Abstract
The 2Z-isomer of malyngamide K has been isolated along with the known compounds malyngamide C, deoxy-C and K, and characterized from a Papua New Guinea field collection of the cyanobacterium Lyngbya majuscula. The planar structure was deduced by 1D and 2D NMR spectroscopic and mass spectral data interpretation. The absolute configurations were determined on the basis of spectroscopic techniques, chemical degradation and DFT theoretical calculations.
Keywords: Lyngbya majuscula, malyngamide, vinyl-chloride, NMR, DFT
1. Introduction
Malyngamides are a predominant class of marine cyanobacterial natural product [1–3] that is generally characterized as N-substituted amides of 7(S)-methoxytetradec-4(E)-enoic acid (lyngbic acid (1), Fig. 1), or in rare cases of its respective C12 [4–6], C16 [7,8], methylated [7] or epimerized 7(R) [9] analogs. In contrast to the fatty acid portion, the amine moiety of the malyngamides shows a remarkable structural diversity, including heterocycles, tripeptides and highly oxidized six-membered rings systems, mostly extended by a vinyl chloride functionality. Regarding the chloromethylene moiety, the majority of the 35 known malyngamides [3,9] were proven to possess 2E geometry. To date, only the naturally occurring isomalyngamides A and B [10] and malyngamides Q and R [11] have been shown to bear 2Z-configured vinyl chloride substructures. Recently, also the synthetically-derived isomalyngamide M [12] was added to this geometrical isomer-series of malyngamides.
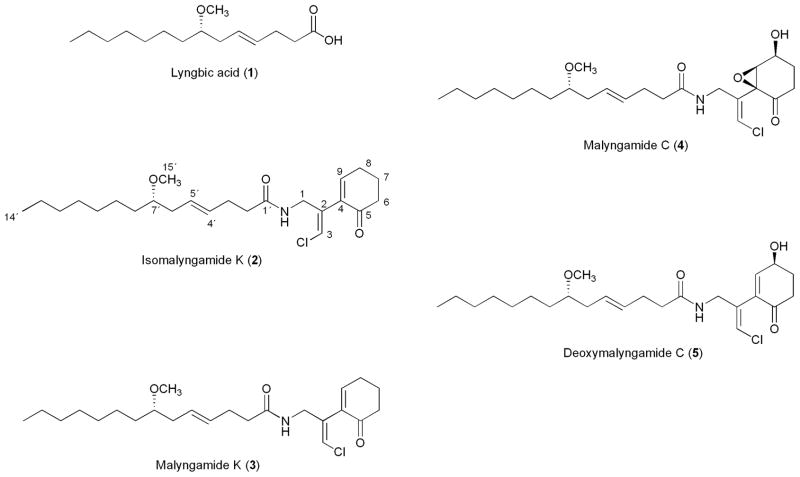
Figure 1.
Malyngamide-based compounds isolated in this report.
In previous papers, we reported the isolation and characterization of several cancer cell toxic lyngbyabellin [13] and aurilide [14] analogs from the chemically rich strain Lyngbya majuscula PNG5-27-02-1, collected from Alotau Bay, Papua New Guinea. Further in-depth analysis of the remaining fractions from this extract led to the isolation of a new stereoisomer (2) of malyngamide K (3) [15] which possesses an exo-chloromethylene moiety in the rare Z configuration, along with the known compounds malyngamides K (3) [15], C (4) [16] and deoxy-C (5) [16] (Fig. 1). This paper describes the isolation and structure elucidation of the new metabolite isomalyngamide K (2) with 2Z stereochemistry.
2. Experimental section
2.1. General experimental procedures
Optical rotation measurements were recorded on a Jasco P-1010 polarimeter. UV and FT-IR spectra were obtained employing Hewlett Packard 8452A and Nicolet 510 instruments, respectively. All NMR spectra were recorded on Bruker Avance DRX300 and DPX400 spectrometers. Spectra were referenced to residual solvent signal with resonances at δH/C 7.26/77.1 (CDCl3) and δH 7.15/128.0 (C6D6). Low resolution ESI-MS spectra were obtained on a Thermo Finnigan LCQ Advantage mass spectrometer. High resolution ESI-TOF and CI mass spectra were recorded on Waters Micromass LCT Classic and JEOL MSRoute mass spectrometers, respectively. HPLC was carried out using a Waters system consisting of a Rheodyne 7725i injector, two 515 pumps, a pump control module and a 996 photodiode array detector. TLC grade (10–40 μm) Si gel was used for vacuum chromatography. All solvents were purchased as HPLC grade.
