Abstract
Corneal Confocal Microscopy (CCM) imaging is a non-invasive surrogate of detecting, quantifying and monitoring diabetic peripheral neuropathy. This paper presents an automated method for detecting nerve-fibres from CCM images using a dual-model detection algorithm and compares the performance to well-established texture and feature detection methods. The algorithm comprises two separate models, one for the background and another for the foreground (nerve-fibres), which work interactively. Our evaluation shows significant improvement (p ≈ 0) in both error rate and signal-to-noise ratio of this model over the competitor methods. The automatic method is also evaluated in comparison with manual ground truth analysis in assessing diabetic neuropathy on the basis of nerve-fibre length, and shows a strong correlation (r = 0.92). Both analyses significantly separate diabetic patients from control subjects (p ≈ 0).
1 Introduction
Diabetic Peripheral Neuropathy (DPN) is one of the most common long-term complications of diabetes. The accurate detection and quantification of DPN are important for defining at-risk patients, anticipating deterioration, and assessing new therapies. Current methods of detecting and quantifying DPN, such as neurophysiology, lack sensitivity, require expert assessment and focus only on large nerve-fibres whereas the earliest signs of neuropathy are likely to be found among small nerve-fibres. On the other hand, small nerve-fibre damage is currently assessed using skin/nerve biopsy, which is highly invasive and is not suitable for repeated investigations.
However, recent research [15,10,8] using Corneal Confocal Microscopy (CCM) suggests that this non-invasive, and hence reiterative, test might be an ideal surrogate endpoint for human diabetic neuropathy. These studies demonstrate that measurements made by CCM accurately quantify corneal nerve fibre morphology. The measurements reflect the severity of DPN and relate to the extent of intra-epidermal nerve-fibre loss seen in skin biopsy. However, the major limitation preventing extension of this technique to wider clinical practice is that analysis of the images using interactive image analysis is highly labour-intensive and requires considerable expertise to quantify nerve-fibre pathology. To be clinically useful as a diagnostic tool, it is essential that the measurements be extracted automatically.
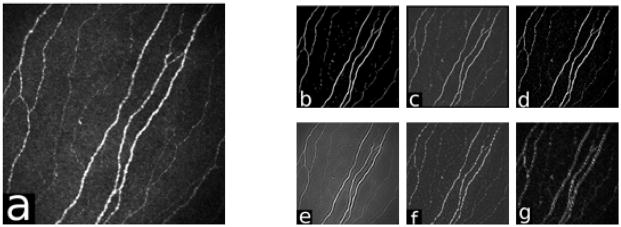
The first critical stage in analysis of CCM images (an example is shown in Figure 1(a)) is the detection of nerve-fibres. This is challenging as the nerve-fibres often show poor contrast in the relatively noisy images. The literature on this topic is not extensive, although the problem has a superficial similarity to other, more widely investigated, applications, such as detection of blood-vessels in retinal images. Ruggeri et al. [17] describe a heuristic method that was adapted from retinal analysis. In [2] we conducted a preliminary comparison of methods for contrast enhancement of nerve-fibres, comparing a Gabor wavelet with a well-established line detector.
Fig. 1.
An illustration of the methods’ responses. (a) the CCM image, (b) Dual-model, (c) Linop, (d) Hessian, (e) 2D Gabor, (f) Monogenic and (g) DTCWT.
This paper presents a dual-model algorithm for automatic detection and measurement of nerve-fibres in CCM images. Using a 2D Gabor wavelet and a Gaussian envelope, the dual-model of foreground (nerve-fibres) and background is constructed and applied to the original CCM image. The detection relies on estimating the correct local and dominant orientation of the nerve-fibres. Identifying low-contrast fibrous structures is a commonly encountered problem in several areas of investigation. Examples include mammography, retinopathy, angiography and detection of asbestos fibres. A number of methods have been developed and successfully applied in these applications. We evaluate our dual-model in comparison with some of these methods and with appropriate well-established feature detectors. While our analysis focuses on CCM images, our results suggest that the this may be an appropriate contrast enhancement method in other application domains. In addition to the evaluation of the nerve-fibre detection responses, we have also evaluated the clinical utility of the method by a comparison with manual analysis.
2 Linear-Structure and Feature Detection
A method of linear structure detection (Line Operator - Linop), originally developed for detection of asbestos fibres [4] has also been shown to be effective in detecting ducts in mammograms [18]. Linop exploits the linear nature of the structures to enhance their contrast by computing the average intensity of pixels lying on a line passing through the reference pixel for multiple orientations and scales. The largest values are chosen to be corresponding to the line, the strength of which is determined by the difference with the average intensity of the similarly oriented square neighbourhood.
