Abstract
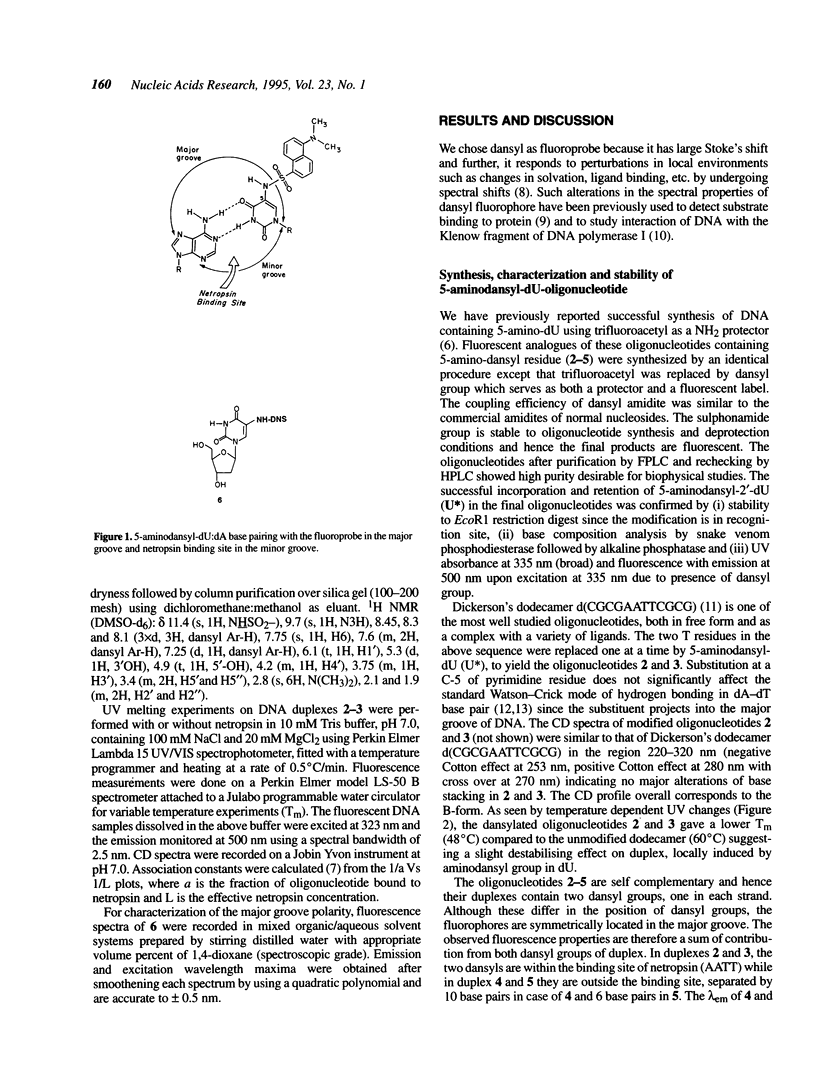
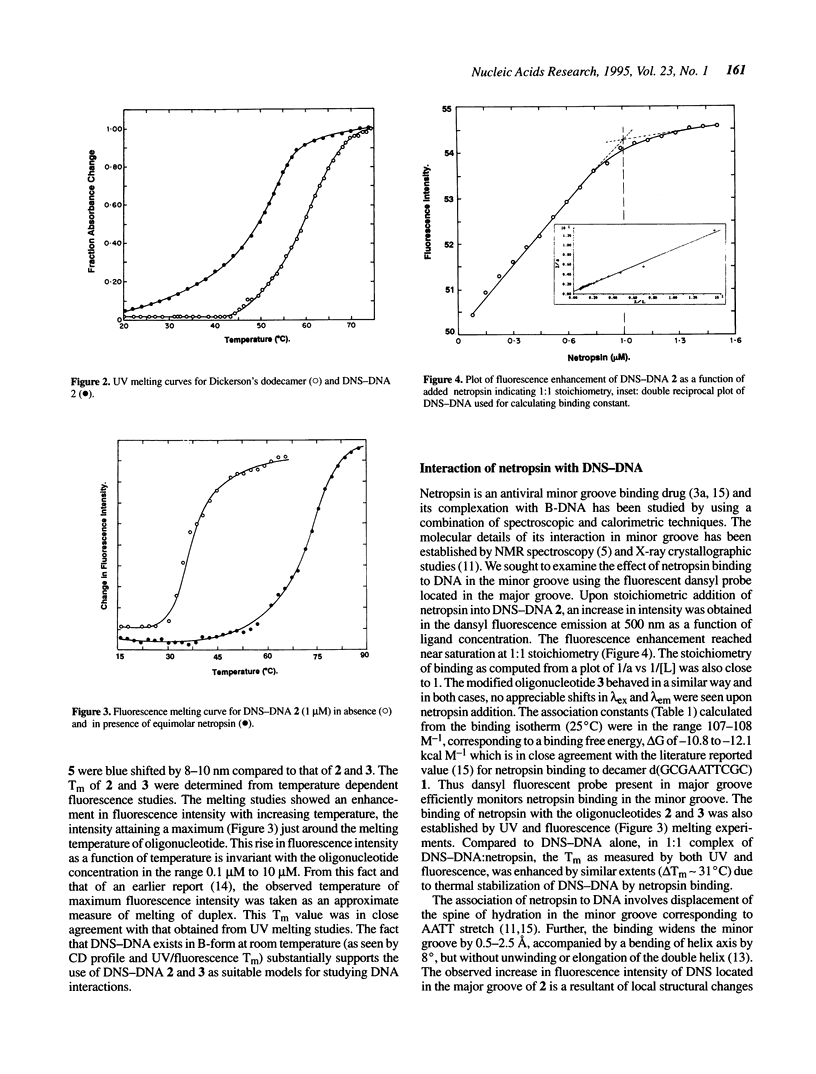
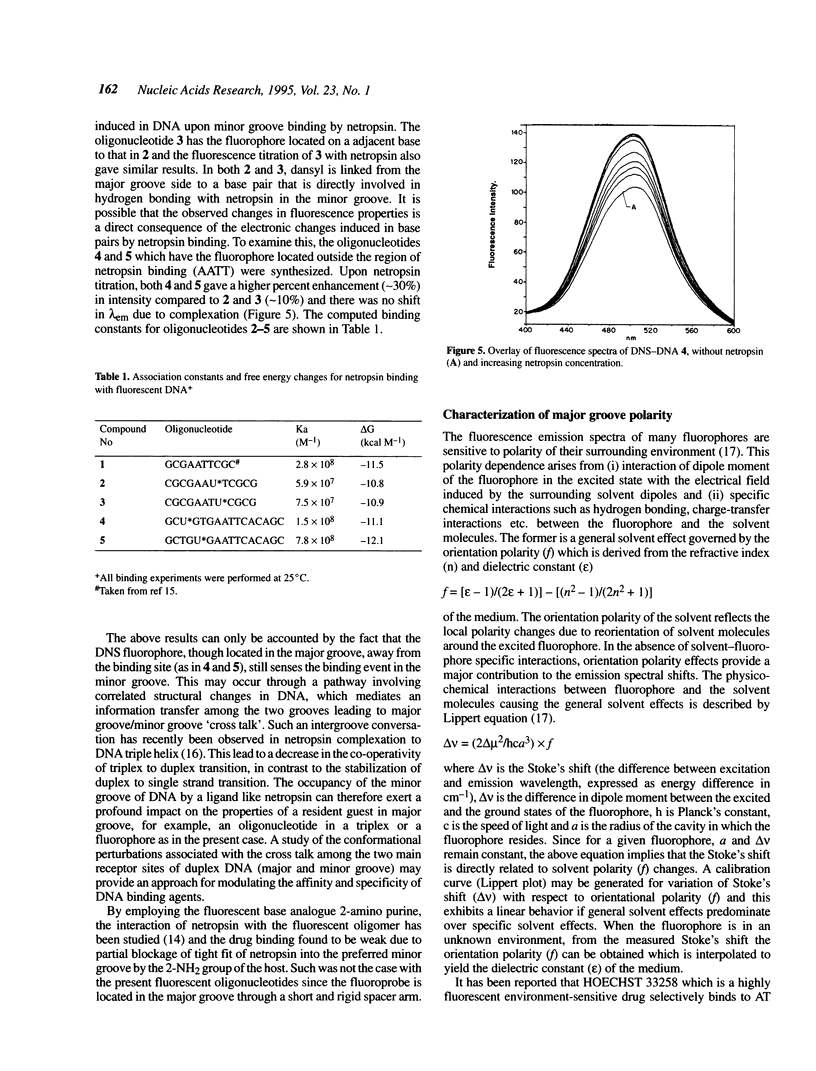
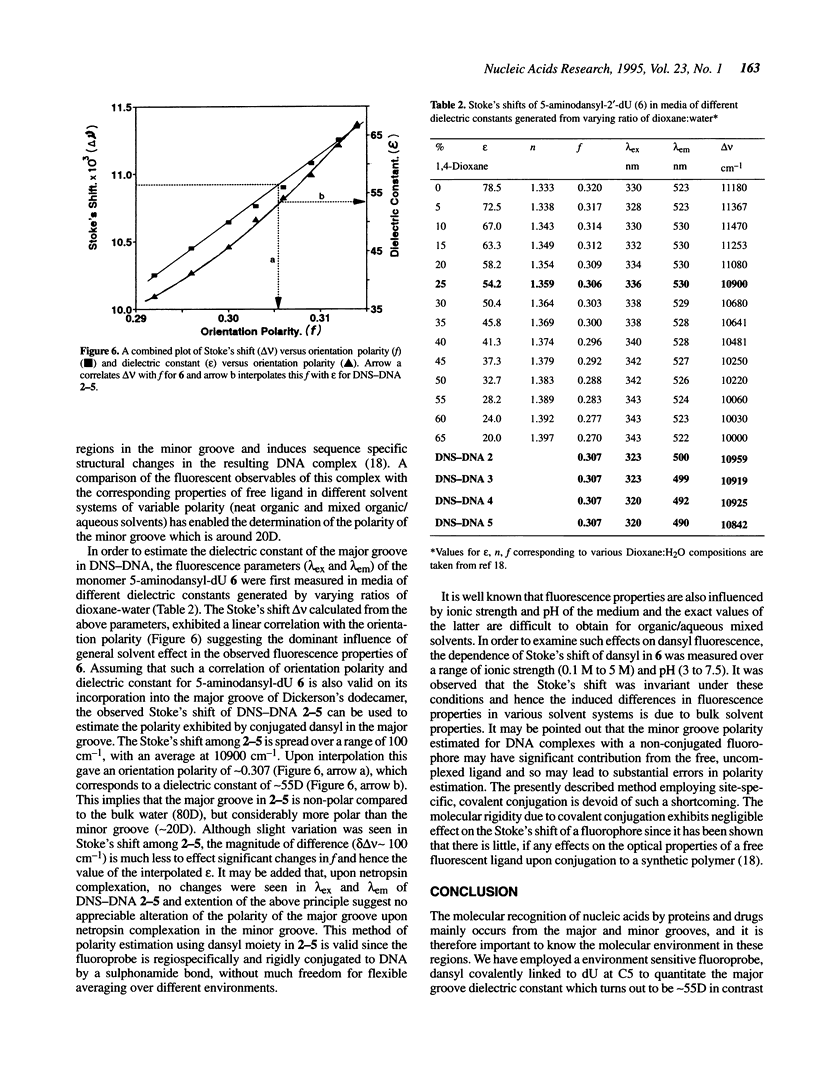
The major and minor groove in duplex DNA are sites of specific molecular recognition by DNA-binding agents such as proteins, drugs and metal complexes and have functional significance. In view of this, understanding of the inherent differences in their environment and the allosteric information transfer between them induced by DNA-binding agents assumes importance. Site-specific incorporation of 5-aminodansyl-dU, (U*) in oligonucleotides d(CGCGAAU*TCGCG) and d(CGCGAATU*CGCG) leads to fluorogenic nucleic acids, in which the reporter group resides in the major groove. The fluorescent observables from such a probe are used to estimate the dielectric constant of the major groove to be approximately 55D, in comparison to the reported non polar environment of the minor groove (approximately 20D) in poly d[AT]-poly d[AT]. An exclusive minor groove event such as DNA-netropsin association can be quantitatively monitored by fluorescence of the dansyl moiety located in the major groove. This suggests existence of an information network among the two grooves. The fluorescent DNA probes as reported here may have potential applications in the study of structural polymorphisms in DNA, DNA-ligand interactions and triple helix structure.