cells
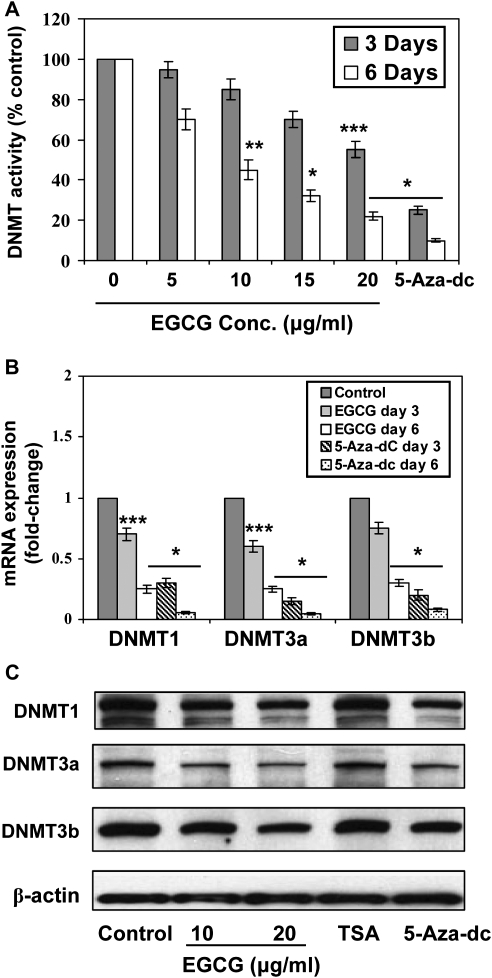
As DNMTs play a crucial role in DNA methylation, we determined the activity of DNMT in A431 cells after treatment of cells with EGCG for 3 and 6 days. EGCG decreased DNMT activity greater after treatment of cells for 6 days than the treatment of cells for 3 days, and this effect of EGCG was concentration dependent (20–75%, P < 0.05–0.001), as shown in Figure 4A. EGCG also significantly decreased DNMT activity (P < 0.01 to P < 0.001) in SCC 13 cells and this effect of EGCG was dose dependent (Figure 3C). The decrease in DNMT activity by EGCG may be due to reduced expression of DNMT. These results were consistent with the quantitative analysis of the mRNA expression of the DNMTs using real-time PCR (Figure 4B). The mRNA levels of DNMT1, DNMT3a and DNMT3b were significantly decreased (P < 0.05 to P < 0.001) in A431 cells after the treatment of cells with EGCG for 3 or 6 days. The decrease in mRNA expression was greater after the treatment of cells with EGCG for 6 days than the treatment of cells with EGCG for 3 days (Figure 4B). There was also a decrease in the protein expression levels of DNMT1, DNMT3a and DNMT3b after the treatment of cells with EGCG for 6 days compared with non-EGCG-treated controls. Treatment of A431 cells with 5-aza-dc for 6 days also decreased the protein expression levels of DNMT1, DNMT3a and DNMT3b, whereas TSA had no effect on the protein levels of DNMTs under identical conditions (Figure 4C). As the effect of EGCG was identical in two different skin cancer cell lines, further detailed mechanistic studies were performed only with A431 cell line.
Fig. 4.
Treatment of A431 cells with EGCG or 5-aza-dc for 3 or 6 days inhibits DNMT activity and decreases the levels of mRNA and protein expressions of DNMT1, DNMT3a and DNMT3b. (A) total DNMT activity in nuclear extracts was determined using the DNMT Activity Assay Kit. Data are presented in terms of percentage versus the results using non-EGCG-treated controls, which was assigned a value of 100%, and as means ± standard deviations; n = 3. (B) Quantitative real-time PCR analysis of mRNA levels of DNMT1, DNMT3a and DNMT3b in cells. The results are presented as the expression of the individual mRNA with normalization to β-actin and as mean values ± standard deviations, n = 3. Significant difference versus non-EGCG-treated controls; ***P < 0.05, **P < 0.01, *P < 0.001. (C) The levels of DNMT1, DNMT3a and DNMT3b in cell lysates were determined using western blot analysis after treating the cells with EGCG for 6 days. The treatment groups of 5-aza-dc and TSA were used as controls.