In a preliminary study [2], we use the 2D Gabor filter [9] to detect nerve-fibres in CCM images. The filter is a band-pass filter that consists of a sinusoidal plane wave with a certain orientation and frequency, modulated by a Gaussian envelope. This spatial domain enhancement is based on the convolution of the image with the even-symmetric Gabor filter that is tuned to the local nerve-fibre orientation.
Frangi et al. [6] used a multiscale decomposition of the Hessian matrix to detect and measure blood vessels in Digital Subtraction Angiography images. They derived a discriminant function based on the eigenvalues and eigenvectors that has maximum response for tube-like structures. The external energy is used to attract the curve towards points which have a high likelihood of lying on a central vessel axis.
The Dual-Tree Complex Wavelet Transform (DTCWT) [11] is an extension of the Discrete Wavelet Transform (DWT), which provides a sparse representation and characterisation of structures and texture of the image at multiresolutions. The DTCWT utilises two DWT decompositions (trees) with specifically selected filters that gives it the properties of approximate shift-invariance and good directionality. The key feature of the DTCWT operation lies in the differences between the filters in the two trees.
The Monogenic signal [5] (a variant of a 2D analytic signal) is an extension of the analytic signal using quaternionic algebra in an attempt to generalise the method so it is capable of analysing intrinsically 2D signals e.g. structures within images. The Monogenic signal is based on the Riesz transform, which is a 2D generalization of the Hilbert transform used in the conventional analytic signal. The Monogenic signal is defined as the combination of the original signal and the Riesz-transformed one in the algebra of quaternions.
3 Dual-Model Nerve-Fibre Detection
In order to quantify the CCM images the nerve-fibres have to be detected. These captured images of nerve-fibre structures could suffer from several types of corruption due to some acquisition conditions, and nerve-fibres may appear faint due to small size or being only partly in the focus plane. Therefore, a nerve-fibre contrast enhancement algorithm is needed to exploit the linear structure of the nerve-fibres and distinguish them from the background noise. All of the methods described in the previous section are capable of providing this enhancement. In the next section we describe our approach.
3.1 Nerve-Fibre Contrast Enhancement
For this purpose the foreground model 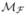 is an an even-symmetric and real-valued Gabor [9,3] wavelet and the background model
is an an even-symmetric and real-valued Gabor [9,3] wavelet and the background model 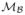 is a two-dimensional Gaussian envelope.
is a two-dimensional Gaussian envelope.
| (1) |
| (2) |
| (3) |
| (4) |
The x and y axes of the dual-model coordinate frame xθ and yθ are defined by a rotation of θ, which is the dominant orientation of the nerve-fibres in a particular region within the image (see Section 3.2). This dual-model is used to generate the positive response 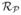 =
= 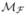 +
+ 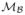 and the negative response
and the negative response 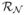 =
= 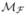 −
− 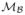 that are applied to the original CCM image and can be represented as in Equations (5) and (6) respectively.
that are applied to the original CCM image and can be represented as in Equations (5) and (6) respectively.
| (5) |
| (6) |
The equations of 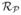 and
and 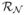 assume that the Gaussian envelope of both responses are identical i.e. they have the same variances
and the same aspect ratio γ. The magnitude of the Gaussian envelope α defines the threshold in which a nerve-fibre can be distinguished from the background image. The value of α can be set empirically to control sensitivity and accuracy of detection. The wavelength λ defines the frequency band of the information to be detected in the CCM image. Its value might be computed for a subregion within the image that has significant variability of nerve-fibre width. However for simplicity, λ is chosen to be a global estimate of the entire image based on empirical results.
assume that the Gaussian envelope of both responses are identical i.e. they have the same variances
and the same aspect ratio γ. The magnitude of the Gaussian envelope α defines the threshold in which a nerve-fibre can be distinguished from the background image. The value of α can be set empirically to control sensitivity and accuracy of detection. The wavelength λ defines the frequency band of the information to be detected in the CCM image. Its value might be computed for a subregion within the image that has significant variability of nerve-fibre width. However for simplicity, λ is chosen to be a global estimate of the entire image based on empirical results.
