Abstract
Published genome-wide association studies (GWASs) have identified few variants in the known biological pathways involved in lung cancer etiology. To mine the possibly hidden causal single nucleotide polymorphisms (SNPs), we explored all SNPs in the extrinsic apoptosis pathway from our published GWAS dataset for 1154 lung cancer cases and 1137 cancer-free controls. In an initial association analysis of 611 tagSNPs in 41 apoptosis-related genes, we identified only 10 tagSNPs associated with lung cancer risk with a P value <10−2, including four tagSNPs in DAPK1 and three tagSNPs in TNFSF8. Unlike DAPK1 SNPs, TNFSF8 rs2181033 tagged other four predicted functional but untyped SNPs (rs776576, rs776577, rs31813148 and rs2075533) in the promoter region. Therefore, we further tested binding affinity of these four SNPs by performing the electrophoretic mobility shift assay. We found that only rs2075533T allele modified levels of nuclear proteins bound to DNA, leading to significantly decreased expression of luciferase reporter constructs by 5- to –10-fold in H1299, HeLa and HCT116 cell lines compared with the C allele. We also performed a replication study of the untyped rs2075533 in an independent Texas population but did not confirm the protective effect. We further performed a mini meta-analysis for SNPs of TNFSF8 obtained from other four published lung cancer GWASs with 12 214 cases and 47 721 controls, and we found that only rs3181366 (r2 = 0.69 with the untyped rs2075533) was associated to lung cancer risk (P = 0.008). Our findings suggest a possible role of novel TNFSF8 variants in susceptibility to lung cancer.
Introduction
Although smoking is a major risk factor for lung cancer, numerous studies have suggested that genetic variants in genes involved in various biological pathways have an impact on lung cancer susceptibility (1–4). Recently, genome-wide association studies (GWASs), as a powerful high-throughput approach to identify common and low-penetrance risk alleles involved in the etiology of diseases, are a novel approach to identify possible causal genetic variants for lung cancer (5). Several GWASs have reported some novel loci associated with lung cancer risk, and subsequent studies have supported these new findings (6–9). However, these findings are only the tip of the iceberg in the etiology of lung cancer, and the most significant single nucleotide polymorphisms (SNPs) identified in GWAS explain only a small proportion of lung cancer risk.
Why is so much of the heritability unexplained by initial GWAS findings and where is the missing heritability? Many explanations for these questions have been suggested. Genetic variants that are not well covered by available genotyping chips, such as rare genetic variants and structural variants, have been suggested to account for the missing heritability (10,11). It means that the missing heritability may hide among markers of low significance (11,12). To further mine GWAS data to identify the role of additional novel causal variants in the development of lung cancer, a potential and feasible strategy is to focus on those variants in the known canonical pathways that have been widely suggested to play a critical role in the cancer etiology (13,14).
Apoptosis is programmed cell death that is an essential mechanism of maintaining tissue homeostasis in organisms (15). A deregulation of apoptosis contributes to a wide variety of conditions including cancer (16). Apoptosis in cells occurs through two major molecular signaling pathways: extrinsic and intrinsic pathways, which play different roles in cancer development. The intrinsic pathway can be triggered by cell stresses mainly through the mitochondria in response to DNA damage and plays a key role in mediating nicotine effects on normal epithelial lung cells (17). In contrast, the extrinsic pathway is initiated when ligands, such as FAS ligand and tumor necrosis factor (TNF), bind to membrane receptors, such as FAS (also known as APO-1 or CD95) and the TNF receptor, respectively, which induce the cascade of procaspase activation (18). The extrinsic pathway has been suggested to play a role in the immune response against cancer cells.
Polymorphisms in genes involved in the apoptosis pathway have been extensively studied in the candidate gene or pathway approaches and suggested to contribute to the etiology of lung cancer (19–21). However, most of published candidate-gene studies have investigated only a few genes and a few variants in this pathway. Published data from lung cancer GWAS provide a unique opportunity for us to elucidate the impact of variants in the whole genome on lung cancer risk and to re-evaluate the possible impact of common variants in the extrinsic apoptosis pathway on the development of the disease.
We first selected top significant tagSNPs in the extrinsic apoptosis pathway for a replication study, using an independent but similar study population. We then searched for putative functional SNPs in strong linkage disequilibrium (LD) with these identified tagSNPs by bioinformatics approaches and further validated the most promising functional variants by laboratory approaches, including the electrophoretic mobility shift assay (EMSA) and the luciferase reporter assay. Finally, we reassessed associations of the identified SNPs in TNFSF8 in a mini GWAS meta-analysis using the other published GWAS datasets.
Methods and materials
Study populations
The Texas lung cancer GWAS population has been described previously (6). Briefly, this study included 1154 non-small cell lung cancer patients and 1137 cancer-free controls who were non-Hispanic white ever smokers frequency matched by age (±5 years) and sex. The replication study included an additional 622 lung cancer cases and 632 cancer-free healthy controls that were similarly recruited from MD Anderson Cancer Center and the multispecialty physician practice, respectively. Demographic characteristics of subjects in the two populations are shown in Table I. All cases in the two study populations were newly diagnosed with histology confirmed and untreated non-small cell lung cancer. All subjects signed a written informed consent before providing a 30-ml blood sample and information about environmental exposure history including tobacco use. The research protocol was approved by the MD Anderson Institutional Review Board.
Table I.
Top 10 hits of genetic variants in the extrinsic apoptosis pathway and their associations with lung cancer risk based on data from the Texas lung cancer GWAS
| SNPs | Gene | Minor allele (A1) | Major allele (A2) | MAF | Case A1/A2 | Control A1/A2 | Pa |
| rs10124291 | DAPK1 | A | G | 0.31 | 816/1492 | 699/1575 | 0.0009 |
| rs2181033 | TNFSF8 | G | A | 0.39 | 806/1502 | 898/1376 | 0.0014 |
| rs721936 | DAPK1 | A | G | 0.39 | 793/1515 | 880/1394 | 0.0023 |
| rs3128477 | DAPK1 | A | G | 0.39 | 792/1516 | 878/1394 | 0.0023 |
| rs1322058 | TNFSF8 | A | G | 0.42 | 876/1432 | 956/1318 | 0.0048 |
| rs3181366 | TNFSF8 | A | G | 0.42 | 874/1418 | 953/1305 | 0.0051 |
| rs1905776 | TNFRSF11B | A | G | 0.20 | 387/1917 | 452/1810 | 0.0055 |
| rs7046290 | DAPK1 | G | A | 0.05 | 147/1951 | 105/1997 | 0.0061 |
| rs6557634 | TNFRSF10A | G | A | 0.49 | 1150/1158 | 1039/1229 | 0.0066 |
| rs168259 | TNFSF7 | A | G | 0.12 | 333/1975 | 267/2007 | 0.0070 |
χ2 test for the difference in the distributions of alleles.
