Abstract
The XPC gene is involved in repair of bulky DNA adducts formed by carcinogenic metabolites and oxidative DNA damage, both known bladder cancer risk factors. Single nucleotide polymorphisms (SNPs) in XPC have been associated with increased bladder cancer risk. Recently, rarer genetic variants have been identified but it is difficult to ascertain which are of functional importance. During a mutation screen of XPC in DNA from 33 bladder tumour samples and matched blood samples, we identified five novel variants in the patients’ germ line DNA. In a case–control study of 771 bladder cancer cases and 800 controls, c.905T>C (Phe302Ser), c.1177C>T (Arg393Trp), c.*156G>A [3′ untranslated region (UTR)] and c.2251-37C>A (in an intronic C>G SNP site) were found to be rare variants, with a combined odds ratio of 3.1 (95% confidence interval 1.0–9.8, P = 0.048) for carriage of one variant. The fifth variant was a 2% minor allele frequency SNP not associated with bladder cancer. The two non-synonymous coding variants were predicted to have functional effects using analytical algorithms; a reduced recruitment of GFP-tagged XPC plasmids containing either c.905T>C or c.1177C>T to sites of 408 nm wavelength laser-induced oxidative DNA damage was found in vitro. c.*156G>A appeared to be associated with reduced messenger RNA stability in an in vitro plasmid-based assay. Although the laser microbeam assay is relevant to a range of DNA repair genes, our 3′ UTR assay based on Green fluorescent protein(GFP) has widespread applicability and could be used to assess any gene. These assays may be useful in determining which rare variants are functional, prior to large genotyping efforts.
Introduction
Transitional cell carcinoma of the bladder is the fourth most common cancer in Western men (1) with cigarette smoking and exposure to industrial chemicals being major risk factors. Carcinogen metabolism generates bulky DNA adducts, repaired by nucleotide excision repair (NER), which also repairs some endogenously generated oxidative DNA lesions (2).
Xeroderma pigmentosum (XP), a rare autosomal recessive disorder characterized by a high incidence of skin cancer, involves mutations in NER genes (3,4). One such gene, XPC, on chromosome 3p25, encodes a protein forming a heterotrimeric complex with RAD23B and centrin 2 (4,5), which recognizes and binds to helix-distorting DNA adducts (6). XP-C patients have a 1000-fold increased incidence of skin tumours but, as few patients survive beyond early adulthood, internal malignancies have only rarely been reported (7). The estimated 1 in 500 individuals heterozygous for an XPC mutation are also at increased risk of malignancy (8), as reported for heterozygous mutations in other recessive DNA repair disease genes (9,10).
XPC knockout mice have an increased risk of skin tumours following ultraviolet (UV) B radiation, and also develop liver and lung tumours following exposure to the chemical carcinogen acetylaminofluorene (11) reflecting defective NER. They have a 100% incidence of spontaneous lung tumours (12,13), attributed to defective repair of endogenous oxidative lesions. It is now clear that the XPC protein is involved in repair of oxidative DNA lesions not only via its NER function, repairing bulky oxidative lesions including 8,5′-cyclopurine 2′-deoxynucleosides (14,15), but also through XPC-RAD23B acting as a cofactor both in the base excision repair of 8-hydroxyguanine, by stimulating OGG1 activity (15), and in base excision repair of T/G mismatches, by stimulating thymine DNA glycosylase activity (16).
During a mutation screen of XPC in DNAs extracted from 33 bladder tumours, we found five previously unidentified XPC variants in both tumour and matched germ line blood DNAs. We performed a case–control study, and one variant (c.621 + 22G>A) was revealed as a 2% minor allele frequency single nucleotide polymorphism (SNP) not associated with bladder cancer risk. The remaining four variants, namely c.905T>C (rs121965091), c.1177C>T (rs1211965090), c.*156G>A (rs121965092) and c.2251-37C>A (rs2470353), were rare in the case–control analysis. As we hypothesized that the rare coding and 3′ untranslated regions (UTR) variants might have a functional impact, we assessed their effects on XPC protein recruitment to focal DNA damage and on messenger RNA (mRNA) stability.
Materials and methods
Study population and clinical materials
Local ethical approval was granted by the Leeds Teaching Hospitals Local Research Ethics Committee and informed consent obtained. DNA from 33 fresh-frozen bladder transitional cell carcinomas (16 pTaG2/G3, 10 pT1 at least G2/G3, six pT2 at least G3 and one pTxG3) was extracted using a QIAamp DNA Micro kit (Qiagen, Crawley, UK), and DNA extracted from 32 matched bloods (one tumour having no available matched blood) using the Puregene modified salt precipitation method (Flowgen, Nottingham, UK). The case–control study population has been described previously (17).
Cell lines
RT112M bladder cancer cells were grown at 37°C in a 5% CO2 humidified atmosphere, cultured in RPMI 1640, 10% fetal bovine serum, 1% L-glutamine and passaged for fewer than 6 months. This cell line has been authenticated in the MK laboratory by extensive genomic analysis (microsatellite typing, conventional karyotypic analysis, MFISH and array-based copy number analysis).
Mutation detection
All 16 exons and the 5′ and 3′ UTR of the XPC gene were amplified from tumour DNA by polymerase chain reaction (PCR) (supplementary Table I is available at Carcinogenesis Online). PCR reactions were carried out in a total volume of 20 μl containing 20 ng DNA, 0.5 U Thermostat (Abgene, Epsom, UK), 2 μl of manufacturer’s buffer, 0.5 mM forward and reverse primers, 10 mM deoxynucleoside triphosphates, 3 mM MgCl2 and 5% dimethyl sulfoxide, on a PTC200 DNA engine (GRI, Braintree, UK). Thermal cycling parameters were: 95°C for 12 min, 35 cycles of 95°C for 30 s, optimal primer annealing temperature for 30 s, 72°C for 30 s, followed by a final extension of 72°C for 10 min.