This in turn enhances the nerve-fibres that are oriented in the dominant direction, and decreases anything that is oriented differently by increasing the contrast between the foreground and the noisy background, whilst effectively reducing noise around the nerve-fibre structure as shown in Figure 1(b). This pixel-wise operation adjusts the models to suit the local neighbourhood characteristics of the reference pixel at f(i,j) by modifying the parameters of the foreground and background models. The dot products of the models and the reference pixel’s neighbourhood (Equations 7 and 8) are then combined to generate the final enhanced value of this particular reference pixel g(i,j) (Equation 9).
| (7) |
| (8) |
| (9) |
The neighbourhood area of the reference pixel is defined by the width ω. The sharpness of the transition of the enhanced image value at a particular pixel g(i,j) is controlled by k. A larger k amounts to a sharper transition when Γn = 0.
3.2 Nerve-Fibre Orientation Estimation
In CCM images, the nerve-fibres flow in locally consistent orientations everywhere. In addition, there is a global orientation that dominates the general flow. This orientation field describes the coarse structure of nerve-fibres. Using the least mean square (LMS) algorithm [7], the local orientation of the block centred at certain pixel is computed as in [16].
Since the orientations vary at a slow rate, a low-pass Gaussian filter is applied globally in order to further reduce errors at near-nerve-fibre and non-nerve-fibre regions. The LMS produces a stable smooth orientation field in the region of the nerve-fibres; however when applied on the background of the image, i.e. between fibres, the estimate is dominated by noise due to the lack of structure and uniform direction.
4 Experimental Results and Analysis
The evaluation has been conducted on a database of 525 CCM images captured using the HRT-III1 microscope from 69 subjects (20 controls and 49 diabetic patients). The resolution is 1.0417μm and the field of view is 400 × 400μm2 of the cornea. For each individual, several fields of view are selected manually from near the centre of the cornea that show recognisable nerve-fibres. Using the Neuropathy Disability Score (NDS) [1], 48 patients were categorised into four groups according to severity of neuropathy (asymptomatic: 0 ≥NDS≤ 2 (n = 26), mild: 3 ≥NDS≤ 5 (n = 9), moderate: 6 ≥NDS≤ 8 (n = 10) and severe: 9 ≥NDS≤ 10 (n = 3)).
The performance of all methods is obtained by validating the extracted nerve-fibres in comparison with an expert manual delineation using CCMetrics2. Only the raw response of each method is taken into account without any further post-processing operations or shade correction methods as shown in Figure (1). Binary images are obtained by a simple uniform thresholding operation that is followed by a thinning operation to achieve a one-pixel-wide skeleton image.
4.1 Comparison of Nerve-Fibre Detection Methods
Three measures have been used in order to quantify the evaluation: the false-positive (FPR), the true-positive (TPR) and the equal-error rate (EER), which is the average of optimal FPR and false-negative rate at minimal difference between both. The measurements are taken by comparing the generated skeleton at different threshold intervals of the methods’ responses with the manually delineated “ground-truth”. A tolerance of ±3.141μm (3 pixels) was allowed in determining coincidence between the ground-truth and the detected nerve-fibres. The Peak Signal to Noise Ratio (PSNR) is also used to evaluate the performance of all methods. The PSNR is computed with respect to the mean squared error of the detected nerve-fibres from the manual delineation. The practical implementations of the Hessian, the DTCWT and the Monogenic signal were obtained from public domain sources [12,14,13], while the rest are implemented by our research group.
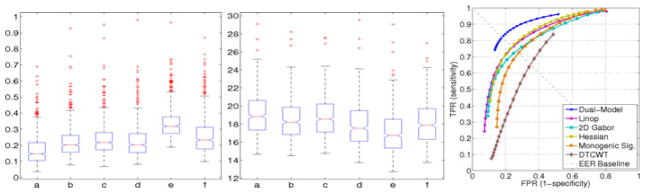
The EER and PSNR values for all the methods are presented in the box-plots in Figure 2 and Table 1. Each data point in Figure 2 corresponds to the evaluation on one of the 525 CCM images in the database. The dual-model shows lower EER and higher PSNR than all other methods (Table 1). These improvements are statistically significant (p ≈ 0 using three different non-parametric tests). The table also shows that the standard deviations of both EER and PSNR are low for the dual-model, which indicates a more stable and robust behaviour. The closest competitor is Linop. The methods designed for linear structures perform rather better on this test than the more generic DTCWT and Monogenic signal methods.
Fig. 2.