Genotyping
For the Texas lung cancer GWAS, genotyping procedures of Illumina HumanHap300 v1.1 BeadChips with the genomic DNA and quality control have been described elsewhere (6). In the replication study, Taqman assays or restriction fragment length polymorphism polymerase chain reaction (PCR) assays (when Taqman probes were not available) were used to validate the most significant SNPs of the apoptosis genes identified in the GWAS discovery dataset.
Gene and SNP selection
In the reanalysis of the Texas lung cancer GWAS data, genes involved in the extrinsic apoptosis pathway were selected based on the following criteria: genes that have been reported to be involved in the extrinsic apoptosis pathway; genes that have been included in the extrinsic apoptosis pathway as described in the cell signaling pathways (http://www.cellsignal.com/pathways/apoptosis-signaling.jsp) and classified as in the extrinsic apoptosis pathway (22); and genes have been covered by the Illumina Human Hap300 v1.1 BeadChips (Illumina, San Diego, CA). This chip contains 317 503-tagging SNPs derived from the International HapMap project phase I data. There were a total of 611 tagSNPs in 41 genes in the extrinsic apoptosis pathway available from the GWAS database. The assignation of a tagSNPs to a gene was defined by Illumina annotation file ‘HumanHap317K_annotation.txt’, which annotates all tagSNPs to their closest gene regardless how far a tagSNP is away from the gene. Three tagging SNPs (rs10124291, rs3128477 and rs7046290) in DAPK1 and one tagging SNP rs2181033 in TNFSF8 were also genotyped in the replication study.
Functional SNP searching
To search for putative functional SNPs in high LD with the tagging SNPs, we used SNPinfo (23) (http://snpinfo.niehs.nih.gov/snpfunc.htm) to identify any putative functional SNPs with r2 of 0.7 or higher, based on the HapMap phase II data in CEU (Utah residents with Northern and Western European ancestry from the CEPH collection) population. For any putative functional findings, we further examined their function by using the relevant bioinformatics software or databases. For example, for SNPs that were predicted to affect transcription factor-binding sites in the promoter region, we further evaluated their effects on the transcription factor binding in TFSEARCH (24) (http://molsun1.cbrc.aist.go.jp/research/db/TFSEARCH.html). For SNPs that were predicted to affect a microRNA-binding site in the 3′ untranslated region, we then evaluated their function in microRNA databases miRanda (http://www.microrna.org) and miRBase (http://www.mirbase.org). To obtain more confident prediction, only those SNPs that were identified to be functional in two or more bioinformatics softwares or databases were further validated by the laboratory functional assays.
Electrophoretic mobility shift assay
Lung cancer cell line H1299 nuclear extracts were prepared according to the method of Andrews and Faller (25). Complementary single-stranded oligonucleotides for four SNPs of TNFSF8 (rs726656, 5′-GATAGAGACACGGAGCACAGTTGAGAAGAG-3′ for the C allele and 5′-GATAGAGACACGGAGTACAGTTGAGAAGAG-3′ for the T allele; rs726657, 5′-TTGAAGGTCAACCCTACATATATGACCAGC-3′ for the A allele and 5′-TTGAAGGTCAACCCTGCATATATGACCAGC-3′ for the G allele; rs3181348, 5′-GATTAACATTTTAAACGGTATTTTGAAATG-3′ for the C allele and 5′-GATTAACATTTTAAATGGTATTTTGAAATG-3′ for the T allele; rs2075533, 5′-TCAATTATAGTAGTCACATACACACACAACACAT-3′ for the C allele and 5′-TCAATTATAGTAGTCATATACACACACAACACAT-3′ for the T allele) were biotin labeled using the 3′-end biotin labeling kit (Thermo Scientific, Rockford, IL) and reannealed before performing the DNA-binding assays using the LightShift Chemiluminescent EMSA kit (Thermo Scientific, Rockford, IL). The binding of DNA and protein was performed by incubation of 10 μg of nuclear extracts with 3′ biotin-labeled double strand oligonucleotides at room temperature for 30 min using the LightShift Chemiluminescent EMSA kit (Thermo Scientific). The DNA–protein complexes were separated on 6% polyacrylamide gel, and the products were detected by stabilized streptavidin–horseradish peroxidase conjugate (Thermo Scientific). The competition assays were performed with 50-fold excess of unlabeled wild-type and mutant oligonucleotides,respectively.
Construction of luciferase reporter plasmids
Since SNP rs2075533C>T was suggested to be the most effective variant in the promoter of TNFSF8 in EMSA, we further tested its effect on the promoter activity of the gene by the luciferase assay. PCR fragments containing either C or T allele of SNP rs2075533 were amplified from genomic DNA isolated from homozygous C or homozygous T carriers using the following primers: forward primer 5′-AAGGTACCGGAGGTGGAAGTGGAATGAA-3′, reverse primer 5′-AAGCTAGCTGCCTGGTGGAGAAACTCTT-3′. PCRs were carried out as follows: the initial denaturation at 95°C for 5 min was followed by 35 cycles of denaturing at 95°C for 30 s, annealing at 59°C for 45 s and extension at 72°C for 1 min. The PCR products were then cloned into the pGL3-Basic vector (Promega, Madison, WI) between KpnI and NheI sites. The sequences of the cloned PCR fragments containing the expected C or T allele of rs2075533 were verified by direct sequencing.
Transient transfection and luciferase assays
Cancer cell lines H1299 (non-small cell lung cancer), HeLa (cervical cancer) and HCT116 (colon cancer) were placed on 24-well plates at 1.0 × 105 cells per well with ×1 RPMI 1640 or Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum and allowed to grow for 1 day prior to transfection (50–70% confluence). Transfection experiments were performed using FuGENE HD (Invitrogen, Carlsbad, CA). For each well, 100 μl transfection mixture was made by mixing 95 μl serum-free medium, 6 μl FuGENE HD, 1 μg of pGL-3 constructs (containing C or T allele) and 20 ng of renilla vector (pRLTK, Promega). The resulting mixtures were incubated for 5 min at room temperature prior to their addition to cells. Each transfection was performed in triplicates. After 48 h, cell lysates were made using the passive lysis buffer provided in the Dual Luciferase Kit (Promega), and firefly luciferase and Renilla luciferase activities were measured in the reporter microplate luminometer (Turner Designs, Sunnyvale, CA) using 10 μl of lysate, 100 μl of the luciferase assay reagent II (Promega) and 100 μl stop and glo reagent (Promega).