Ten microlitres of PCR products were denatured at 95°C for 5 min and allowed to cool to 65°C for the formation of heteroduplexes. Denaturing high-performance liquid chromatography (DHPLC) was carried out using a Transgenomic WAVE Nucleic Acid Fragment Analysis system and DNASep column (Transgenomic Ltd, Glasgow, UK). The composition of buffer A was 0.1 M trimethylammonium acetate; buffer B contained 0.1 M trimethylammonium acetate and 25% acetonitrile. Analysis was carried out at a flow rate of 0.9 ml/min−1 and a buffer B gradient increase of 2%/min−1 for 4 min. Start and end concentrations of buffer B were determined empirically for each fragment. If the results were not conclusive, then the DHPLC was conducted at two temperatures. DHPLC data analysis was by visual inspection of sample and reference chromatograms, with results scored independently by two researchers.
Sequencing in bladder tumours
PCR reactions were carried out in a total volume of 20 μl containing 20 ng of DNA, 10 μl HotstarTaq Master Mix (Qiagen), 0.5 μM forward and reverse primers (supplementary Table I is available at Carcinogenesis Online). Thermal cycling parameters were 15 min at 95°C, followed by 45 cycles of 95°C for 30 s, optimal primer annealing temperature for 30 s and 72°C for 30 s, followed by a final extension of 72°C for 10 min. The PCR products (5 μl) were treated with 1 μl of exonuclease I and 2 μl of Shrimp alkaline phosphatase (USB Corporation, Cleveland, Ohio) at 37°C for 30 min to remove unincorporated nucleotides followed by heat inactivation at 80°C for 15 min. Three microlitre of treated PCR products were forward and reverse sequenced using HalfBD with the ABI BigDye Terminator reaction and analysed on a 3130×1 Genetic Analyser (Applied Biosystems, Foster City, CA). Sequences were analysed using the reference sequence, GenBank accession number AC090645.
Genotyping by Taqman and pyrosequencing
Variants c.621+22 G>A, c.905T>C (rs121965091) and c.*156 G>A (rs121965092) were genotyped using the allelic discrimination 5′ nuclease assay (Taqman; supplementary Table II is available on Carcinogenesis Online), as described previously (17,18), but with 50 cycles for c.621+22 G>A. Known-mutant DNA samples were included in a test run to help identify clusters. Assays for c.1177 C>T (rs121965090) and c.2251-37C>A (rs2470353) were designed using the Pyrosequencing Assay Design Software (Biotage, Uppsala, Sweden and Qiagen): c.1177 C>T biotinylated forward primer (5′TGCCAAAGGGAAGAGGAACA3′), reverse primer (5′GTCTCCTGGGCCCTCATCTT3′) and sequencing primer (5′TGGAGGAGGGCTTGC3′). c.2251-37C>A biotinylated forward primer (5′GAGGTACACATTCCCAAACTCG3′), reverse primer (5′GAAGGTAAGGGCAGCATCAGA3′) and sequencing primer (5′CAGGCCGCCTTGTTCC3′). PCR reactions were carried out in 96-well plates on a PTC-225 Thermocycler (Bio-Rad Laboratories, Hercules, CA), using a total volume of 20 μl containing 20 ng of DNA, 10 μl of ×2 Hotstar Taq Master Mix (Qiagen), and final concentrations of 0.2 μM each of forward and reverse primers, and 0.5 mM additional MgCl2. Thermal cycling parameters were 15 min at 95°C, followed by 35 cycles of 30 s each at 94°C, 63.4°C and 72°C, followed by 10 min at 72°C. PCR products were run on the Pyromark ID system (Biotage) according to the manufacturer’s standard protocol. Five percent of samples were re-genotyped with 100% concordance.
3′UTR plasmid reporter system and FACS analysis
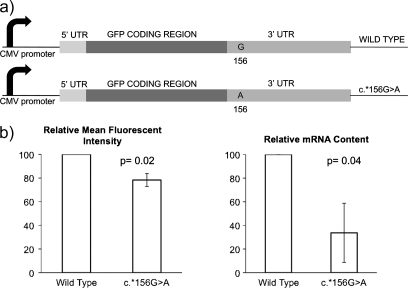
Plasmid pTH-GFPa (19), a generous gift from Thomas Hughes, Leeds Institute of Molecular Medicine, University of Leeds, UK, consists of GFP cloned into pcDNA3MycHis (Invitrogen, Paisley, UK). The 5′- and 3′-UTR regions of XPC were cloned into pTH-GFPa, flanking the GFP coding region and replacing the plasmid viral 5′- and 3′-UTR regions to minimize extraneous DNA. The change c.*156G>A was introduced into the 3′-UTR region by site-directed mutagenesis (Quik Change Mutagenesis kit II; Stratagene, Stockport, UK), (Figure 1a).
Fig. 1.
3′ UTR plasmid reporter system and FACS analysis (n = 3, duplicate wells analysed in each experiment, two-tailed t-test, error bars standard deviation). (a) Schematic diagram of pTH-GFPa constructs used in mRNA translational efficiency assays. (b) Mean fluorescence intensity of RT112 cells transfected with GFP-flanked by mutant 3′ UTR sequences relative to that of GFP-flanked by wild-type XPC UTR sequences, and relative levels of GFP mRNA analysed by quantitative reverse transcription–PCR using the ΔΔCt.