2.2. Algal material
The marine cyanobacterium L. majuscula (voucher specimen available from WHG as collection number PNG5-27-02-1) was collected by hand from shallow waters (1–3 m) in Alotau Bay, Papua New Guinea, on May 7, 2002. The material was stored in 2-propanol at −20 °C until extraction.
2.3. Extraction and isolation
Approximately 138 g (dry wt) of the collected cyanobacterial mat was extracted repeatedly with CH2Cl2/MeOH (2:1) to produce 3.05 g of crude organic extract. The extract (3.0 g) was fractionated by silica gel vacuum liquid chromatography using a stepwise gradient solvent system of increasing polarity starting from 10% EtOAc in hexanes to 100% MeOH. The fractions eluting with 90% and 100% EtOAc showed proton resonances in the 1H NMR spectrum, indicative for malyngamide-based compounds and peptidic metabolites. Both fractions were separately further chromatographed on Mega Bond RP18 solid-phase extraction (SPE) cartridges using a stepwise gradient solvent system of decreasing polarity starting with 80% MeOH in H2O and ending with 100% MeOH. 1H NMR profiling revealed malyngamide-containing fractions which were subsequently purified by HPLC (Phenomenex Sphereclone 5μ ODS, 250×10 mm, 9:1 MeOH/H2O, detection at 211 nm), yielding isomalyngamide K (2) (4.2 mg), malyngamide K (3) (3.0 mg), malyngamide C (4) (2.3 mg) and deoxy-malyngamide C (5) (2.1 mg).
2.4. Basic hydrolysis of isomalyngamide K (2)
Compound 2 (4.0 mg) was dissolved in 2 mL solution of 10% KOH in EtOH-H2O (4:1) and the stirred mixture refluxed for 15 h. The hydrolysate was concentrated in vacuo and partitioned between H2O and CH2Cl2. The H2O layer was isolated, acidified, and extracted with CH2Cl2 to yield 1.1 mg of lyngbic acid (1): brownish oil; [α]D −8° (c 0.5, CHCl3) [lit. −5° (c 0.22, CHCl3) [10]; −10° (c 0.5, CHCl3), [17]; HRCIMS m/z 257.2110 [M+H]+ (calcd. for C15H29O3, 257.2117); 1H NMR (300 MHz, CDCl3): δ 0.88 (3H, t, J=6.9 Hz, H-14′), 1.27 m (10H, m, H-9′, H-10′, H-11′, H-12′ and H-13′), 1.42 (2H, m, H-8′), 2.19 (2H, m, H-6′), 2.36 (2H, m, H-2′), 2.42 (2H, d, J=5.9 Hz, H-3′), 3.15 (1H, t, J=5.8 Hz, H-7′), 3.32 (3H, s, H-15′), 5.49 (2H, m, H-4′and H-5′).
2.5. Computational procedures
All structures were built with Gaussian03 [18]. Initial optimizations were done on the B3LYP/6-311+g(2d,p) level of theory. Subsequent relaxed scans were done with AM1. Low energy conformations (1 kcal/mole cut-off) were further geometry optimized on the B3LYP/6-31G(d) level of theory. The chemical shifts were calculated on the B3LYP 6-31g(2d,p)/IEFPCM level of theory with chloroform as solvent.