Statistical analysis
Differences in the distributions of demographic variables and the known risk factors between lung cancer patients and controls were assessed by the χ2 test. The associations between genotypes of each tagSNP and lung cancer risk were primarily evaluated using the allelic test in PLINK1.07 (26). The Hardy–Weinberg equilibrium for each tagSNP was tested in controls using the χ2 test with one degree of freedom. LD patterns among tagSNPs were estimated by Haploview (27). The multivariable logistic regression method was used to assess the associations between genotypes of tagSNPs and lung cancer risk in codominant, additive, dominant and recessive models. Odds ratios and 95% confidence intervals were estimated with adjustment for the known risk factors for lung cancer, such as age, sex, smoking status and pack-years. For the luciferase assay, the relative light units were calculated using the firefly luciferse activity divided by the Renilla luciferase activity for each sample, setting the reporter gene activity of construct with the C allele as the baseline. Significant differences between groups were determined by Student’s t test. For the mini meta-analysis, we evaluated the top five significant tagSNPs (rs2181033, rs1322058, rs3181366, rs3181348 and rs2295800) of TNFSF8 available in the Texas lung cancer GWAS as well as from other lung cancer GWASs published by deCODE (28), National Cancer Institute (29), International Agency for Research on Cancer (IARC) (8) and UK (7). We assessed the between-study heterogeneity by using the Cochran’s Q-test with P < 0.05 as the significance level. We performed initial analyses with a fixed effect model and confirmatory analyses with a random effect model, if there was significant heterogeneity between studies. Unless specified otherwise, all other statistical analyses were performed using SAS 9.1.3 (SAS Institute Inc, Cary, NC) and STATA 10.0 (for the meta-analysis), and all statistical tests were two sided, with a P < 0.05 set as the level of statistical significance. For controlling the effect of multiple tests, P values were adjusted by Bonfferoni correction.
Results
The characteristics of the Texas GWAS discovery dataset and the replication dataset are presented in supplementary Table S1, available at Carcinogenesis Online. In general, the two study populations were similar in terms of the distributions of the subgroups by age, sex, smoking status, pack-years smoked except that the fraction of adenocarcinoma was higher in the GWAS dataset.
Association between genetic variation in the extrinsic apoptosis pathway and lung cancer risk
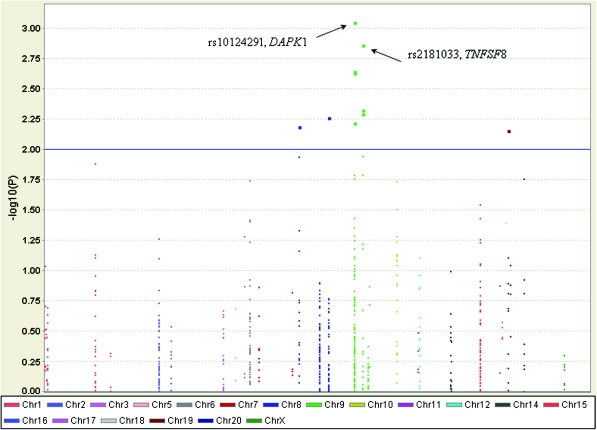
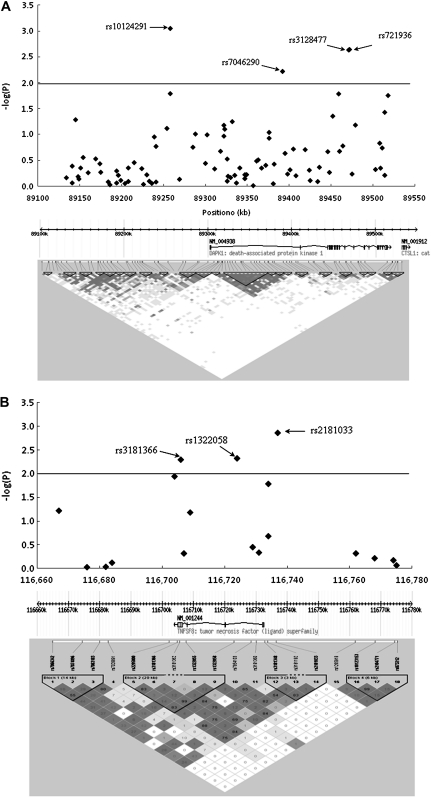
The associations between alleles and genotypes of the selected variants in the extrinsic apoptosis pathway and lung cancer risk are shown in Figure 1 (the Manhattan plot) and supplementary Table S2, available at Carcinogenesis Online. Of these, 10 SNPs in five genes (i.e. PAPK1, TNFSF78, TNFSF8, TNFSF10A and TNFSF11B) were associated with risk of lung cancer at P < 0.01 level in allelic tests (Table I), in which the most significant SNPs were located in two genes (i.e. DAPK1 and TNFSF8) on chromosome 9. The top significant hit was rs10124291, which is in the DAPK1 (MIM# 600831) gene region, and the second most significant SNP was rs2181033, which is in the 5′ flanking region of TNFSF8 (MIM# 603875). Further evaluation of associations between genetic variants in these two genes and lung cancer risk is shown in Figure 2A and 2B. There were 92 tagSNPs around the DAPK1 gene that covered the region between 89175300 and 89347200 bp on chromosome 9, of which only four tagSNPs (rs10124291, rs7046290, rs3128477 and rs721936) had a P value <0.01. In contrast, fewer variants (18 tagSNPs) were around TNFSF8, covering the region between 116 660 000 and 116 780 000 bp on chromosome 9, of which only three SNPs (rs2181033, rs1322058 and rs3181366) had a P value <0.01. Further LD analyses among these selected SNPs suggested that two SNPs (rs3128477 and rs721936 in DAPK1) and all three SNPs (rs2181033, rs1322058 and rs3181366) in TNFSF8 were in high LD (r2 > 0.8; supplementary Figure S1 is available at Carcinogenesis Online). As a result, we genotyped three tagSNPs (rs10124291, rs7046290 and rs3128477) in DAPK1 and one tagSNP rs2181033 in TNFSF8 in the replication study.
Fig. 1.
Association between genetic variants in the extrinsic apoptosis pathway and lung cancer risk based on the data from the Texas lung cancer GWAS.
Fig. 2.