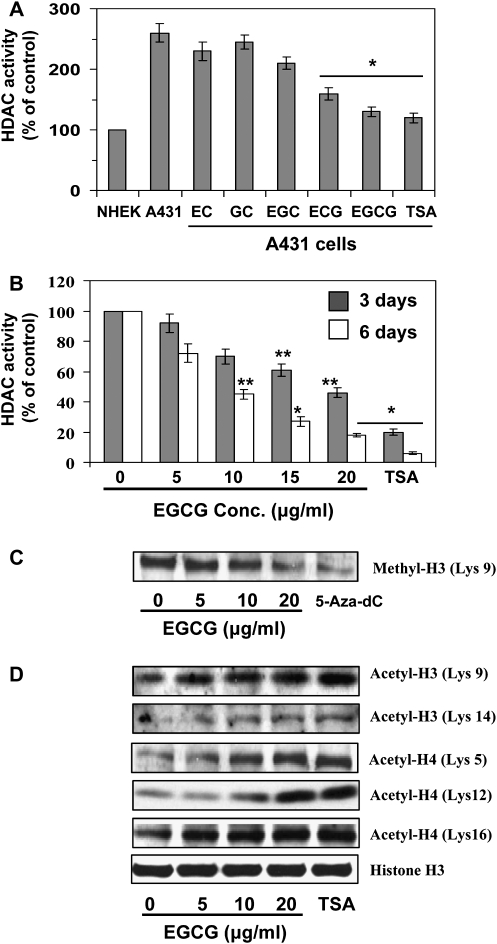
Effect of green tea catechins and TSA on HDAC activity in A431 cells
First, we have examined comparative effects of various catechins, such as EC, GC, EGC, ECG and EGCG, on HDAC activity in A431 cells and data were compared with NHEK. For this purpose, A431 cells were treated with equimolar concentrations (25 μM) of EC, GC, EGC, ECG and EGCG for 6 days. NHEK were used as a control. At the termination of the experiments, cells were harvested and HDAC activity was determined as detailed under Materials and Methods. As shown in Figure 5A, the level of HDAC activity in A431 cells was >2-fold higher compared with NHEK. The inhibitory effect of ECG and EGCG on HDAC activity was significantly greater (P < 0.001) than other catechins such as EC, GC and EGC. The treatment of A431 cells with TSA, a well-known inhibitor of HDAC activity, also significantly decreased HDAC activity and used as a positive control. As histone acetylation, an important mechanism of regulation of gene expression, is in part regulated by HDAC, we determined further whether EGCG treatment altered the expression and activity of HDAC in A431 cells using two different time points. As shown in Figure 5B, treatment of A431 cells with EGCG for 3 days resulted in reduction of HDAC activity; however, this decreasing effect of EGCG on HDAC activity was significantly greater (P < 0.01 to P < 0.001) when cells were incubated with EGCG for 6 days. Similar to EGCG, TSA also decreased HDAC activity and this effect was greater than EGCG at the 0.2 μM concentration used in this study (Figure 5B) and served as a control.
Fig. 5.
Effect of green tea catechins and TSA on HDAC activity and acetylation of histones in A431 cells. (A) Comparative effect of equimolar concentration of EC, GC, EGC, ECG, EGCG (25 μM) and TSA on HDAC activity in A431 cells and NHEK. Cells were treated with different catechins for 6 days and then harvested. HDAC activity was assessed with proteins of nuclear extracts prepared from untreated or treated cells using HDAC Activity Assay Kit following the manufacturer’s instructions. (B) Effect of various concentrations of EGCG (0, 5, 10, 15 and 20 μg/ml) or TSA (0.2 μM) on HDAC activity. Cells were treated for 3 and 6 days. Treatment with TSA was used as a positive control. Data are mean ± standard deviation (n = 3) and presented in terms of percentage versus non-EGCG-treated control. Significant difference versus non-EGCG-treated control; *P < 0.001, **P < 0.01. (C) Analysis of the level of methyl H3 (Lys 9) in cell lysates using western blotting. Cells were treated for 6 days with EGCG or 5-aza-dc. Cells treated with 5-aza-dc served as a control. (D) Cells were treated with EGCG or TSA for 6 days, and cell lysates were subjected to the analysis of modifications in histones using western blotting. Representative blots are shown from three experiments.