3. Results and discussion
Compound 2 showed an [M+H]+ peak at m/z 424.2610, consistent with the molecular formula C24H39ClNO3 by HRESITOFMS (calcd for C24H3935ClNO3, 424.2618, Δ − 1.9 ppm). The IR spectrum possessed absorptions due to free OH or amide NH groups (3347 cm−1), aliphatic atoms (2927, 2854 cm−1) and amide carbonyl groups (1725, 1672 cm−1). UV (MeOH) maxima were observed at λmax 208 nm (ε = 4910) and 236 nm (ε = 3025), suggestive of one conjugated enone system. Diagnostic resonances in the 1H NMR spectra with CDCl3 as solvent strongly suggested that compound 2 was a metabolite of the malyngamide class. For example, a methoxy singlet signal (δ 3.31) and its corresponding α-methoxy methine multiplet (δ 3.14), two olefinic signals (δ 5.46), three methylenes between δ 2.17 and 2.32, a methylene envelope (δ 1.25 – 1.42) and a terminal methyl (δ 0.88) were indicative of the lyngbic acid (1) moiety in 2. In addition, the 1H and 13C NMR spectra contained resonances consistent with the presence of an exomethylene functionality possessing a vinyl chloride (δH 6.20/δC 119.8) and an exchangeable amide signal (δ 6.02), revealing further typical structural features of this compound class. Based on its mass and overall NMR features, isolate 2 was initially dereplicated as malyngamide K (3) [15] using the MarinLit database [19]. However, when the spectral data were carefully compared with those reported in the literature, deviations in the 13C NMR spectra between 2 and malyngamide K (3) became apparent (Table 1). Analysis of the 2D-NMR data confirmed that 2 had the same constitution as malyngamide K (3), and hence, the two molecules must differ in the configuration of either the chiral center C-7′ and/or the geometry of the Δ2 and/or Δ4′ double bonds. The geometry of the Δ4′ double bond was inferred from 1H decoupling experiments. The olefinic proton signals of C-4′ and C-5′ were overlapped in CDCl3 (Table 1), but resolved to a distinct pair of multiplets in C6D6. Decoupling of the adjacent methylene protons at C3′ and C6′ allowed the measurement of the 3JH4′-H5′ as 15.4 Hz, confirming an E geometry for Δ4′ in 2, the same configuration as in 3. This E-double bond geometry was also revealed from the chemical shifts of the adjacent C3′ and C6′ carbon atoms (δ 28.7 and δ 36.5, respectively) [20]. Since the optical rotation value obtained for 2 ( −12.9, c = 1.05, CHCl3) was comparable to that of malyngamide K ([α]D −8.4, c = 0.28, CHCl3) in sign and amount, the absolute configuration of the lyngbic acid moiety of 2 was assumed to be 7′S. This hypothesis was verified by base hydrolysis of 2, yielding the fatty acid portion 1, which was identical in all respects to that previously reported for 7(S)-methoxytetradec-4-(E)-enoic acid, thus establishing the 7′S configuration for 2.
Table 1.
1H and 13C NMR spectroscopic data for isomalyngamide K (2) and malyngamide K (3) in CDCl3.
| Position | isomalyngamide K (2)a 2Z configuration |
malyngamide K (3)a,b 2E configuration |
Δ δC |
||
|---|---|---|---|---|---|
| δH | δC | δH | δC | δC(2)− δC(3) | |
| 1 | 4.14 d (5.9) | 39.0 CH2 | 3.94 d (6.1) | 44.0 CH2 | −5.0 |
| 2 | 138.5 qC | 136.7 qC | +1.8 | ||
| 3 | 6.20 brs | 119.8 CH | 6.20 brs | 119.7 CH | +0.1 |
| 4 | 138.2 qC | 136.5 qC | +1.7 | ||
| 5 | 199.2 qC | 198.6 qC | +0.6 | ||
| 6 | 2.49 m | 38.7 CH2 | 2.54 ddd (6.5, 13.5) | 38.6 CH2 | +0.1 |
| 7 | 2.01 ddd (6.1, 13.1,10.4) | 22.6 CH2 | 2.08 ddd (6.5, 13.2, 10.3) | 22.9 CH2 | −0.3 |
| 8 | 2.43 ddd (10.4, 4.2) | 26.3 CH2 | 2.50 ddd (10.2, 4.3) | 26.3 CH2 | 0.0 |
| 9 | 6.94 t (4.2) | 149.5 CH | 6.95 t (4.1) | 151.5 CH | −2.0 |
| 1′ | 172.0 qC | 172.5 qC | −0.5 | ||
| 2′ | 2.19 m (5.8, 7.5) | 36.4 CH2 | 2.24 dd (5.8, 7.5) | 36.6 CH2 | −0.2 |
| 3′ | 2.31 m | 28.7 CH2 | 2.32 m | 28.8c CH2 | −0.1 |
| 4′ | 5.46 m | 130.7 CH | 5.47 m | 130.9 CH | −0.2 |
| 5′ | 5.46 m | 127.5 CH | 5.47 m | 127.9 CH | −0.4 |
| 6′ | 2.19 m | 36.5 CH2 | 2.20 m | 36.6 CH2 | −0.1 |
| 7′ | 3.14 ddd (5.5, 11.1) | 80.7 CH | 3.15 dddd (5.8, 11.4) | 80.9 CH | −0.2 |
| 8′ | 1.41 t 6.2 | 33.3 CH2 | 1.43 m | 33.6 CH2 | −0.3 |
| 9′ | 1.26 m | 31.8 CH2 | 1.28 m | 32.0 CH2 | −0.2 |
| 10′ | 1.26 m | 25.3 CH2 | 1.28 m | 25.5 CH2 | −0.2 |
| 11′ | 1.26 m | 29.3 CH2 | 1.28 m | 29.5c CH2 | −0.2 |
| 12′ | 1.26 m | 29.7 CH2 | 1.28 m | 29.9c CH2 | −0.2 |
| 13′ | 1.26 m | 22.7 CH2 | 1.28 m | 22.9 CH2 | −0.2 |
| 14′ | 0.88 t (7.0) | 14.1 CH3 | 0.88 t | 14.3 CH3 | −0.2 |
| 15′ | 3.32 s | 56.5 CH3 | 3.34 s | 56.7 CH3 | −0.2 |
| NH | 6.02 brt (5.3) | 6.10 m | |||
The coupling constants (J) are in parentheses and reported in Hz; chemical shifts are given in ppm.