From left to right, the box-plots of the EER and the PSNR are shown for all methods. The ROC curves are presented at the far right. The box-plots indicate the upper and the lower quartiles as well as the median (the bar) of the EER and PSNR values respectively; whiskers show the extent of the rest of the data while crosses indicate outliers for (a) dual-model, (b) Linop, (c) 2D Gabor, (d) Hessian, (e) DTCWT and (f) Monogenic.
Table 1.
A comparison of mean EER and PSNR and their standard deviations
| Dual-Model | Linop [4] | 2D Gabor [2,9] | Hessian [6] | DTCWT [11] | Monog. [5] | |
|---|---|---|---|---|---|---|
| EER[%] | 17.79± 10.58 | 22.65± 10.76 | 24.15± 10.74 | 23.14± 11.53 | 34.17± 10.43 | 26.50± 12.58 |
| PSNR[dB] | 19.08± 2.16 | 18.51± 2.09 | 18.80± 2.11 | 17.93± 2.27 | 17.00± 2.23 | 18.11± 2.20 |
The superior performance of the dual-model is borne out by the ROC curves of Figure 2, in which the dual model shows improved detection at all operation points.
4.2 Assessment of Clinical Utility Results
In previous studies, using manual measurement of nerve-fibres, several features have been used to quantify the CCM images, including nerve-fibre length (NFL): the total length of nerve-fibres measured in an image, nerve-fibre density: the total number of nerve-fibres per unit area and branch density: the number of fibre branches per unit area. Of these nerve-fibre length proved to be the most discriminating, and we use this measure here to compare automated with manual measurement of the nerve-fibre images.
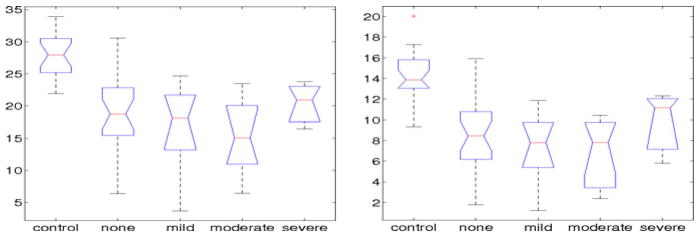
The box-plots in Figure 3 show a strong similarity between the manual and the automated analysis. However the scale of the NFL has slightly changed from (3.68–33.91) for the manual analysis to (5.67–26.53) for the automated analysis. ANOVA analysis results in a p-value for discrimination among these groups which is slightly higher for the automated than the manual analysis, though both are significant (p ≈ 0). The automated NFL measurements show a very strong correlation (r = 0.92) with the manual NFL values, which indicates that the automated system is successfully identifying the correct nerve-fibres. The coeffient of variation of the manual analysis is 0.34, reducing for the automated analysis to 0.29, which indicates more reliability and robustness of the results.
Fig. 3.
Box-plots showing the NFL scores for each of the NDS groups calculated manually (left) and automatically (right)
5 Conclusion
The analysis of CCM images requires the identification of fibre-like structures with low contrast in noisy images. This is a requirement shared by a number of imaging applications in biology, medicine and other fields. A number of methods have been applied in these applications, and we have compared some of these, and more generic methods with a dual-model detection algorithm devised for this study. The comparison used a large set of images with manual ground truth. In terms of both error-rates (pixel misclassification) and signal-to-noise ratio, the dual model achieved highest performance. It seems reasonable to propose that this filter is likely to prove equally useful in applications of a similar nature. The question of the clinical utility of the method was also addressed in this paper. The evaluation has shown that the automatic analysis is consistent with the manual ground truth with a correlation of (r = 0.92). Similarity in grouping control and patient subjects between manual and automated analysis was also achieved with (p ≈0). Therefore, it is sound to conclude that the automated analysis, which can be much quicker, is a potentially more reliable and practical alternative to manual analysis due to its consistency and immunity to the inter/intra-observer variabilities.
Footnotes
Heidelberg Engineering Inc., modified to acquire corneal confocal images.
CCMetrics is a purpose built interactive graphical interface which helps experts to manually delineate nerve-fibres in CCM images.
Contributor Information
M.A. Dabbah, Email: m.a.dabbah@manchester.ac.uk.
J. Graham, Email: jim.graham@manchester.ac.uk.
I. Petropoulos, Email: ioannis.petropoulos@manchester.ac.uk.
M. Tavakoli, Email: mitra.tavakoli@manchester.ac.uk.
R.A. Malik, Email: rayaz.a.malik@manchester.ac.uk.