Association between genetic variants of DAPK1 and TNFSF8 and lung cancer risk based on the data from the Texas lung cancer GWAS. (A) A total of 92 SNPs were assigned to DAPK1, which covered the gene region between 89 175 300 and 89 347 200 bp on chromosome 9. Only four SNPs had a P value <0.01 and two SNPs rs3128477 and rs721936 were in high LD (r2 = 0.99). LD patterns among these SNPs were inferred in the control subjects by Haploview. (B) A total of 18 SNPs were assigned to TNFSF8, which covered the gene region between 116 660 000 and 116 780 000 bp on chromosome 9. Only three SNPs had a P value <0.01 and three SNPs were in high LD to each other (r2 > 0.8). LD patterns among these SNPs were inferred in the control subjects by Haploview.
Replication study
The genotype distribution of four tagSNPs in GWAS, replication and pooled datasets are summarized in Table II. The genotype distribution for most of the tagSNPs in controls of GWAS, replication and pooled datasets were consistent with Hardy–Weinberg equilibrium, except that the genotypes of rs2181033 had a slight deviation from Hardy–Weinberg equilibrium in the replication dataset (P = 0.023). Association analysis showed that there were no overall significant associations between the four tagSNPs with lung cancer risk in the replication population. The replication dataset also suggested that the rs2181033 seemed to be a protective variant for lung cancer, which was similar to the finding in the GWAS data. Overall, there was an association between rs2181033 and lung cancer risk in the pooled dataset, which did not change even after Bonfferoni correction (P for homozygote variant group was 0.008).
Table II.
Association of the top significant SNPs of DAPK1 and TNFSF8 with risk of lung cancer in the Texas Lung cancer GWAS and replication study
| SNPs | GWAS set |
Replication set |
Pooled |
||||||
| Ca/Con | OR (95% CI) | Pa | Ca/Con | OR (95% CI) | Pa | Ca/Con | OR (95% CI) | Pa | |
| DAPK1 | |||||||||
| rs10124291 | |||||||||
| GG | 492/555 | 1.00 | 266/279 | 1.00 | 758/834 | ||||
| AG | 508/465 | 1.22 (1.02–1.45) | 0.031 | 288/282 | 1.03 (0.81–1.30) | 0.830 | 796/747 | 1.15 (1.00–1.33) | 0.051 |
| AA | 154/117 | 1.45 (1.11–1.91) | 0.007 | 68/71 | 0.96 (0.65–1.40) | 0.814 | 222/188 | 1.27 (1.02–1.58) | 0.036 |
| Additive | –– | 1.21 (1.07–1.37) | 0.003 | –– | 0.99 (0.84–1.18) | 0.942 | –– | 1.13 (1.03–1.25) | 0.013 |
| Dominant | –– | 1.26 (1.07–1.49) | 0.006 | –– | 1.01 (0.81–1.27) | 0.916 | –– | 1.17 (1.03–1.34) | 0.018 |
| Recessive | –– | 1.32 (1.02–1.71) | 0.034 | –– | 0.94 (0.66–1.35) | 0.747 | –– | 1.18 (0.96–1.46) | 0.118 |
| rs7046290 | |||||||||
| AA | 910/950 | 1.00 | 544/555 | 1.00 | 1454/1505 | 1.00 | |||
| AG | 131/97 | 1.36 (1.03–1.80) | 0.029 | 72/73 | 1.05 (0.73–1.50) | 0.780 | 203/170 | 1.23 (0.99–1.53) | 0.062 |
| GG | 8/4 | 2.25 (0.67–7.56) | 0.190 | 4/1 | 3.42 (0.38–30.9) | 0.274 | 12/5 | 2.51 (0.88–7.18) | 0.087 |
| Additive | –– | 1.42 (1.10–1.84) | 0.007 | –– | 1.12 (0.80–1.56) | 0.512 | –– | 1.30 (1.06–1.59) | 0.012 |
| Dominant | –– | 1.44 (1.09–1.89) | 0.010 | –– | 1.09 (0.77–1.54) | 0.643 | –– | 1.29 (1.04–1.59) | 0.022 |
| Recessive | –– | 2.26 (0.67–7.59) | 0.188 | –– | 3.39 (0.38–30.6) | 0.277 | –– | 2.51 (0.88–7.18) | 0.087 |
| rs3128477 | |||||||||
| GG | 488/427 | 1.00 | 244/261 | 1.00 | 732/688 | 1.00 | |||
| AG | 540/540 | 0.88 (0.74–1.06) | 0.176 | 298/279 | 1.15 (0.91–1.47) | 0.251 | 838/819 | 0.97 (0.84–1.12) | 0.659 |
| AA | 126/169 | 0.66 (0.50–0.86) | 0.002 | 80/89 | 0.97 (0.68–1.38) | 0.852 | 206/258 | 0.76 (0.61–0.94) | 0.010 |
| Additive | –– | 0.83 (0.73–0.94) | 0.003 | –– | 1.02 (0.86–1.20) | 0.834 | –– | 0.89 (0.81–0.99) | 0.023 |
| Dominant | –– | 0.83 (0.70–0.98) | 0.028 | –– | 1.09 (0.87–1.38) | 0.441 | –– | 0.91 (0.80–1.05) | 0.184 |
| Recessive | –– | 0.70 (0.55–0.90) | 0.005 | –– | 0.89 (0.64–1.24) | 0.493 | –– | 0.77 (0.63–0.94) | 0.009 |
| TNFSF8 | |||||||||
| rs2181033 | |||||||||
| AA | 491/431 | 1.00 | 213/203 | 1.00 | 704/634 | 1.00 | |||
| AG | 520/514 | 0.89 (0.75–1.07) | 0.216 | 304/332 | 0.86 (0.67–1.10) | 0.237 | 824/846 | 0.88 (0.76–1.01) | 0.072 |
| GG | 143/192 | 0.65 (0.51–0.84) | 0.001 | 96/93 | 0.98 (0.69–1.38) | 0.904 | 239/285 | 0.76 (0.62–0.93) | 0.008 |
| Additive | –– | 0.83 (0.73–0.93) | 0.002 | –– | 0.98 (0.83–1.15) | 0.763 | –– | 0.88 (0.79–0.96) | 0.007 |
| Dominant | –– | 0.83 (0.70–0.98) | 0.027 | –– | 0.91 (0.71–1.15) | 0.424 | –– | 0.85 (0.74–0.98) | 0.022 |
| Recessive | –– | 0.69 (0.55–0.88) | 0.002 | –– | 1.08 (0.79–1.45) | 0.628 | –– | 0.82 (0.68–0.99) | 0.036 |
CI, confidence interval; OR, odds ratio.
Adjusted for age, sex, smoking status and pack-years.