NMR values taken from reference Wu et al. 1997 [15].
The 13C NMR values of carbons C-3′, C-11′ and C-12′ deviate from the original report. Re-examination of the original 13C spectrum of malyngamide K (See Supplementary data) revealed that these carbons were not correctly assigned. Table 1 shows the corrected values.
These results focused our efforts on elucidation of the geometry of the chlorinated exomethylene functionality of 2, the Δ2 double bond. Moreover, this was the part of the molecule where the majority of 13C NMR chemical shift differences were observed between 2 and malyngamide K (Table 1). Compound 2 decomposed soon after it was defined, such that it precluded the determination of its relative configuration by measurement of key diagnostic 3JCH couplings utilizing HSQMBC [11] or editing-HETLOC and phase sensitive HMBC [10] NMR experiments. However, the geometry of the chlorinated exomethylene group was suggested by comparison of 13C NMR data of 2 with those of regular and isomeric forms of malyngamides A, B [10] and M [12]. This analysis revealed that compared to the E-configured vinyl chloride malyngamides, in the Z-configured vinyl chloride malyngamides the 13C NMR shift value of C-1 usually experiences a significant upfield-shift in the range of 2.8 – 3.4 ppm while for carbons C-2, C-3 and C-4 a corresponding downfield-shift in the range 0.3–2.9 ppm is observed. The observed shifts in 13C NMR spectrum of 2 (Table 1) for C-1 (−5.0 ppm, upfield), C-2 (+1.8 ppm, downfield), C-3 (+0.1 ppm, downfield) and C-4 (+0.6 ppm, downfield) are therefore consistent with the 2Z configuration. Thus we propose for compound 2 the trivial name isomalyngamide K. In order to consolidate this hypothesis, the NMR analysis was complemented by the comparison of quantum-mechanically calculated 13C NMR chemical shifts [21–23] with experimental ones. To increase the reliability of this comparison, the most indicative differences in chemical shifts were chosen that occur between the published data for malyngamide K (3) and the putative isomalyngamide K (2): C-1 (−5.0 ppm) and C-2 (+1.8 ppm).
To derive the stereochemistry two simplified model compounds with E or Z configuration were constructed, respectively. These models represent cut-outs of the original structures and contain the relevant vinyl chloride substructure up to carbon C-2′ regarding the western end of the molecule and up to C-4 concerning the eastern part of the molecule (Fig. 2). In this way, the resulting model compounds contain the relevant carbons to answer the stereoisomeric question and were free from complicating conformers originating from the remaining long fatty acid side chain. Prior to the reduction of the cyclohexenone-ring down to a methyl group, DFT calculations were initiated to probe whether the pucker of the ring system had a significant influence on calculated 13C chemical shifts for C-1 and C-2. Since only a minor influence by the orientation of the cyclohexenone ring on the calculated 13C NMR shift of C-1 and C-2 was observed (< 0.2 ppm), the simplified model was considered as acceptable.
Figure 2.

Model compounds utilized for DFT calculations.