RT112M bladder cancer cells were seeded into six-well plates (4 × 105 cells) and 24 h later transfected with pcDNA3MycHis and a GFP-containing test plasmid (at 10:1 ratio to prevent saturation of the system with GFP) using Fugene (1 μ DNA: 3 μl transfection reagent). Following overnight incubation, duplicate wells were analysed by Fluorescence activated cell sorting (FACS) (LSRII and FACSDiva software; BD, Oxford, UK) using mean fluorescence intensity to determine GFP protein expression levels.
RNA was isolated from the remaining two wells using a GenElute total RNA kit (Sigma, Dorset, UK) as per the manufacturers’ protocol and used to synthesize complementary DNA (cDNA) using random hexamers and M-MuLV reverse transcriptase (New England Biolabs, Hitchin, UK). Quantitative real-time reverse transcription–PCR was performed in triplicate using 1% of this cDNA as template and SYBR green I as the fluorescent reporter. GFP mRNA was quantified relative to expression of the constitutively expressed housekeeping gene 36B4 (supplementary Table III is available on Carcinogenesis Online). The Ct value was adjusted to account for differences in transfection efficiency (number of GFP protein expressing cells as determined by FACS analysis, doubling of mRNA for change in Ct value of 1). mRNA levels were normalized to the wild-type mRNA expression using the ΔΔCt method.
Preparation of XPC mutant plasmids and site-directed mutagenesis
Plasmid pLGR224-XPC-GFP was a generous gift Dr Jaime Angulo-Mora, Commissariat à L'Energie Atomique, Fontenay aux Roses, France. This contained a mutation c.1399C>A, coding Gln467Lys, predicted to be silent by SIFT (http://sift.jcvi.org/), Polyphen (http://tux.embl-heidelberg.de/ramensky/), Pfam (http://pfam.sanger.ac.uk) and NPS@ (http://pbil.ibcp.fr). However, this variant was converted back to wild-type (c.1399C) using a QuikChange Multi Site-Directed Mutagenesis Kit (Stratagene). Variants were then introduced using the same kit (primer sequences available on request), and the changes in the DNA sequence confirmed by direct sequencing.
Laser microbeam irradiation experiments
RT112M cells were seeded onto 35 mm glass-bottomed dishes (8 × 105 cells) and 24 h later transfected with the pLGR224-XPC-GFP-based plasmids using FuGene HD transfection reagent (2 μg DNA:6 μl Fugene; Roche, Burgess Hill, UK). Twenty-four to 30 h posttransfection, the growth medium was replaced with Optimem (Gibco, Paisley, UK), containing 10% fetal bovine serum. Live cell imaging experiments were performed on a Nikon Eclipse TE2000E confocal laser scanning microscope system (×60/1.4 objective) at 37°C. Microirradiation was carried out using a 17 mW 408 nm diode laser set to maximum power. Cells with intermediate fluorescence levels were selected for treatment, on the assumption that they would have similar protein expression levels. Cells were focally damaged using the Zoom FRAP 2.1.848 macro (to detect fluorescence recruitment after photodamage), with one laser pulse (duration 1.06 s) to a nuclear spot 176 nm in diameter. Recruitment of fluorescent protein was recorded using time-lapse microscopy with a calibrated 488 nm argon laser (10% power to reduce photobleaching), with one pre-irradiation image and at subsequent 5 s intervals for 4 min postirradiation. Five to ten G1 phase cells were treated per experiment. All treated cells were analysed at the same magnification with a zoom factor of 8, frame size 512 × 512 and pixel size 51.8 nm. All images were processed using ImageJ software [see (20) for details] to analyse the recruitment kinetics.
Statistical analysis
All statistical analyses were performed using Excel and SPSS software. Analysis of genotyping data was as described previously (18). Student’s t-test was used for comparison of means. Pairwise correlations between the relative expressions levels were analysed by the Pearson correlation coefficient. Odds ratios (OR) and 95% confidence intervals (CIs) were calculated from logistic regression models to assess the effect of the three rare variants and the combined effect of the three variants (≥1 rare allele versus no rare alleles) on bladder cancer risk. P values < 0.05 were considered statistically significant.
For the combined genetic effect, assuming a binary variable (≥1 rare allele versus no rare alleles) with a prevalence of 0.5% amongst controls, the power is 13% when the OR is 2.0 and 40% when the OR is 3.0.
Results
No XPC mutations found in bladder tumours
Internal malignancies have been reported in XP-C patients (7) and are thought to arise from unrepaired oxidative damage resulting in TP53 mutations (21). As TP53 mutations are commonly seen in muscle invasive bladder tumours (22), we looked for XPC mutations in 33 human sporadic bladder tumour specimens. At least one variant in each of XPC exons 1, 2, 5.1, 7, 8, 10–15, the 3′ UTR and flanking regions was detected by DHPLC screening. The exact nature of each variant was determined by direct sequencing of both tumour and matched germ line DNA. No somatic mutations were identified in the bladder tumour samples. However, in matched germ line DNA, we identified 19 known SNPs. Comparison of tumour and germ line DNA revealed allelic losses at 11 polymorphic sites (loss of heterozygosity), in two muscle invasive tumours; this has also been described in human lung and ovarian tumours (12,23).