References
- 1.Abbott CA, Carrington AL, Ashe H, Bath S, Every LC, Griffths J, Hann AW, Hussein A, Jackson N, Johnson KE, Ryder CH, Torkington R, Ross EREV, Whalley AM, Widdows P, Williamson S, Boulton AJM. The north-west diabetes foot care study: incidence of, and risk factors for, new diabetic foot ulceration in a community-based patient cohort. Diabetic Medicine. 2002;19(5):377–384. doi: 10.1046/j.1464-5491.2002.00698.x. [DOI] [PubMed] [Google Scholar]
- 2.Dabbah MA, Graham J, Tavakoli M, Petropoulos Y, Malik RA. Nerve fibre extraction in confocal corneal microscopy images for human diabetic neuropathy detection using gabor filters. Medical Image Understanding and Analysis (MIUA) 2009 July;:254–258. [Google Scholar]
- 3.Daugman JG. Two-dimensional spectral analysis of cortical receptive field profiles. Vision Research. 1980;20(10):847–856. doi: 10.1016/0042-6989(80)90065-6. [DOI] [PubMed] [Google Scholar]
- 4.Dixon RN, Taylor CJ. Machine Aided Image Analysis. Institute of Physics; London: 1979. Automated asbestos fibre counting; pp. 178–185. [Google Scholar]
- 5.Felsberg M, Sommer G. The monogenic signal. IEEE Transactions on Signal Processing. 2001;49(12):3136–3144. [Google Scholar]
- 6.Frangi AF, Niessen WJ, Vincken KL, Viergever MA. Multiscale vessel enhancement filtering. In: Wells WM, Colchester ACF, Delp SL, editors. MICCAI 1998. LNCS. Vol. 1496. Springer; Heidelberg: 1998. pp. 130–137. [Google Scholar]
- 7.Hong L, Wan Y, Jain A. Fingerprint image enhancement: algorithm and performance evaluation. IEEE Transactions on Pattern Analysis and Machine Intelligence. 1998;20(8):777–789. [Google Scholar]
- 8.Hossain P, Sachdev A, Malik RA. Early detection of diabetic peripheral neuropathy with corneal confocal microscopy. The Lancet. 2005;366(9494):1340–1343. doi: 10.1016/S0140-6736(05)67546-0. [DOI] [PubMed] [Google Scholar]
- 9.Jain AK, Farrokhnia F. Unsupervised texture segmentation using gabor filters. Pattern Recognition. 1991;24(12):1167–1186. [Google Scholar]
- 10.Kallinikos P, Berbanu M, O’Donnell C, Boulton A, Efron N, Malik R. Corneal nerve tortuosity in diabetic patients with neuropathy. Investigative Ophthalmology & Visual Science. 2004;45(2):418–422. doi: 10.1167/iovs.03-0637. [DOI] [PubMed] [Google Scholar]
- 11.Kingsbury N. Complex wavelets for shift invariant analysis and filtering of signals. Applied and Computational Harmonic Analysis. 2001;10(3):234–253. [Google Scholar]
- 12.Kingsbury N. Dual-Tree Complex Wavelet Transform Pack. 2002 June; http://www-sigproc.eng.cam.ac.uk/~ngk/
- 13.Kovesi P. An implementation of Felsberg’s monogenic filters. 2005 August; http://www.csse.uwa.edu.au/~pk/research/matlabfns/
- 14.Kroon DJ, Schrijver M. Hessian based Frangi Vesselness filter. 2009 October; http://www.mathworks.co.uk/
- 15.Malik RA, Kallinikos P, Abbott CA, van Schie CHM, Morgan P, Efron N, Boulton AJM. Corneal confocal microscopy: a non-invasive surrogate of nerve fibre damage and repair in diabetic patients. Diabetologia. 2003;46(5):683–688. doi: 10.1007/s00125-003-1086-8. [DOI] [PubMed] [Google Scholar]
- 16.Rao AR. A taxonomy for texture description and identification. Springer; New York: 1990. [Google Scholar]
- 17.Ruggeri A, Scarpa F, Grisan E. Analysis of corneal images for the recognition of nerve structures. IEEE Conference of the Engineering in Medicine and Biology Society (EMBS); September 2006; pp. 4739–4742. [DOI] [PubMed] [Google Scholar]
- 18.Zwiggelaar R, Astley S, Boggis C, Taylor C. Linear structures in mammo-graphic images: Detection and classification. IEEE Transactions on Medical Imaging. 2004;23(9):1077–1086. doi: 10.1109/TMI.2004.828675. [DOI] [PubMed] [Google Scholar]