Effect of EGCG, TSA and 5-aza-dc on histone acetylation and methylation
Deacetylation and/or methylation of histone proteins H3 and H4 are known to be involved in modification of chromatin structure and silencing of tumor suppressor genes. To further verify the effect of EGCG on HDAC activity in A431 cells, we examined the effect of EGCG on acetylation of histone H3 and H4. As the effect of EGCG on HDAC activity was greater after 6 days of the treatment of A431 cells than the treatment of cells for 3 days, we selected the time point 6 days for further studies. For this purpose, A431 cells were treated with EGCG, 5-aza-dc or TSA for 6 days. Cells were harvested, cell lysates prepared and subjected to the analysis of histone methylation and histone acetylation using western blot analysis. It has been suggested that deacetylation and methylation of H3-Lys 9 are associated with silencing of gene expression, whereas acetylation of H3-Lys 9 is involved in activation of gene expression (16). Therefore, we selected to analyse H3-Lys 9 to determine whether EGCG would alter its acetylation and methylation states using western blot analysis. As shown in Figure 5C, treatment of EGCG decreased the levels of methylated H3-Lys 9, whereas increased the levels of acetylated H3-Lys 9 (Figure 5D). Treatment of 5-aza-dc also decreased levels of methylated H3-Lys 9 (Figure 5C) but did not change the level of acetylated H3-Lys 9 (data not shown). In contrast, treatment of cells with TSA increased the level of acetylated H3-Lys 9 but did not alter the level of methylated H3-Lys 9 (data not shown). Treatment of EGCG, TSA or 5-aza-dc however, did not change protein levels of total histone 3 (H3). Furthermore, the effect of EGCG was also evaluated on acetylation status of H3 at Lys 14, and acetylation status of H4 on different lysine residues. As shown in Figure 5D, EGCG treatment enhanced the levels of acetylated H3-Lys 14 and acetylated H4 at Lys 5, Lys 12 and Lys 16. Furthermore, the treatment of EGCG or TSA did not change the protein levels of total histone H3.
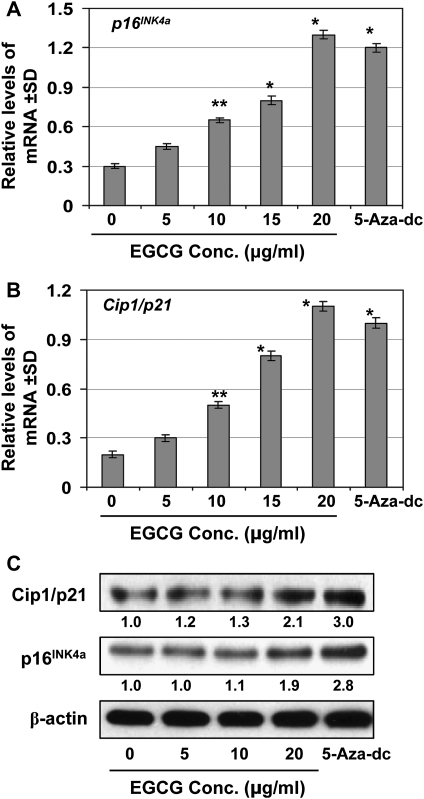
EGCG and 5-aza-dc reactivate tumor suppressor genes and proteins, p16INK4a and Cip1/p21 in A431 cells
We further determined if DNA demethylation and reduction in HDAC activity in A431 cells by EGCG leads to the reactivation of tumor suppressor genes in this skin cancer cell line which was used as an in vitro model. For this purpose, A431 cells were treated with various concentrations of EGCG and 5-aza-dc for 6 days. Cellular RNA from different treatment groups was isolated and subjected to real-time PCR as detailed under Materials and Methods. As shown in Figure 6A and B, real-time PCR analysis revealed that treatment of A431 cells with EGCG resulted in a significant reactivation of silenced mRNA expression of p16INK4a and Cip1/p21 (P < 0.01 and P < 0.001) in a concentration-dependent manner. 5-Aza-dc treatment also significantly reactivated the mRNA expression of both tumor suppressor genes. Similar to mRNA expression, protein reactivation levels were also determined using western blot analysis (Figure 6C). EGCG treatment reactivated the silenced protein levels of tumor suppressor genes, p16INK4a and Cip1/p21, in a concentration-dependent manner, and similar reactivation effect on tumor suppressor proteins was also observed when cells were treated with 5-aza-dc under identical conditions.