Five novel XPC variants in germ line DNA from bladder cancer patients
Five additional variants not annotated in dbSNP (www.ncbi.nlm.nih.gov/projects/SNP/) were identified (supplementary Figure 1a is available on Carcinogenesis Online). Thirty-two controls were randomly selected from our case–control series (18) and sequenced but none of the five new variants was detected. Two variants were present in two patients. c.621 + 22G>A was genotyped by Taqman (95% success rate) and found to have a minor allele frequency of 0.02 in the control population. The OR for individuals heterozygous for the rare allele compared with those carrying the common homozygous genotype was 0.9 (95% CI 0.5–1.5); therefore, this SNP was not studied further.
The variants c.905T>C, c.*156G>A, c.1177C>T and c.2251-37>A were genotyped in germ line DNA from 771 bladder cancer cases and 800 controls with >96% success rates. They were present in only 0.3, 0.1, 0.9 and 0.3% of cases, respectively. No individual carried more than one of these rare variants. When the genotypes were combined, the OR for carriage of at least one rare allele was 3.1 (95% CI 1.0–9.8, P = 0.048, Table I). The genotypes of the 16 individuals carrying one of the four rare variants were compared with our previous genotyping data for the three SNPs we previously found to be associated with bladder cancer risk, namely rs2228000, rs2470352 and rs2470458 (17). The carriage of the rare variants was largely associated with the rs2228000 wild-type genotype, which we previously found associated with bladder cancer risk (Table II).
Table I.
Genotyping of the three rare variants
| Variant | Amino acid substitution | Allele frequency amongst controls | Genotype | Controls (%) | Cases (%) | OR (95% CI) |
| c.2251-37 rs2470353 Triallelic: C>G SNP, C>A rare variant | Intron 11 | 0 | CC | 201 (27.2) | 183 (25.4) | 1 |
| CG | 358 (48.4) | 347 (48.1) | 1.1 (0.8–1.4) | |||
| GG | 181 (24.5) | 190 (26.3) | 1.2 (0.9–1.5) | |||
| CA | 0 (0) | 2 (0.28) | ND | |||
| AA | 0 (0) | 0 (0) | ND | |||
| c.1177C>T rs121965090 | Arg393Trp | 0.002 | CC | 790 (99.6) | 750 (99.1) | 1.0 |
| CT | 3 (0.4) | 7 (0.9) | 2.5 (0.6–9.5) | |||
| TT | 0 (0) | 0 (0) | ND | |||
| c.905T>C rs121965091 | Phe302Ser | 0.0006 | TT | 787 (99.9) | 757 (99.7) | 1.0 |
| TC | 1 (0.1) | 2 (0.3) | 2.1 (0.2–23.0) | |||
| CC | 0 (0) | 0 (0) | ND | |||
| c.*156G>A rs121965092 | 3′ UTR | 0 | GG | 785 (100) | 759 (99.9) | 1.0 |
| GA | 0 (0) | 1 (0.1) | ND | |||
| AA | 0 (0) | 0 (0) | ND | |||
| c.2251-37C>A, c.1177C>T c.905T>C and c.*156G>A combined | No rare allele | 771 (99.5) | 735 (98.4) | 1.0 | ||
| ≥1 rare allele | 4 (0.5) | 12 (1.6) | 3.1 (1.0–9.8)a |
Statistically significant with P-value < 0.05.
Table II.
Haplotypes for the individuals carrying the rare variants
| Case/control | Age | Smoking | New rare variants |
SNPs associated with bladder cancer risk |
||||
| c.2251-37 C>A Intron 11 rs2470353 | c.1177 C>T Arg393Trp rs121965090 | c.905 T>C Phe302Ser rs121965091 | c.*156G>A 3′ UTR rs121965092 | c.1496C>T Ala499Val (rs2228000) | c.*611 T>A, Ex15-184 (rs2470352) and c.*618 A>Ga, Ex15-177 (rs2470458) | |||
| Cases | ||||||||
| 1 | 86 | 48years | wt | het | wt | wt | wt | wt |
| 2 | 82 | 16 years | wt | het | wt | wt | wt | wt |
| 3 | 79 | non-smoker | bwt | wt | het | wt | het | het |
| 4 | 75 | 31 years | wt | het | wt | wt | wt | wt |
| 5 | 76 | 40 years | bwt | wt | wt | het | wt | wt |
| 6 | 63 | 4 years | wt | wt | het | wt | wt | wt |
| 7 | 63 | 37 years | wt | het | wt | wt | wt | wt |
| 8 | 62 | non-smoker | bwt | het | wt | wt | het | het |
| 9 | 80 | 49 years | wt | het | wt | wt | cwt | wt |
| 10 | 73 | 27 years | bwt | het | wt | wt | cwt | wt |
| 11 | 76 | non-smoker | het | wt | wt | wt | wt | het |
| 12 | 78 | 20 years | het | wt | wt | wt | cwt | het |
| Controls | ||||||||
| 1 | 62 | non-smoker | bwt | het | wt | wt | het | het |
| 2 | 66 | non-smoker | bwt | het | wt | wt | wt | wt |
| 3 | 73 | 37 years | bwt | het | wt | wt | wt | wt |
| 4 | 79 | 23 years | bwt | wt | het | wt | wt | wt |
These are compared with three SNPs associated with increased bladder cancer risk in Sak et al. (17). The 2% minor allele frequency SNP c.621 + 22G>A was wild-type in all 16 individuals.
rs2470352 and rs2470458 are in complete linkage.
Heterozygote for common SNP rs2470353 C>G.
SNP genotypes in cases 9, 10 and 12 were established by direct sequencing, as they were not included in the original 547 cases:579 control study.
Effects of the 3′UTR rare variant c.*156 on transcription and mRNA stability
GFP-expressing plasmids containing either wild-type or the 3′UTR variant cDNA were transfected into RT112M cells, and protein expression assayed by FACS and mRNA content assayed by quantitative real-time reverse transcription–PCR (Figure 1b). There was a significant reduction in protein and mRNA expression in the presence of the 3′UTR variant.