Searching for functional SNPs
To search all possible functional SNPs in strong LD with the significant tagSNPs, SNPs with r2 of 0.7 or higher based on the HapMap phase II CEU population data were evaluated in SNPinfo (23). No putative functional SNP was found to be in high LD with the three top tagSNPs in DAPK1 (supplementary Table S2, available at Carcinogenesis Online). However, four SNPs in the 5′ flanking region and five SNPs in the 3′ untranslated region of TNFSF8 were predicted to be functional SNPs and were in high LD with rs2181033 (Table III). For the four SNPs (rs776576, rs776577, rs3181348 and rs2075533) that were predicted to affect putative transcription factor-binding sites in the promoter region of TNFSF8, we further evaluated their functions by TFSEARCH (24). All of four SNPs were predicted to modify the binding efficiency of possible transcription factors. For five SNPs that were at putative microRNA-binding sites in the 3′ untranslated region, we further examined their function in microRNA databases, miRanda (http://www.microrna.org/) and miRBase (http://www.mirbase.org/). Only one SNP rs3181370 G>A was identified to be located at the microRNA hsa-miR-549-binding site in two databases, but further examination suggested that this SNP did not strikingly change the binding efficiency of hsa-miR-549 (data not shown). Taken together, we selected four SNPs in the promoter region of TNFSF8 for functional assay.
Table III.
Putative functional SNPs in the TNFSF8 region that are in high LD with rs2181033, based on the genotype information in the CEU population in the HapMap
| SNP | Gene region | LD SNP | r2 | Allele change | Allele | Allele frequency | Putative function changea |
| rs927375 | 5′ flanking | rs2181033 | 0.764 | A/G | A | 0.417 | NA |
| rs2181033 | 5′ flanking | rs2181033 | 1.000 | A/G | A | 0.656 | NA |
| rs1322060 | 5′ flanking | rs2181033 | 0.857 | C/G | C | 0.621 | NA |
| rs1322059 | 5′ flanking | rs2181033 | 0.924 | G/T | C | 0.633 | NA |
| rs726656 | 5′ flanking, −3678 bp | rs2181033 | 0.924 | A/C | A | 0.633 | TFBS |
| rs726657 | 5′ flanking, −3566 bp | rs2181033 | 0.809 | A/G | C | 0.607 | TFBS |
| rs3181348 | 5′ flanking, −1414 bp | rs2181033 | 0.794 | A/C | A | 0.401 | TFBS |
| rs2075533 | 5′ flanking, −861 bp | rs2181033 | 0.795 | T/C | A | 0.397 | TFBS |
| rs1006027 | Intron | rs2181033 | 0.814 | G/A | T | 0.617 | NA |
| rs1006026 | Intron | rs2181033 | 0.763 | G/A | G | 0.405 | NA |
| rs1006025 | Intron | rs2181033 | 0.814 | T/C | C | 0.617 | NA |
| rs7036962 | Intron | rs2181033 | 0.814 | G/A | G | 0.617 | NA |
| rs10982455 | Intron | rs2181033 | 0.869 | G/A | C | 0.634 | NA |
| rs4979472 | Intron | rs2181033 | 0.751 | G/A | G | 0.411 | NA |
| rs1322058 | Intron | rs2181033 | 0.869 | G/A | C | 0.634 | NA |
| rs10982451 | Intron | rs2181033 | 0.814 | T/C | C | 0.617 | NA |
| rs7872878 | Intron | rs2181033 | 0.772 | G/C | C | 0.434 | NA |
| rs10982450 | Intron | rs2181033 | 0.812 | T/G | G | 0.617 | NA |
| rs3789879 | Intron | rs2181033 | 0.751 | T/C | C | 0.411 | NA |
| rs4978611 | Intron | rs2181033 | 0.751 | C/T | C | 0.411 | NA |
| rs927373 | Intron | rs2181033 | 0.869 | A/G | G | 0.634 | NA |
| rs2208640 | Intron | rs2181033 | 0.814 | A/G | A | 0.617 | NA |
| rs12337739 | Intron | rs2181033 | 0.751 | C/G | T | 0.411 | NA |
| rs17292087 | Intron | rs2181033 | 0.829 | A/G | C | 0.643 | NA |
| rs3181195 | Intron | rs2181033 | 0.849 | A/G | C | 0.638 | NA |
| rs3181363 | Intron | rs2181033 | 0.774 | C/T | C | 0.625 | NA |
| rs3181364 | Intron | rs2181033 | 0.769 | A/T | A | 0.627 | NA |
| rs3181366 | Intron | rs2181033 | 0.774 | G/A | G | 0.625 | NA |
| rs3181367 | Intron | rs2181033 | 0.730 | A/C | A | 0.406 | NA |
| rs3181369 | 3′ UTR | rs2181033 | 0.832 | C/T | G | 0.634 | microRNA-binding site |
| rs3181370 | 3′ UTR | rs2181033 | 0.839 | G/T | T | 0.638 | microRNA-binding site |
| rs3181372 | 3′ UTR | rs2181033 | 0.730 | C/T | G | 0.406 | microRNA-binding site |
| rs1126711 | 3′ UTR | rs2181033 | 0.742 | C/T | G | 0.401 | microRNA-binding site |
| rs3181374 | 3′ UTR | rs2181033 | 0.744 | A/G | G | 0.402 | microRNA-binding site |
| rs2295800 | 3′ flanking | rs2181033 | 0.723 | C/T | C | 0.397 | NA |
| rs3181200 | 3′ flanking | rs2181033 | 0.744 | T/C | T | 0.402 | NA |
| rs3181201 | 3′ flanking | rs2181033 | 0.744 | G/A | C | 0.402 | NA |
| rs7025577 | 3′ flanking | rs2181033 | 0.725 | G/A | A | 0.405 | NA |
| rs1322067 | 3′ flanking | rs2181033 | 0.730 | G/A | A | 0.594 | NA |
| rs10982445 | 3′ flanking | rs2181033 | 0.709 | C/G | C | 0.402 | NA |
| rs1407309 | 3′ flanking | rs2181033 | 0.723 | A/C | A | 0.397 | NA |
NA, not applicable; TFBS, transcription factor-binding site; UTR, untranslated region.