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bramhall J. Phospholipid packing asymmetry in curved membranes detected by fluorescence spectroscopy. Biochemistry. 1986 Jun 3;25(11):3479–3486. doi: 10.1021/bi00359a057. [DOI] [PubMed] [Google Scholar]
- Durand M., Thuong N. T., Maurizot J. C. Binding of netropsin to a DNA triple helix. J Biol Chem. 1992 Dec 5;267(34):24394–24399. [PubMed] [Google Scholar]
- Guest C. R., Hochstrasser R. A., Dupuy C. G., Allen D. J., Benkovic S. J., Millar D. P. Interaction of DNA with the Klenow fragment of DNA polymerase I studied by time-resolved fluorescence spectroscopy. Biochemistry. 1991 Sep 10;30(36):8759–8770. doi: 10.1021/bi00100a007. [DOI] [PubMed] [Google Scholar]
- Jin R., Breslauer K. J. Characterization of the minor groove environment in a drug-DNA complex: bisbenzimide bound to the poly[d(AT)].poly[d(AT)]duplex. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8939–8942. doi: 10.1073/pnas.85.23.8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopka M. L., Yoon C., Goodsell D., Pjura P., Dickerson R. E. Binding of an antitumor drug to DNA, Netropsin and C-G-C-G-A-A-T-T-BrC-G-C-G. J Mol Biol. 1985 Jun 25;183(4):553–563. doi: 10.1016/0022-2836(85)90171-8. [DOI] [PubMed] [Google Scholar]