Two relaxed scans were conducted with the E and Z-versions of the vinyl chloride model compound: the first one around the C-1-C-2 bond, the second around C-1 and NH, each with a stepsize of 36°. From the calculated energy differences, Boltzmann-derived populations for different conformations were calculated with a 1 kcal/mole cut-off. For the Z configuration 10 conformations were obtained, for the E configuration 43. These conformations were geometry optimized on the B3LYP/6-31g(d) level of theory, and subsequently the 13C chemical shifts of C-1 and C-2 were calculated on the B3LYP/6-31g(2d,p)/IEFPCM level of theory. This procedure was successfully applied even for the calculation of proton chemical shifts which are even more difficult to predict compared to 13C chemical shifts [24]. The averaged calculated 13C chemical shifts of C-1 and C-2 for the Z model are 38.4 and 129.1 ppm, whereas for the E model 43.2 ppm and 126.8 ppm were found, respectively (Fig 2). Consequently, the two isomers were predicted to differ for C-1 by −4.8 ppm (experimental: −5.0 ppm) and for C-2 by +2.3 ppm (experimental: +1.8 ppm). In conclusion, the quantum-mechanical calculated data showed an excellent fit in direction and strength with the experimental data and thus independently corroborated the assignment of the Z-geometry for the vinyl chloride moiety in isomalyngamide K (2).
The identity of compounds 3–5 was established by direct comparison of the HR-EIMS, 1H NMR and 13C data with literature data [15,16].
4. Conclusions
The isolation of the new L. majuscula metabolite isomalyngamide K (2), in addition to eleven known compounds (3–5, [13,14]) from this single collection, demonstrates the capacity of this organism to produce a remarkable diversity if structures. Compound 2 represents a rare type of malyngamide, in which the conformation of the chloromethylene group is opposite from the majority of previously reported malyngamides.
Speculating on the reasons for the different double bond geometry, a possible photochemical isomerization process of the vinylchloride moiety [25] was considered but excluded since the interconversion from E to the Z isomer was never observed with our pure E-configured malyngamides. Furthermore, also the Z-configured malyngamides Q and R give support the hypothesis of a subtle yet fundamental difference in the biosynthetic pathway giving rise to the Z-vinylchloride isomers.
The detailed biosynthesis of malyngamides is to date unknown, however, the NH, C-1 and C-2 carbons of malyngamide K derivatives are likely derived from glycine, while C-3 is most probably generated by an HMGCoA synthase-like reaction as determined for the jamaicamides [26]. Extension by three malonyl-CoA derived acetate units and further processing produce the basic skeleton of the peptidic moiety of malyngamide K. From a recent study on vinylchloride formation in the cyanobacterial metabolite jamaicamide A [27], it can be concluded that the geometry of the double bond may be determined either by the configuration of the radical halogenase-introduced chlorine atom, the ensuing dehydratase and/or the decarboxylase which act on the halogenated β-branching product. Due to the presence of 2E and 2Z configured malyngamide K in this single cyanobacterial extract, the existence of enzymes with different chiral specificities in this reaction manifold, or showing a lower regiochemical control, is postulated.
Supplementary Material
Acknowledgments
We gratefully acknowledge the government of Papua New Guinea for permission and L. Matainaho, University of Papua New Guinea for assistance in making these collections, the NMR facility of the Department of Chemistry at Oregon State University, and the OSU mass spectrometry facility. H.G. acknowledges fellowship support from the German Research Foundation (GR 2673/1-1). U.M.R. acknowledges support of the Max-Planck-Society. W.H.G. acknowledges support from NIH NS 053398.