Function assay for SNP in the promoter region of TNFSF8
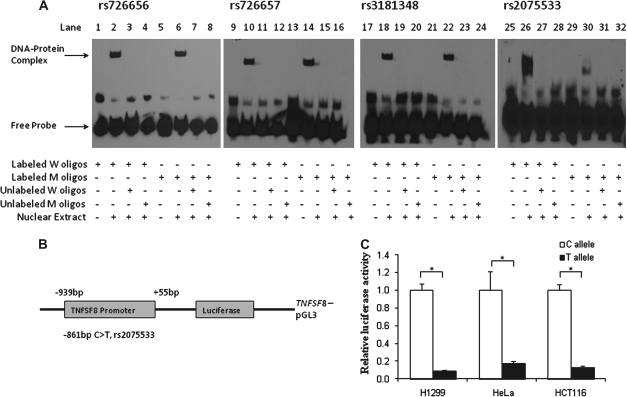
First, we examined whether the four SNPs (rs776576, rs776577, rs3181348 and rs2075533) could change the binding patterns of transcriptional factors by EMSA. As shown in Figure 3A, nuclear proteins prepared from H1299 cells were able to bind to both the oligo probes containing wild-type and variant alleles of these SNPs (Figure 3A, lanes 2 and 6, 10 and 14, 18 and 22 and 26 and 30). Competition assays further showed that the addition of unlabeled probes with the wild-type or variant allele to the reaction mixture completely eliminated these DNA–protein complexes, indicating that the binding was sequence specific (Figure 3A, lanes 3, 4 and lanes 7, 8 for rs776576; lanes 11, 12 and 15, 16 for rs776577; lanes 19, 20 and 23, 24 for rs3181348; lanes 27, 28 and 31, 32 for rs2075533). Among these SNPs, however, only rs2075533 showed an allele-specific difference in the DNA–protein-binding pattern (Figure 3A, lane 26 versus lane 30).
Fig. 3.
Function assays for SNPs in the promoter of TNFSF8. (A) EMSA with biotin-labeled oligo contained the wild- type (W) allele, variant (M) allele for four putative functional SNPs and nuclear extracts from H1299 cells. (B) Schematic of the position of rs2075533 SNP relative to the transcription start site. (C) Luciferase expression of constructs in H1299, HeLa and HCT116 cell lines. Values represent fold change of luciferase activity relative to the wild-type constructs as one. Each bar represents the mean of triplicate transfected plates plus standard deviation. *P < 0.001, compared with the wild-type construct.
To test the effect of rs2075533 on the promoter activity of the TNFSF8 gene, we cloned ∼1.0 kb of the promoter sequence from individuals homozygous for the C or T allele into the pGL3 vector (Figure 3B). The promoter activities were determined in H1299, HeLa and HCT116 cell lines. As shown in Figure 3C, the transcriptional activities of the TNFSF8 promoter fragments with the T allele of this SNP significantly decreased expression levels of the luciferase reporter constructs by 5- to 10-folds, compared with the C allele (P < 0.001). Since the T allele of SNP rs2075533 putatively disrupts the transcript factor activator protein 1 binding site at the promoter region of TNFSF8, the decreased promoter activity of TNFSF8 is likely attributable to the disruption of an the activator protein 1 binding site by the SNP.
Associations between SNPs in TNFSF8 and lung cancer risk
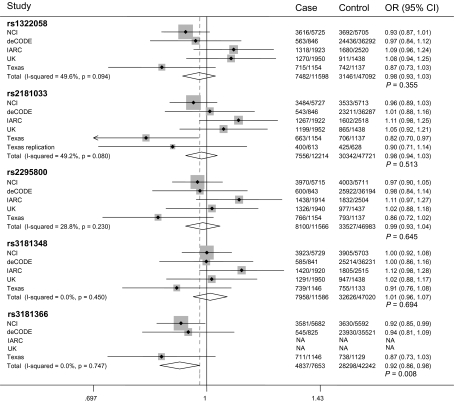
We further assessed the association between rs2075533 and lung cancer risk in the Texas replication population. Although analyses in all models suggested that the T allele of rs207553 may be associated with a decrease in lung cancer risk, the result did not reach the significance level (supplementary Table S4 is available at Carcinogenesis Online). We further selected the five top significant tagSNPs (rs2181033, rs1322058, rs3181366, rs2295800 and rs3181348) of TNFSF8 identified in the Texas Lung cancer GWAS and assessed their associations with lung cancer risk in a mini GWAS meta-analysis using data from deCODE, National Cancer Institute, UK and IARC lung cancer GWASs. The characteristics of National Cancer Institute, deCODE, UK and IARC lung cancer GWAS are shown in supplementary Table S5, available at Carcinogenesis Online. There were a total of 12 214 cases and 47 721 controls in the combined dataset. The Q-test showed that there was no significant heterogeneity between studies (all Q-test P values were >0.05) based on a dominant genetic model. When the fixed effect model was used, the most significant SNP was rs3181366 (P = 0.008), which is in high LD with rs2181033 (r2 = 0.846 in CEU) and rs2075533 (r2 = 0.694 in CEU) (supplementary Figure S1 is available at Carcinogenesis Online). The results for rs3181366 did not include data from the IARC and UK GWASs because genotypes of this SNP were not available in these two datasets. P values for other tagSNPs were >0.05 in the combined populations of the four GWASs (Figure 4). Conversely, the heterogeneity was observed among studies (the Q-tests P value was <0.05 or close to 0.05) based on a recessive model, and all tagSNPs were not associated with lung cancer risk by the random effects model (Data not shown). We also tested the associations between TNFSF8 SNPs in current smokers and ever smokers in the Texas lung cancer GWAS dataset and the Texas replication population (supplementary Table S6 is available at Carcinogenesis Online), and no difference was observed between two subgroups. Further analyses by different histology subgroups showed that the associations of SNPs in TNFSF8 with lung cancer risk seemed to be stronger in adenocarcinoma than that in other two histology groups (supplementary Table S7 is available at Carcinogenesis Online), which was not observed in the Texas replication population (supplementary Tables S6 and S7 are available at Carcinogenesis Online.). We were not able to validate this finding in other four lung cancer GWASs for lack of the needed information.
Fig. 4.
Meta-analysis for the five top significant SNPs in TNFSF8 based on fixed effect, dominant model using genotypes data from deCODE, National Cancer Institute, IARC, Texas lung cancer GWAS and Texas replication study.
Discussion
In this study, we analyzed the associations between genetic variants in the extrinsic apoptosis pathway and lung cancer risk, using the existing genotyping data from the Texas lung cancer GWAS. We found that SNP rs2181033 in the TNFSF8 promoter region was associated with lung cancer risk, and the replication study also indicated its potential protective role in the development of lung cancer. By bioinformatics approaches, we found that the untyped SNP rs2075533 located in the promoter region of TNFSF8 was in high LD with SNP rs2181033, which may be the most informative functional variant as evidenced by the luciferase reporter and EMSA assays. Further mini meta-analysis using genotyping data obtained from other four published lung cancer GWASs with 7653 cases and 42 242 controls identified a significant TNFSF8 SNP rs3181366 that is in LD with the untyped rs2075533.