- Kopka M. L., Yoon C., Goodsell D., Pjura P., Dickerson R. E. The molecular origin of DNA-drug specificity in netropsin and distamycin. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1376–1380. doi: 10.1073/pnas.82.5.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lycksell P. O., Gräslund A., Claesens F., McLaughlin L. W., Larsson U., Rigler R. Base pair opening dynamics of a 2-aminopurine substituted Eco RI restriction sequence and its unsubstituted counterpart in oligonucleotides. Nucleic Acids Res. 1987 Nov 11;15(21):9011–9025. doi: 10.1093/nar/15.21.9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marky L. A., Breslauer K. J. Origins of netropsin binding affinity and specificity: correlations of thermodynamic and structural data. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4359–4363. doi: 10.1073/pnas.84.13.4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidle S., Pearl L. H., Skelly J. V. DNA structure and perturbation by drug binding. Biochem J. 1987 Apr 1;243(1):1–13. doi: 10.1042/bj2430001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y. W., Breslauer K. J. Drug binding to higher ordered DNA structures: netropsin complexation with a nucleic acid triple helix. Proc Natl Acad Sci U S A. 1992 Jul 15;89(14):6653–6657. doi: 10.1073/pnas.89.14.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D. J. Antibiotic-DNA interactions: intermolecular nuclear Overhauser effects in the netropsin-d(C-G-C-G-A-A-T-T-C-G-C-G) complex in solution. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6424–6428. doi: 10.1073/pnas.79.21.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel N., Berglund H., Nilsson L., Rigler R., McLaughlin L. W., Gräslund A. Thermodynamics of interaction of a fluorescent DNA oligomer with the anti-tumour drug netropsin. Eur J Biochem. 1992 Feb 1;203(3):361–366. doi: 10.1111/j.1432-1033.1992.tb16558.x. [DOI] [PubMed] [Google Scholar]
- Schleif R. DNA binding by proteins. Science. 1988 Sep 2;241(4870):1182–1187. doi: 10.1126/science.2842864. [DOI] [PubMed] [Google Scholar]
- Skorka G., Shuker P., Gill D., Zabicky J., Parola A. H. Fluorescent substrate analogue for adenosine deaminase: 3'-O-[5-(dimethylamino)naphthalene-1-sulfonyl]adenosine. Biochemistry. 1981 May 26;20(11):3103–3109. doi: 10.1021/bi00514a018. [DOI] [PubMed] [Google Scholar]
- Travers A. A. DNA conformation and protein binding. Annu Rev Biochem. 1989;58:427–452. doi: 10.1146/annurev.bi.58.070189.002235. [DOI] [PubMed] [Google Scholar]
- Zimmer C., Wähnert U. Nonintercalating DNA-binding ligands: specificity of the interaction and their use as tools in biophysical, biochemical and biological investigations of the genetic material. Prog Biophys Mol Biol. 1986;47(1):31–112. doi: 10.1016/0079-6107(86)90005-2. [DOI] [PubMed] [Google Scholar]