Fig. 6.
Treatment of cells with EGCG or 5-aza-dc for 6 days reactivates silenced tumor suppressor genes, p16INK4a and Cip1/p21. (A and B) RNA was isolated from the cells of different treatment groups and subjected to the quantification of mRNA expression levels of p16INK4a and Cip1/p21 by real-time PCR using the procedure detailed in Materials and Methods. Data were normalized to housekeeping gene (β-actin) and are presented as relative change in mRNA levels in terms of mean values ± standard deviation (n = 3). Significant difference versus non-EGCG-treated control; *P < 0.001, **P < 0.01. (C) The protein levels of Cip1/p21 and p16INK4a were determined in cell lysates using western blotting under identical conditions. The relative density (arbitrary) of each band after normalization for β-actin is determined and shown as a fold-change compared with non-EGCG-treated control, which was assigned an arbitrary unit 1 in each case. Data under immunoblots show fold-change. Representative blots are shown from three experiments.
Discussion
Green tea polyphenols have been extensively studied for their anticarcinogenic effects in various tumor models including the prevention of skin cancers using multiple in vitro and in vivo systems (17,18). Intake of green tea prevents skin cancer through their anti-inflammatory, antioxidative, DNA repair and various other mechanisms (17,18). Extensive studies have shown that green tea polyphenols prevent skin cancer in laboratory animals whether they are applied topically or given in drinking water of laboratory animals (17–19). Moreover, cancer is a manifestation of both genetic and epigenetic modifications, and less attention has been paid on the epigenetic modifications by green tea in prevention of skin cancer risk. Epigenetic gene regulations have been recognized to play a crucial role in the etiology of cancer. DNA methylation and posttranslational histone modifications are important epigenetic events in regulation of gene expression and maintenance of cellular function and that may contribute to cancer development (6,7,10). Abnormal methylation in DNA is a hallmark of cancer and often leads to silencing of tumor suppressor genes, which leads to cancer development and progression. Dietary phytochemicals have been shown to be involved in epigenetic modifications to regulate cellular function and to modify the risk of cancer.
In this study, we investigated epigenetic regulation by tea catechins and elucidated the underlying mechanisms using the A431 human epidermoid carcinoma cell line as an in vitro model system. Also, we used 5-aza-dc, a well-characterized DNA methylation inhibitor, and TSA, a well-known HDAC inhibitor, as controls. In the beginning of experiments, we have characterized that EC, GC and EGC are the weaker epigenetic modulators, whereas ECG and EGCG are better epigenetic modulators than other tea EC derivatives. Our study reveals that EGCG significantly decreased the levels of global DNA methylation and decreased the levels of 5-mC in DNA of A431 cells, and these effects were concentration dependent. However, the effect of EGCG on epigenetic modulation was slow as significant epigenetic alterations were observed 6 days after treatment of A431 cells with EGCG. Similar effect of EGCG was also found in another human SCC 13 cell line. We also examined the effect of EGCG on epigenetic modifications in melanoma cell lines (A375 and Hs294) under identical conditions. EGCG showed promising effect on DNA demethylation and HDAC activity in melanoma cancer cell lines (Singh, T. and Katiyar, S.K. unpublished results). Our results indicate that 5-aza-dc also significantly decreased the levels of global DNA methylation and the activity as well as mRNA and protein expressions of DNMT1, DNMT3a and DNMT3b in A431 cells. The inhibitory effect of EGCG on the DNMT activity, mRNA and protein expressions of DNMT1, DNMT3a and DNTM3b were also dose and time dependent. These results indicate that the action of EGCG is similar to 5-aza-dc, which is a well-characterized DNA methylation inhibitor. We also observed that treatment of cells at higher concentrations of EGCG-induced cell death and that mask the epigenetic effects of EGCG in A431 cells (data not shown).