TNFSF8, also known as CD30L or CD153, is the only known ligand for CD30 and a member of the TNF superfamily. It has a highly similar protein structure to that of TNFα, TNFβ, CD40 ligand and FAS ligand. In non-pathologic conditions, TNFSF8 is mainly expressed in T cells, B cells, neutrophils, mast cells, monocytes and macrophages. TNFSF8 has been also found to be expressed and upregulated in some tumor cells such as mast cells and basal cell carcinoma (30). TNFSF8 binds exclusively to CD30 and activates CD30 receptor by the member of the tumor necrosis factor receptor-associated factor family (31). Activated CD30 then engages the MAP kinase in the nuclear factor-kappaB pathway and involves cell differentiation, apoptosis and immune response (32). This interaction between CD30 and TNFSF8 suggests that they may have different roles. For example, thymocytes of transgenic mice overexpressing CD30 had normal survival and responses to apoptotic stimuli in the absence of CD30 ligation, but when TNFSF8 was presented, overexpression of CD30 resulted in an increase in thymocyte apoptosis (33).It has been suggested that CD30 function depends upon the availability of TNFSF8. Although ligand-independent CD30 expression was found in several cell lines (34), TNFSF8 has been still viewed as a main regulator for the function of CD30 (35). Until now, there are very few studies that have investigated CD30/TNFSF8 and risk of lung cancer. However, recent evidence indicated that CD30-deficient mice delayed recruitment of lymphocytes into the lungs and that CD30/TNFSF8 interactions were involved in lung immune-mediated inflammation (36), whereas lung inflammation was significantly diminished in CD30-deficient mice (37). Such evidence suggests that CD30/TNFSF8 may play a role in the development of lung cancer by inducing apoptosis in immune cells in lung tissue. Our study showed that the functional SNP rs2075533, which led to low expression of TNFSF8, was associated with decreased risk of lung cancer.
TNFSF8 is located at 9q33 extending 27.65 kb, including four exons and three introns (38). There are 199 SNPs that have been reported in the gene region (dbSNP database), of which only one SNP in intron 3 of TNFSF8 was recently reported to be associated with bone diseases in myeloma, but it was still short of functional evidence for such an association (39). Our study suggests that SNP rs2075533 located in the promoter region of TNFSF8 had a striking effect on the promoter activity, and this SNP was in strong LD with SNP rs2181033 that was associated with lung cancer risk as identified in the Texas lung cancer GWAS.
Taken all data together, the present study further indicates a novel role of TNFSF8 in the development of lung cancer. However, there were some limitations in our study. First, we did not observe a significant association in the replication population, although the association observed was in the right direction. Second, we observed only a weak association in the pooled GWAS populations. Ideally, replication of GWAS findings is reassuring. However, whether the replication is sufficient or necessary as a gold standard for true causal variants in association studies is still questionable (40). In fact, positive findings in a GWAS have been difficult to replicate in other GWASs or follow-up studies due to the modest effects of genetic variants on disease risk (41). The meta-analysis in our study also suggests that it is difficult, if not impossible, to validate the positive finding in the original Texas GWAS, when study populations were different. For example, although all subjects in IARC GWAS were Caucasians, they were Central Europeans who may have a different exposure pattern than US participants [most of the cases had occupational exposures (42)]. As an alternative, the supportive evidence from functional assays might be as or more relevant than replication for finding a true causal variant (43). Second, although we had evidence that rs2075533 probably contributed to the observed association, our study did not confirm whether there were any other functional variants in strong LD with the causal ones or the combined risk alleles or genotypes on lung cancer risk, possibly due to small sample size of our replication study. Third, SNP coverage of Illumina HumanHap300 v1.1 BeadChips is relatively low for tagSNPs that were selected based on the HapMap phase I data. So, the potential causal variants around this region may be missed in our study. A higher density array or sequencing data are needed to search for possible causal variants in this region.
In summary, to search potential hidden causal genetic variants in the Texas GWAS study, we initially identified the significant rs2181033 in the discovery dataset. By additional bioinformatics approaches and functional assays, we identified the SNP rs2075533, located in the promoter region of TNFSF8, which had a striking effect on the promoter activity of TNFSF8. Rs2075533 in turn is in LD with rs3181366, a significant SNP identified in the mini meta-analysis of published lung cancer GWASs. Our findings indicated a possible novel role of TNFSF8 in the etiology of lung cancer. More functional studies are needed to explore possible mechanisms of how polymorphisms in TNFSF8 may contribute to lung cancer risk. Our study also supports the notion that the missing heritability may hide in the low significant hits in GWAS.
Supplementary material
Supplementary Tables S1–S7 and Figure 1 can be found at http://carcin.oxfordjournals.org/
Funding
This study was supported in part by National Institutes of Health grants (R01ES011740 and R01CA131274 to Q.W., R01CA055769 and R01CA127219 to M.R.S. and R01CA121197 to C.I.S.) and Cancer Center Core Grant (P30 CA016672 to MD Anderson Cancer Center).