Recruitment of XPC to sites of 408 nm laser-induced damage is reduced in the presence of rare coding variants c.905T>C and c.1177C>T
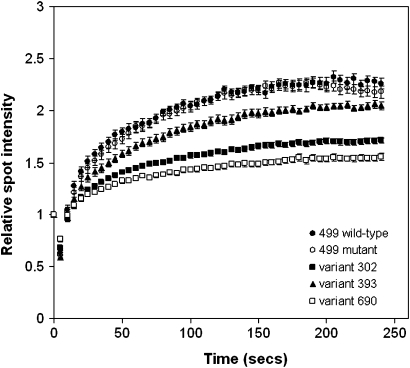
We attempted unsuccessfully to perform UV-host cell reactivation assays in stably transfected XPC-deficient GM15983 cells. Exogenous XPC protein was very unstable when expressed in XPC-deficient cells using a strong SV40 promoter, consistent with a previous report (24) and mRNA levels decreased with increasing cellular exposure times to the selection agent Zeocin. Therefore, we adopted a different approach. RT112M bladder cancer cells transiently transfected with plasmids containing XPC cDNA fused with GFP (Table III) were exposed to focal laser microbeam damage, using a 408 nm (near UVA) laser microbeam (20), to generate oxidative damage, including DNA single and double-strand breaks (supplementary Figure 2 is available on Carcinogenesis Online). We specifically selected cells of intermediate fluorescence intensity for study, and the tight error bars on these data are supportive of the fact that such cells have similar protein expression. XPC recruitment to the sites of damage was tracked in real time. Recruitment of the c.2059G>C mutant was low, whereas the c.499C>T variant (rs2228000) had no effect on XPC recruitment. We found intermediate levels of recruitment for proteins containing the c.905T>C and c.1177C>T variants (Figure 2).
Table III.
Constructs containing XPC variants used for laser microbeam experiments and predicted changes for variants based on software tools and amino acid chemistry
| Coding variants | c.905T>C (rs121965091) | c.1177C>T (rs121965090) | c.1496C>T (rs2228000) | c.2059G>C |
| Exons | Exon 7 | Exon 8 | Exon 8 | Exon 10 |
| Amino acid change | Phe302Ser | Arg393Trp | Ala499Val | Trp690Ser |
| Constructs | ||||
| Wild-type XPC | − | − | − | − |
| c.905T>C | + | − | − | − |
| c.1177C>T | − | + | − | − |
| c.1496C>T | − | − | + | − |
| c.2059G>C | − | − | − | + |
| Prediction tools | ||||
| Amino acid chemistry | Hydrophobic to small polar | Positively charged to hydrophobic aromatic | Both have similar chemical properties | Aromatic hydrophobic to small polar |
| Polyphen | Probably damaging | Possibly damaging | Benign | Probably damaging |
| SIFT | Not tolerated | Not tolerated | Tolerated | Not tolerated |
| Pfam | No change | No change | No active domain change | Active domain change |
| NPS@ | No change in secondary structure | No change in secondary structure | Unclear—half of models predict formation of Beta sheet, half no change in secondary structure | Possible shortening of helical structure |
Mutation nomenclature based on A of start codon ATG as nucleotide 1. Using site-directed mutagenesis, c.1399A was converted back to the wild-type C allele.
Fig. 2.
Relative spot intensity versus time following 408 nm laser damage. Plots show means and standard error of the means of 21–25 samples treated over three independent experiments for each variant. Comparisons were made using two-sided t-test: 499 wild-type (499 wt) and 499 mutant (c.1496C>T), P = 0.357, variant 302 (c.905T>C) and 499 wt, P < 0.001, variant 393 (c.1177C>T) and 499 wt, P = 0.004, variant 690 (c.2059G>C) and 499 wt, P < 0.001.
Discussion
We have identified four new rare variants in the NER gene XPC. We established that these were rare using a case–control study of 771 bladder cancer cases and 800 controls. A striking feature in comparing the new rare variants with known SNP genotypes (Table II) is that carriage of the rare variants detected in this study was largely associated with the rs2228000 wild-type genotype, which we previously found associated with bladder cancer risk. The combined OR in our case–control study for the carriage of any one of these four variants was 3.1 (95% CI 1.0–9.8, P = 0.048) for bladder cancer risk.
There was a significant reduction in protein and mRNA expression in the presence of the 3′ UTR variant, suggestive of reduced mRNA stability and/or transcription. Associations have been reported for 3′ UTR SNPs with alterations in mRNA stability (25–27) although coding SNPs can also have an effect via interactions with RNA-binding proteins (28). The intrinsic stability of mRNA is determined by cis-acting sequences located within the 3′ UTR as well as trans-acting RNA-binding proteins (25); 3′ UTR variants may alter the binding of these proteins or alter the secondary structure of the 3′ UTR. Therefore, it is feasible that the c.*156 3′ UTR variant has functional effects.
The p.Trp690Ser substitution (c.2059G>C), a known XP mutation, destabilizes XPC and causes functional defects, including severely impaired repair of 6-4 photoproducts and lack of XPC binding to undamaged or damaged DNA (5,29). Recruitment of the c.2059G>C mutant was low, reflecting both its effects on protein stability and its relative lack of binding affinity for damaged DNA. The c.1496C>T (rs2228000) variant had no effect on XPC recruitment, as was expected based on the bioinformatics analysis using the different prediction softwares. We found intermediate levels of recruitment for proteins containing the c.905T>C and c.1177C>T variants (Figure 2). These variants are located within the XPA-interaction domain (154-331) and the OGG1-interaction domain of XPC (in the region of codon 334) (30,31) and are located in conserved regions, suggesting that these two rare variants might alter protein–protein interactions (supplementary Figure 3 is available on Carcinogenesis Online).