Histone modifications are epigenetic marks linked to transcriptional activation and repression of genes (20). Normally, histone acetylation results in an open chromatin structure associated with transcriptional activation, whereas histone deacetylation leads to a closed chromatin structure with transcriptional repression (20). Posttranscriptional modifications of histone methylation and acetylation may contribute to cancer development by modulation of the expression of tumor suppressor genes and oncogenes. HDACs play a key role in regulation of histone deacetylation. Deacetylation and methylation of H3-Lys 9 are the most common histone modifications involved in epigenetic repression of genes (16). Our data demonstrate that EGCG significantly decreases HDAC activity after the treatment of skin cancer cells for 6 days time period. EGCG decreased the levels of methylated H3-Lys 9, but increased the levels of acetylated H3-Lys 9 and H3-Lys 14. Similarly, TSA also decreases HDAC activity and increases acetylated H3-Lys 9 and H3-Lys 14. These observations suggest that EGCG decreases HDAC activity to maintain H3-Lys 9 at a high level of acetylation and a low level of methylation, which may favor transcriptional activation of genes. The effects of 5-aza-dc were similar to EGCG in methylation of H3-Lys 9 but have no detectable effects on histone acetylation in cells (data not shown). In contrast, TSA has no effect on methylated H3-Lys 9. These results also suggest that EGCG and 5-aza-dc may inhibit methyltransferase to prevent histone methylation. Treatment of cells with EGCG also increases acetylation of H4-Lys 5, H4-Lys 12 and H4-Lys 16, which may also favor transcriptional activation of tumor suppressor genes. Unlike 5-aza-dc and TSA, EGCG has dual actions involving both DNA demethylation and modifications of histone acetylation and methylation. Similar to EGCG, other dietary phytochemicals, such as diallyl disulfide and genistein, have showed epigenetic alterations on DNA methylation and histone modifications in colon and prostate cancer cells (21,22). Lycopene also demethylated the GSTP1 promoter and reactivated GSTP1 expression in human breast cancer cells (23). EGCG has been shown to induce epigenetic modifications in human prostate, colon and esophageal cancer cell lines (24). Our study also revealed that EGCG reactivates or re-expresses mRNA expression of tumor suppressor genes and proteins of p16INK4a and Cip1/p21 in skin cancer cells. Similar to EGCG, 5-aza-dc also reactivated the mRNA expression of both tumor suppressor genes in A431 cells. The reactivation of silenced tumor suppressor genes by EGCG is associated with DNA demethylation and histone modifications in cancer cells.
In summary, we are reporting for the first time that EGCG can restore or reactivate the expression of the DNA hypermethylation-silenced genes, p16INK4a and Cip1/p21, in human skin cancer cells by downregulation of DNMT and HDAC activity. Our data suggest that epigenetic regulation of these tumor suppressor genes by EGCG may play a role in skin cancer chemoprevention. These findings are of importance for understanding the anticancer mechanisms and clinical applications of green tea polyphenols. In vivo animal experiments are underway in our laboratory to assess the skin cancer chemopreventive mechanism of green tea polyphenols through epigenetic events, including DNA methylation, posttranslational histone modifications and reactivation of tumor suppressor genes.
Funding
National Institutes of Health (CA140832 to S.K.K.); Veterans Administration Merit Review Award (S.K.K.).
Acknowledgments
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- 5-Aza-dC
5-aza-2′-deoxycytidine
- DNMT
DNA methyltransferase
- EC
(−)-epicatechin
- ECG
(−)-epicatechin-gallate
- EGC
(−)-epigallocatechin
- EGCG
(−)-epigallocatechin-3-galate
- GC
(−)-gallocatechin
- HDAC
histone deacetylase
- 5-mC
5-methylcytosine
- mRNA
messenger RNA
- NHEK
normal human epidermal keratinocytes
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- SCC
squamous cell carcinoma
- TSA
trichostatin A
References