Acknowledgments
We thank Min Zhao, Jianzhong He, Kejing Xu and Hongxia Ma for their laboratory assistance, Hongping Yu, Yujing Huang, Ming Yin and Hongliang Liu for their help in data analysis and Dakai Zhu for his technical support.The study’s contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- EMSA
electrophoretic mobility shift assay
- GWAS
genome-wide association study
- IARC
International Agency for Research on Cancer
- LD
linkage disequilibrium
- PCR
polymerase chain reaction
- TNF
tumor necrosis factor
References
- 1.Spitz MR, et al. Genetic susceptibility to lung cancer: the role of DNA damage and repair. Cancer Epidemiol. Biomarkers Prev. 2003;12:689–698. [PubMed] [Google Scholar]
- 2.Neumann AS, et al. Nucleotide excision repair as a marker for susceptibility to tobacco-related cancers: a review of molecular epidemiological studies. Mol Carcinog. 2005;42:65–92. doi: 10.1002/mc.20069. [DOI] [PubMed] [Google Scholar]
- 3.Wang L, et al. Association of genetic variants of O6-methylguanine-DNA methyltransferase with risk of lung cancer in non-Hispanic Whites. Cancer Epidemiol. Biomarkers Prev. 2006;15:2364–2369. doi: 10.1158/1055-9965.EPI-06-0437. [DOI] [PubMed] [Google Scholar]
- 4.Wang L, et al. Polymorphisms of cytosolic serine hydroxymethyltransferase and risk of lung cancer: a case-control analysis. Lung Cancer. 2007;57:143–151. doi: 10.1016/j.lungcan.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Easton DF, et al. Genome-wide association studies in cancer. Hum. Mol. Genet. 2008;17:R109–R115. doi: 10.1093/hmg/ddn287. [DOI] [PubMed] [Google Scholar]
- 6.Amos CI, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat. Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat. Genet. 2008;40:1407–1409. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKay JD. Lung cancer susceptibility locus at 5p15.33. Nat. Genet. 2008;40:1404–1406. doi: 10.1038/ng.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hung RJ. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 10.Manolio TA. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eichler EE, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat. Rev. Genet. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donnelly P. Progress and challenges in genome-wide association studies in humans. Nature. 2008;456:728–731. doi: 10.1038/nature07631. [DOI] [PubMed] [Google Scholar]
- 13.Hong MG, et al. Strategies and issues in the detection of pathway enrichment in genome-wide association studies. Hum. Genet. 2009;126:289–301. doi: 10.1007/s00439-009-0676-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ioannidis JP, et al. Validating, augmenting and refining genome-wide association signals. Nat. Rev. Genet. 2009;10:318–329. doi: 10.1038/nrg2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elmore S. Apoptosis: a review of programmed cell death. Toxicol. Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shivapurkar N, et al. Apoptosis and lung cancer: a review. J. Cell Biochem. 2003;88:885–898. doi: 10.1002/jcb.10440. [DOI] [PubMed] [Google Scholar]
- 17.Catassi A, et al. Multiple roles of nicotine on cell proliferation and inhibition of apoptosis: implications on lung carcinogenesis. Mutat. Res. 2008;659:221–231. doi: 10.1016/j.mrrev.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 18.Cotter TG. Apoptosis and cancer: the genesis of a research field. Nat. Rev. Cancer. 2009;9:501–507. doi: 10.1038/nrc2663. [DOI] [PubMed] [Google Scholar]
- 19.Wang LE, et al. Fas A670G polymorphism, apoptotic capacity in lymphocyte cultures, and risk of lung cancer. Lung Cancer. 2003;42:1–8. doi: 10.1016/s0169-5002(03)00276-9. [DOI] [PubMed] [Google Scholar]
- 20.Ter-Minassian M, et al. Apoptosis gene polymorphisms, age, smoking and the risk of non-small cell lung cancer. Carcinogenesis. 2008;29:2147–2152. doi: 10.1093/carcin/bgn205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, et al. Functional polymorphisms in cell death pathway genes FAS and FASL contribute to risk of lung cancer. J. Med. Genet. 2005;42:479–484. doi: 10.1136/jmg.2004.030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeiss CJ. The apoptosis-necrosis continuum: insights from genetically altered mice. Vet. Pathol. 2003;40:481–495. doi: 10.1354/vp.40-5-481. [DOI] [PubMed] [Google Scholar]
- 23.Xu Z, et al. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009;37:W600–W605. doi: 10.1093/nar/gkp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heinemeyer T, et al. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andrews NC, et al. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrett JC, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 28.Thorgeirsson TE, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landi MT. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am. J. Hum. Genet. 2009;85:679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diaconu NC, et al. Increase in CD30 ligand/CD153 and TNF-alpha expressing mast cells in basal cell carcinoma. Cancer Immunol. Immunother. 2007;56:1407–1415. doi: 10.1007/s00262-007-0290-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duckett CS, et al. CD30-dependent degradation of TRAF2: implications for negative regulation of TRAF signaling and the control of cell survival. Genes Dev. 1997;11:2810–2821. doi: 10.1101/gad.11.21.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oflazoglu E, et al. Targeting CD30/CD30L in oncology and autoimmune and inflammatory diseases. Adv. Exp. Med. Biol. 2009;647:174–185. doi: 10.1007/978-0-387-89520-8_12. [DOI] [PubMed] [Google Scholar]
- 33.Chiarle R, et al. CD30 overexpression enhances negative selection in the thymus and mediates programmed cell death via a Bcl-2-sensitive pathway. J. Immunol. 1999;163:194–205. [PubMed] [Google Scholar]
- 34.Horie R, et al. Ligand-independent signaling by overexpressed CD30 drives NF-kappaB activation in Hodgkin-Reed-Sternberg cells. Oncogene. 2002;21:2493–2503. doi: 10.1038/sj.onc.1205337. [DOI] [PubMed] [Google Scholar]
- 35.Kennedy MK, et al. Deciphering CD30 ligand biology and its role in humoral immunity. Immunology. 2006;118:143–152. doi: 10.1111/j.1365-2567.2006.02354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Florido M, et al. Delayed recruitment of lymphocytes into the lungs of CD30-deficient mice during aerogenic Mycobacterium avium infections. Immunobiology. 2009;214:643–652. doi: 10.1016/j.imbio.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 37.Nam SY, et al. CD30 supports lung inflammation. Int. Immunol. 2008;20:177–184. doi: 10.1093/intimm/dxm130. [DOI] [PubMed] [Google Scholar]
- 38.Croager EJ, et al. Characterisation of the human CD30 ligand gene structure. Biochim. Biophys. Acta. 1997;1353:231–235. doi: 10.1016/s0167-4781(97)00108-5. [DOI] [PubMed] [Google Scholar]
- 39.Durie BGM, et al. Genetic polymorphisms of EPHX1, Gsk3[beta], TNFSF8 and myeloma cell DKK-1 expression linked to bone disease in myeloma. Leukemia. 2009;23:1913–1919. doi: 10.1038/leu.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu YJ, et al. Is replication the gold standard for validating genome-wide association findings? PLoS One. 2008;3:e4037. doi: 10.1371/journal.pone.0004037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cazier JB, et al. General lessons from large-scale studies to identify human cancer predisposition genes. J. Pathol. 2010;220:255–262. doi: 10.1002/path.2650. [DOI] [PubMed] [Google Scholar]
- 42.Scelo G, et al. Occupational exposure to vinyl chloride, acrylonitrile and styrene and lung cancer risk (Europe) Cancer Causes Control. 2004;15:445–452. doi: 10.1023/B:CACO.0000036444.11655.be. [DOI] [PubMed] [Google Scholar]
- 43.Khoury MJ, et al. Genome-wide association studies, field synopses, and the development of the knowledge base on genetic variation and human diseases. Am. J. Epidemiol. 2009;170:269–279. doi: 10.1093/aje/kwp119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.