The reductions in recruitment for c.1177C>T, c.905T>C and c.2059G>C were in the same rank order as the severity of changes, in terms of functional effects, domain preservation, secondary structure sequence homology and physical properties of amino acids, predicted by the software tools Polyphen, SIFT, Pfam and NPS@ (Figure 2 and Table III). Although we confirmed the low damage recognition capability of the c.2059G>C variant (5) in our assay, we do not yet know the underlying mechanism for reduced recruitment of the c.905T>C and c.1177C>T variants. Other plasmid assays, performed in XPC-deficient GM15983 cells, showed that full-length XPC protein was expressed in cells transfected with the variants c.*156A, c.905T>C and c.1177C>T, respectively, although it is not possible to comment on absolute XPC expression levels as all were overexpressed relative to wild-type cells on western blotting (data not shown). Further studies are needed to elucidate the specific role of the rare variants in modulating DNA binding and DNA repair function, as protein recruitment to focal laser damage represents only a limited part of the DNA repair functions of XPC.
Our work on transfected mutant plasmids mimics the homozygous mutant phenotype in humans, so the question remains as to what effect the presence of a rare variant on only one allele might have on XPC expression and function. Ideally, we would have wished to perform quantitative PCR and western blot analysis of the relevant patient samples, to determine to what extent the rare variants affect mRNA and protein expression in the presence of a wild-type allele. Unfortunately, no cell lines were made from these patients’ lymphocytes, and ethical constraints prevent us from reapproaching these patients for a further blood sample to help answer this question. However, Khan et al.. (32), using quantitative reverse transcription–PCR, found reduced XPC mRNA levels (∼59% of normal controls) and reduced XPC protein levels in cells from clinically normal parents of XP-C patients, where the patients’ cells had premature termination codons, nonsense-mediated mRNA decay and absence of full length or truncated XPC protein on western blotting. They proposed that a half dose of the XPC gene product may be insufficient for full XPC function in the maintenance of genomic integrity, thus, giving rise to an increased mutation rate. In support of this, germ line XP heterozygosity has been identified as a risk factor for lung cancer (33), including Arg671Cys in XPC which is predicted to be probably damaging by SIFT and Polyphen. Also, Xpc+/− mice develop skin tumours after daily UVB exposure although at an older age than Xp-−/− mice. The great majority of skin tumours are mutated in one or both Trp53 alleles, suggesting that p53 function may influence the increased cancer risk. In support of this, Xpa−/−/Trp53+/− mice are even more prone to bladder tumour formation than Trp53+/− mice following 2-acetylaminofluorene exposure, whereas Xpa−/− mice do not develop bladder tumours (34).
It is currently uncertain what relative contribution common and rare genetic variants make to cancer predisposition. The ‘common disease-multiple rare variants’ hypothesis proposes that ‘a significant proportion of the inherited susceptibility to cancer may be due to the summation of the effects of a series of low frequency dominantly and independently acting variants of a variety of different genes, each conferring a moderate but readily detectible increase in the relative risk’ (35). Fearnhead et al. (36), in 124 multiple colorectal adenoma patients and 483 controls, found 24.9% of cases but only 11.5% of controls carried potentially pathogenic variants in five candidate genes implicated in colorectal tumorigenesis. It is also possible that rare variants function as compound heterozygotes, with a different rare variant present on each allele. In colorectal cancer, Tenesa et al. (37), found a strong bilallelic effect for two MUTYH rare variants, coding for Tyr165Cys (c.494A>G and rs34612342) and Gly382Asp (c.1145G>A and rs36053993), which included compound heterozygotes (relative risk 117 (95% CI 74–184) compared with the small but significant mono-allelic effect [genotype relative risk 1.27 (95% CI 1.01–1.61)].
In an appraisal of XPD polymorphisms and DNA repair capacity and cancer susceptibility, Clarkson et al. (38) argued against undertaking genotyping studies for variants where their functional effect is yet to be established. Rather, they suggested a combination of several approaches, including use of analytical algorithms, host cell reactivation assays in an isogenic background and estimation of protein expression levels. The study of rare variants also requires a similar approach, and Bodmer et al. (35) propose that candidate genes should be selected which are known to be involved in the biology of the disease, and where severe disruption of function gives rise to a severe version of the disease. These genes then need to be sequenced in patients with the disease to identify rare variants, which are assessed for their potential consequences based on prediction software and on biochemical and physiological studies, and subsequently assessed for their frequency in an appropriate control population. We are largely following such an approach, using XPC as a candidate gene. However, severe disruption of XPC function has not been associated with bladder cancer in XP-C patients, although these patients die at a young age of their skin tumours, and to our knowledge, bladder tumours have not been reported in XPC knockout mice. However, XPC knockout mice develop more bladder tumours than wild-type mice after exposure to 2-acetylaminofluorene, a metabolite of which forms bulky DNA adducts (39). It is thus probable that in bladder cancer rare XPC variants act by influencing environmental or occupational exposures, as mechanistic studies have shown that bulky DNA adducts formed by aromatic amines are repaired by NER, in epidemiological studies these are associated with increased bladder cancer risk, and case–control studies have demonstrated increased bladder cancer risk in the presence of XPC SNPs.