- 1.Katiyar S, et al. Green tea and skin cancer: photoimmunology, angiogenesis and DNA repair. J. Nutr. Biochem. 2007;18:287–296. doi: 10.1016/j.jnutbio.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 2.Katiyar SK, et al. Tea consumption and cancer. In: Simopoulos A.P., editor. World Review Nutrition and Diet. Vol. 79. Basel: Karger; 1996. pp. 154–184. [PubMed] [Google Scholar]
- 3.Katiyar SK, et al. Tea in chemoprevention of cancer: epidemiologic and experimental studies. Int. J. Oncol. 1996;8:221–238. doi: 10.3892/ijo.8.2.221. [DOI] [PubMed] [Google Scholar]
- 4.Meeran SM, et al. (-)-Epigallocatechin-3-gallate prevents photocarcinogenesis in mice through interleukin-12-dependent DNA repair. Cancer Res. 2006;66:5512–5520. doi: 10.1158/0008-5472.CAN-06-0218. [DOI] [PubMed] [Google Scholar]
- 5.Hegi ME, et al. Epigenetic deregulation of DNA repair and its potential for therapy. Clin. Cancer Res. 2009;15:5026–5031. doi: 10.1158/1078-0432.CCR-08-1169. [DOI] [PubMed] [Google Scholar]
- 6.Jones PA, et al. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 7.Jones PA. DNA methylation and cancer. Oncogene. 2002;21:5358–5360. doi: 10.1038/sj.onc.1205597. [DOI] [PubMed] [Google Scholar]
- 8.Antequera F, et al. CpG islands. EXS. 1993;64:169–185. doi: 10.1007/978-3-0348-9118-9_8. [DOI] [PubMed] [Google Scholar]
- 9.Herman JG, et al. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 10.Counts JL, et al. Alterations in DNA methylation may play a variety of roles in carcinogenesis. Cell. 1995;83:13–15. doi: 10.1016/0092-8674(95)90228-7. [DOI] [PubMed] [Google Scholar]
- 11.Davis CD, et al. Frontiers in nutrigenomics, proteomics, metabolomics and cancer prevention. Mutat. Res. 2004;551:51–64. doi: 10.1016/j.mrfmmm.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 12.Davis CD, et al. DNA methylation, cancer susceptibility, and nutrient interactions. Exp. Biol. Med. (Maywood) 2004;229:988–995. doi: 10.1177/153537020422901002. [DOI] [PubMed] [Google Scholar]
- 13.Huang S. Histone methyltransferases, diet nutrients and tumor suppressors. Nat. Rev. Cancer. 2002;2:469–476. doi: 10.1038/nrc819. [DOI] [PubMed] [Google Scholar]
- 14.Meeran SM, et al. Inhibition of UVB-induced skin tumor development by drinking green tea polyphenols is mediated through DNA repair and subsequent inhibition of inflammation. J. Invest. Dermatol. 2009;129:1258–1270. doi: 10.1038/jid.2008.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mantena SK, et al. Orally administered green tea polyphenols prevent ultraviolet radiation-induced skin cancer in mice through activation of cytotoxic T cells and inhibition of angiogenesis in tumors. J. Nutr. 2005;135:2871–2877. doi: 10.1093/jn/135.12.2871. [DOI] [PubMed] [Google Scholar]
- 16.Nakayama J, et al. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 17.Nichols JA, et al. Skin photoprotection by natural polyphenols: anti-inflammatory, anti-oxidant and DNA repair mechanisms. Arch. Dermatol. Res. 2010;302:71–83. doi: 10.1007/s00403-009-1001-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katiyar SK, et al. Green tea polyphenolic antioxidants and skin photoprotection. Int. J. Oncol. 2001;18:1307–1313. doi: 10.3892/ijo.18.6.1307. [DOI] [PubMed] [Google Scholar]
- 19.Yusuf N, et al. Photoprotective effects of green tea polyphenols. Photodermatol. Photoimmunol. Photomed. 2007;23:48–56. doi: 10.1111/j.1600-0781.2007.00262.x. [DOI] [PubMed] [Google Scholar]
- 20.Rice JC, et al. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr. Opin. Cell Biol. 2001;13:263–273. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- 21.Druesne N, et al. Diallyl disulfide (DADS) increases histone acetylation and p21(waf1/cip1) expression in human colon tumor cell lines. Carcinogenesis. 2004;25:1227–1236. doi: 10.1093/carcin/bgh123. [DOI] [PubMed] [Google Scholar]
- 22.Majid S, et al. Genistein induces the p21WAF1/CIP1 and p16INK4a tumor suppressor genes in prostate cancer cells by epigenetic mechanisms involving active chromatin modification. Cancer Res. 2008;68:2736–2744. doi: 10.1158/0008-5472.CAN-07-2290. [DOI] [PubMed] [Google Scholar]
- 23.King-Batoon A, et al. Modulation of gene methylation by genistein or lycopene in breast cancer cells. Environ. Mol. Mutagen. 2008;49:36–45. doi: 10.1002/em.20363. [DOI] [PubMed] [Google Scholar]
- 24.Fang MZ, et al. Tea polyphenol (-)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003;63:7563–7570. [PubMed] [Google Scholar]