Appendix A. Supplementary data
1H and 13C NMR spectra of isomalyngamide K (2) and malyngamide K (3). Supplementary data associated with this article can be found, in the online version, at doi:
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nagle DG, Paul VJ. J Phycol. 1999;35:1412. [Google Scholar]
- 2.Gerwick WH, Tan LT, Sitachitta N. In: The Alkaloids: Chemistry and Biology. Cordell GA, editor. Vol. 57. Academic Press; San Diego: 2001. pp. 75–184. [DOI] [PubMed] [Google Scholar]
- 3.Tan LT. Phytochemistry. 2007;68:954. doi: 10.1016/j.phytochem.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 4.McPhail KL, Gerwick WH. J Nat Prod. 2003;66:132. doi: 10.1021/np0204186. [DOI] [PubMed] [Google Scholar]
- 5.Appleton DR, Sewell MA, Berridge MV, Copp BR. J Nat Prod. 2002;65:630. doi: 10.1021/np010511e. [DOI] [PubMed] [Google Scholar]
- 6.Praud A, Valls R, Piovetti L, Banaigs B. Tetrahedron Lett. 1993;34:5437. [Google Scholar]
- 7.Mynderse JS, Moore RE. J Org Chem. 1978;43:4359. [Google Scholar]
- 8.Wan F, Erickson KL. J Nat Prod. 1999;62:1696. doi: 10.1021/np990291t. [DOI] [PubMed] [Google Scholar]
- 9.Suntornchashwej S, Suwanborirux K, Koga K, Isobe M. Chem Asian J. 2007;2:114. doi: 10.1002/asia.200600219. [DOI] [PubMed] [Google Scholar]
- 10.Kan Y, Sakamoto B, Fujita T, Nagai H. J Nat Prod. 2000;63:1599. doi: 10.1021/np000250t. [DOI] [PubMed] [Google Scholar]
- 11.Milligan KE, Márquez B, Williamson RT, Davies-Coleman M, Gerwick WH. J Nat Prod. 2000;63:965. doi: 10.1021/np000038p. [DOI] [PubMed] [Google Scholar]
- 12.Chen J, Shi ZF, Zhou L, Xie AL, Cao XP. Tetrahedron. 2010;66:3499. [Google Scholar]
- 13.Han B, McPhail KL, Gross H, Goeger DE, Mooberry SL, Gerwick WH. Tetrahedron. 2005;61:11723. [Google Scholar]
- 14.Han B, Gross H, Goeger DE, Mooberry SL, Gerwick WH. J Nat Prod. 2006;69:572. doi: 10.1021/np0503911. [DOI] [PubMed] [Google Scholar]
- 15.Wu M, Milligan KE, Gerwick WH. Tetrahedron. 1997;53:15983. [Google Scholar]
- 16.Ainslie RD, Barchi JJ, Jr, Kuniyoshi M, Moore RE, Mynderse JS. J Org Chem. 1985;50:2859. [Google Scholar]
- 17.Cardellina JH, II, Dalietos D, Marner FJ, Mynderse JS, Moore RE. Phytochemistry. 1978;17:2091. [Google Scholar]
- 18.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Jr, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian 03, Revision B.02. Gaussian Inc; Pittsburgh, PA: 2003. [Google Scholar]
- 19.Blunt JW, Munro MHG. Marinlit Database. University of Canterbury; Christchurch, NewZealand: 2005. [Google Scholar]
- 20.Cateni F, Zilic J, Falsone G, Scialino G, Banfi E. Bioorg Med Chem Letters. 2003;13:4345. doi: 10.1016/j.bmcl.2003.09.044. [DOI] [PubMed] [Google Scholar]
- 21.Bifulco G, Dambruoso P, Gomez-Paloma L, Riccio R. Chem Rev. 2007;107:3744. doi: 10.1021/cr030733c. [DOI] [PubMed] [Google Scholar]
- 22.Fattorusso E, Luciano P, Romano A, Taglialatela-Scafati O, Appendino G, Borriello M, Fattorusso C. J Nat Prod. 2008;71:1988. doi: 10.1021/np8003547. [DOI] [PubMed] [Google Scholar]
- 23.Gross H, Wright AD, Reinscheid U, König GM. Nat Prod Comm. 2009;4:315. [PubMed] [Google Scholar]
- 24.Jain R, Bally T, Rablen PR. J Org Chem. 2009;74:4017. doi: 10.1021/jo900482q. [DOI] [PubMed] [Google Scholar]
- 25.Lemaire P, Balme G, Desbordes P, Vors JP. Org Biomol Chem. 2003;1:4209. doi: 10.1039/b306356a. [DOI] [PubMed] [Google Scholar]
- 26.Edwards DJ, Marquez BL, Nogle LM, McPhail K, Goeger DE, Roberts MA, Gerwick WH. Chem Biol. 2004;11:817. doi: 10.1016/j.chembiol.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 27.Gu L, Wang B, Kulkarni A, Geders TW, Grindberg RV, Gerwick L, Hakansson K, Wipf P, Smith JL, Gerwick WH, Sherman DH. Nature. 2009;459:731. doi: 10.1038/nature07870. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.