The high cost of mutation scanning techniques and conventional ‘Sanger’ sequencing has limited systematic searches for rare variants (40), but the recent emergence of clonal sequencing technology (41,42) makes such work more cost effective. Now that it is possible to examine large numbers of DNA samples for rare variants, it will be important to use prediction software tools combined with functional assays to determine which variants are worthwhile to study further. In this present study, we have validated two relatively straightforward assays that could be used: firstly, the recruitment of GFP-tagged XPC proteins to 408 nm laser damage, an assay which is appropriate for the evaluation of a range of DNA repair genes, and secondly, the 3′ UTR assay which is generic and so could be applied to any gene.
Supplementary material
Supplementary Tables I–III and Figures 1–3 can be found at http://carcin.oxfordjournals.org/
Funding
Cancer Research UK (C6228/A7625, C15140/A7298, C5255/A12678) to A.E.K.; Yorkshire Cancer Research (L304 to A.E.K.).
Acknowledgments
We thank Jo Brown for tumour tissue collection, Carolyn Hurst for tumour DNA extraction, Alan Monk, Kate Passam, Fabrice Cordelières, Matt Adams and Jacqueline Bond for help with live cell imaging and Thomas Hughes for help with the mRNA assays.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- cDNA
complementary DNA
- CI
confidence interval
- DHPLC
denaturing high-performance liquid chromatography
- mRNA
messenger RNA
- NER
nucleotide excision repair
- OR
odds ratio
- PCR
polymerase chain reaction
- SNP
single nucleotide polymorphism
- UTR
untranslated region
- UV
ultraviolet
- XP
xeroderma pigmentosum
References
- 1.Jemal A, et al. Cancer statistics, 2008. CA Cancer J. Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Friedberg EC. How nucleotide excision repair protects against cancer. Nat. Rev. Cancer. 2001;1:22–33. doi: 10.1038/35094000. [DOI] [PubMed] [Google Scholar]
- 3.van Steeg H, et al. Xeroderma pigmentosum and the role of UV-induced DNA damage in skin cancer. Mol. Med. Today. 1999;5:86–94. doi: 10.1016/s1357-4310(98)01394-x. [DOI] [PubMed] [Google Scholar]
- 4.Masutani C, et al. Purification and cloning of a nucleotide excision repair complex involving the xeroderma pigmentosum group C protein and a human homologue of yeast RAD23. EMBO J. 1994;13:1831–1843. doi: 10.1002/j.1460-2075.1994.tb06452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yasuda G, et al. In vivo destabilization and functional defects of the xeroderma pigmentosum C protein caused by a pathogenic missense mutation. Mol. Cell. Biol. 2007;27:6606–6614. doi: 10.1128/MCB.02166-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sugasawa K, et al. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 7.Kraemer KH, et al. Xeroderma pigmentosum. Cutaneous, ocular, and neurologic abnormalities in 830 published cases. Arch. Dermatol. 1987;123:241–250. doi: 10.1001/archderm.123.2.241. [DOI] [PubMed] [Google Scholar]
- 8.Swift M, et al. Cancer in families with xeroderma pigmentosum. J. Natl Cancer Inst. 1979;62:1415–1421. [PubMed] [Google Scholar]
- 9.Olsen JH, et al. Cancer in patients with ataxia-telangiectasia and in their relatives in the nordic countries. J. Natl Cancer Inst. 2001;93:121–127. doi: 10.1093/jnci/93.2.121. [DOI] [PubMed] [Google Scholar]
- 10.Gruber SB, et al. BLM heterozygosity and the risk of colorectal cancer. Science. 2002;297:2013. doi: 10.1126/science.1074399. [DOI] [PubMed] [Google Scholar]
- 11.Friedberg EC, et al. Defective nucleotide excision repair in xpc mutant mice and its association with cancer predisposition. Mutat. Res. 2000;459:99–108. doi: 10.1016/s0921-8777(99)00068-3. [DOI] [PubMed] [Google Scholar]
- 12.Hollander MC, et al. Deletion of XPC leads to lung tumors in mice and is associated with early events in human lung carcinogenesis. Proc. Natl Acad. Sci. USA. 2005;102:13200–13205. doi: 10.1073/pnas.0503133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melis JP, et al. Mouse models for xeroderma pigmentosum group A and group C show divergent cancer phenotypes. Cancer Res. 2008;68:1347–1353. doi: 10.1158/0008-5472.CAN-07-6067. [DOI] [PubMed] [Google Scholar]
- 14.Kuraoka I, et al. Removal of oxygen free-radical-induced 5',8-purine cyclodeoxynucleosides from DNA by the nucleotide excision-repair pathway in human cells. Proc. Natl Acad. Sci. USA. 2000;97:3832–3837. doi: 10.1073/pnas.070471597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Errico M, et al. New functions of XPC in the protection of human skin cells from oxidative damage. EMBO J. 2006;25:4305–4315. doi: 10.1038/sj.emboj.7601277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimizu Y, et al. Xeroderma pigmentosum group C protein interacts physically and functionally with thymine DNA glycosylase. EMBO J. 2003;22:164–173. doi: 10.1093/emboj/cdg016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sak SC, et al. Comprehensive analysis of 22 XPC polymorphisms and bladder cancer risk. Cancer Epidemiol. Biomarkers Prev. 2006;15:2537–2541. doi: 10.1158/1055-9965.EPI-06-0288. [DOI] [PubMed] [Google Scholar]
- 18.Sak SC, et al. The polyAT, intronic IVS11-6 and Lys939Gln XPC polymorphisms are not associated with transitional cell carcinoma of the bladder. Br. J. Cancer. 2005;92:2262–2265. doi: 10.1038/sj.bjc.6602616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes TA, et al. Cross-talk between pRb/E2F and Wnt/beta-catenin pathways: E2F1 induces axin2 leading to repression of Wnt signalling and to increased cell death. Exp. Cell Res. 2005;303:32–46. doi: 10.1016/j.yexcr.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 20.Godon C, et al. PARP inhibition versus PARP-1 silencing: different outcomes in terms of single-strand break repair and radiation susceptibility. Nucleic Acids Res. 2008;36:4454–4464. doi: 10.1093/nar/gkn403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giglia G, et al. p53 mutations in skin and internal tumors of xeroderma pigmentosum patients belonging to the complementation group C. Cancer Res. 1998;58:4402–4409. [PubMed] [Google Scholar]
- 22.Knowles MA. Molecular pathogenesis of bladder cancer. Int. J. Clin. Oncol. 2008;13:287–297. doi: 10.1007/s10147-008-0812-0. [DOI] [PubMed] [Google Scholar]
- 23.Takebayashi Y, et al. Loss of heterozygosity of nucleotide excision repair factors in sporadic ovarian, colon and lung carcinomas: implication for their roles of carcinogenesis in human solid tumors. Cancer Lett. 2001;174:115–125. doi: 10.1016/s0304-3835(01)00690-5. [DOI] [PubMed] [Google Scholar]
- 24.Okuda Y, et al. Relative levels of the two mammalian Rad23 homologs determine composition and stability of the xeroderma pigmentosum group C protein complex. DNA Repair (Amst.) 2004;3:1285–1295. doi: 10.1016/j.dnarep.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 25.Boffa MB, et al. Effect of single nucleotide polymorphisms on expression of the gene encoding thrombin-activatable fibrinolysis inhibitor: a functional analysis. Blood. 2008;111:183–189. doi: 10.1182/blood-2007-03-078543. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, et al. 3'-UTR polymorphism in the human CYP2A6 gene affects mRNA stability and enzyme expression. Biochem. Biophys. Res. Commun. 2006;340:491–497. doi: 10.1016/j.bbrc.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 27.Kamiyama M, et al. Polymorphisms in the 3' UTR in the neurocalcin delta gene affect mRNA stability, and confer susceptibility to diabetic nephropathy. Hum. Genet. 2007;122:397–407. doi: 10.1007/s00439-007-0414-3. [DOI] [PubMed] [Google Scholar]
- 28.Khan SG, et al. Two essential splice lariat branchpoint sequences in one intron in a xeroderma pigmentosum DNA repair gene: mutations result in reduced XPC mRNA levels that correlate with cancer risk. Hum. Mol. Genet. 2004;13:343–352. doi: 10.1093/hmg/ddh026. [DOI] [PubMed] [Google Scholar]
- 29.Chavanne F, et al. Mutations in the XPC gene in families with xeroderma pigmentosum and consequences at the cell, protein, and transcript levels. Cancer Res. 2000;60:1974–1982. [PubMed] [Google Scholar]
- 30.Bernardes de Jesus BM, et al. Dissection of the molecular defects caused by pathogenic mutations in the DNA repair factor XPC. Mol. Cell. Biol. 2008;28:7225–7235. doi: 10.1128/MCB.00781-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bunick CG, et al. Biochemical and structural domain analysis of xeroderma pigmentosum complementation group C protein. Biochemistry. 2006;45:14965–14979. doi: 10.1021/bi061370o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan SG, et al. Reduced XPC DNA repair gene mRNA levels in clinically normal parents of xeroderma pigmentosum patients. Carcinogenesis. 2006;27:84–94. doi: 10.1093/carcin/bgi204. [DOI] [PubMed] [Google Scholar]
- 33.Matakidou A, et al. Evaluation of xeroderma pigmentosum XPA, XPC, XPD, XPF, XPB, XPG and DDB2 genes in familial early-onset lung cancer predisposition. Int. J. Cancer. 2006;119:964–967. doi: 10.1002/ijc.21931. [DOI] [PubMed] [Google Scholar]
- 34.Hoogervorst EM, et al. p53 heterozygosity results in an increased 2-acetylaminofluorene-induced urinary bladder but not liver tumor response in DNA repair-deficient Xpa mice. Cancer Res. 2004;64:5118–5126. doi: 10.1158/0008-5472.CAN-04-0350. [DOI] [PubMed] [Google Scholar]
- 35.Bodmer W, et al. Common and rare variants in multifactorial susceptibility to common diseases. Nat. Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fearnhead NS, et al. Multiple rare variants in different genes account for multifactorial inherited susceptibility to colorectal adenomas. Proc. Natl Acad. Sci. USA. 2004;101:15992–15997. doi: 10.1073/pnas.0407187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tenesa A, et al. Association of MUTYH and colorectal cancer. Br. J. Cancer. 2006;95:239–242. doi: 10.1038/sj.bjc.6603239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clarkson SG, et al. Polymorphisms in the human XPD (ERCC2) gene, DNA repair capacity and cancer susceptibility: an appraisal. DNA Repair (Amst.) 2005;4:1068–1074. doi: 10.1016/j.dnarep.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 39.Hoogervorst EM, et al. 2-AAF-induced tumor development in nucleotide excision repair-deficient mice is associated with a defect in global genome repair but not with transcription coupled repair. DNA Repair (Amst.) 2005;4:3–9. doi: 10.1016/j.dnarep.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 40.Topol EJ, et al. The resequencing imperative. Nat. Genet. 2007;39:439–440. doi: 10.1038/ng0407-439. [DOI] [PubMed] [Google Scholar]
- 41.Johnson DS, et al. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 42.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.