Summary
Ku heterodimer is essential for the repair of DNA double-strand breaks (DSBs) by non-homologous end-joining (NHEJ). Ku recruits XLF, also known as Cernunnos, to DSBs. Here we report domain analyses of Ku-XLF interaction. The heterodimeric domain of Ku was found to be sufficient for the recruitment of XLF to DSBs and for the interaction of Ku with XLF. A small C-terminal deletion of XLF completely abolished recruitment of XLF to DSBs and Ku-XLF interaction. This deletion also led to marked reduction of XLF-XRCC4 interaction although the XRCC4-binding site on the XLF N-terminal domain remained intact. These results demonstrate the significance of Ku-XLF interaction in the molecular assembly of NHEJ factors.
Keywords: Ku, XLF, Cernunnos, Non-homologous end-joining, DNA double-strand breaks
1. Introduction
In mammalian cells, non-homologous end-joining (NHEJ) is a major cellular pathway for the repair of a DNA double-strand break (DSB), which is one of the most cytotoxic lesions [1, 2]. NHEJ requires a set of essential factors, including Ku, a highly abundant nuclear protein in humans, which plays a pivotal role in NHEJ as a DSB sensor [3]. Ku is a heterodimeric protein consisting of Ku70 and Ku80 subunits, each of which contains a dimerization domain and a C-terminal domain (CTD) [3]. Thus, the Ku heterodimer is composed of three distinct structural moieties, namely a heterodimeric domain, Ku70 CTD and Ku80 CTD [4–6]. The heterodimeric domain forms a ring-shaped structure that binds a DNA end with high affinity [4]. Binding to a DNA end elicits structural changes in Ku, allowing it to associate with other NHEJ core factors. The catalytic subunit of DNA-dependent protein kinase (DNA-PKcs) forms a complex with Ku in a DNA-dependent manner, resulting in the formation of a functional DNA-PK [3, 7, 8]. DNA-PK phosphorylates various substrates, including DNA-PK itself, to regulate the timely progression of the NHEJ process [8]. A tight complex of XRCC4 and DNA-ligase IV ligates two DSB ends and completes NHEJ [9].
XLF/Cernunnos (hereafter referred to as XLF) binds to XRCC4 and stimulates the ligation of mismatched and noncohesive DNA ends [10–13]. Deficiency of XLF in humans results in a genetic disorder characterized by immunodeficiency and increased radiosensitivity, which strongly indicate that XLF is an essential factor in NHEJ [11, 14]. We have shown that XLF quickly responds to DSB induction and accumulates at damaged sites in vivo, which was shown by a live cell imaging technique coupled with site-directed generation of DSBs using laser microbeam [15]. We have also shown that Ku is the sole factor essential for recruitment of XLF to DSBs and that XRCC4 augments the accumulation of XLF at DSBs. Using purified proteins of Ku and XLF, we have shown that XLF associates with Ku in the presence of DNA ends [15]. Our previous observations suggested that, in addition to stimulation of DNA ligation, XLF might play a role in protein assembly in the NHEJ pathway [15–17]. Recently, two independent studies have reported crystal structure of XLF [18, 19]. According to these, XLF forms a homodimer consisting of an N-terminal globular head domain and a C-terminal helical tail region. The N-terminal globular head domain has been reported to be the site of the interaction with XRCC4 [20], while the biological functions of the C-terminal helical region are still obscure [21].
In this study, we analyzed protein domains required for Ku-XLF interaction. Our data indicate that XLF associates with the heterodimeric domain of Ku. The most C-terminal region of XLF is essential for the recruitment of XLF to DSBs in vivo as well as for the association with Ku. Furthermore, a small deletion in the XLF C-terminal region affected the interaction of XLF with XRCC4. Our data substantiate the notion that Ku-XLF interaction is critical for protein assembly in the NHEJ pathway and define the C-terminal region of XLF as the module for this interaction.
2. Materials and Methods
2.1. Plasmid construction
cDNAs for full-length and C-terminally deleted human Ku70 and Ku80 were cloned into pcDNA3 carrying a FLAG epitope and pFastBac (Invitrogen) using standard DNA cloning methods. pFastBac plasmids were introduced into DH10Bac (Invitrogen), and recombinant baculoviral plasmids were selected according to the manufacturer's instructions. For expression of XLF C-terminal deletions, a series of XLF cDNA fragments were cloned into either pcDNA3 (Invitrogen) carrying a FLAG epitope and a SV40 nuclear localization signal (NLS) for immunoprecipitation, or into pcDNA3 harboring a FLAG epitope, NLS and a yellow fluorescent protein (YFP) for live cell imaging analyses. Plasmid constructions were confirmed by sequencing. Other baculoviral and mammalian expression plasmids for human XLF have been described previously [15].
2.2. Protein expression and purification
Recombinant baculoviral plasmids containing Ku cDNA were transfected into Sf9 cells using CellFectin (Invitrogen) to produce viruses. Baculovirus was collected from the culture media of the transfected Sf9 cells and amplified by standard procedures. Ku heterodimer was expressed in Sf9 cells by co-infection of two baculoviruses carrying human Ku80 and Ku70 cDNAs and purified as reported previously [22]. His-tagged human XLF was expressed in Sf9 insect cells and purified by sequential column chromatography using Ni-agarose, Superdex200 gel filtration and MonoQ columns [15].
2.3. Electrophoresis mobility shift assay (EMSA)
A 65-bp DNA fragment was amplified from a green fluorescent protein gene using PCR and purified with a PCR purification kit (QIAGEN). The 5' ends of the DNA fragment were labeled with 32P using T4 polynucleotide kinase. The 32P-labeled probe was combined with the purified Ku heterodimer and XLF proteins in a solution containing 10 mM Tris-Cl (pH 8.0), 150 mM NaCl, 5 mM EDTA, and 10% glycerol at room temperature for 20 min. DNA-protein complexes were resolved by nondenaturing polyacrylamide gel electrophoresis and detected by autoradiography.
2.4. Cell culture, transfection and live cell imaging
Embryonic fibroblasts derived from a Ku70 knockout mouse (Ku70−/− SV40) were kindly provided by Hatsumi Nagasawa, Colorado State University. Ku70−/− SV40, Ku80-defective xrs6, U2OS and 293T cells were maintained in αMEM supplemented with 10% fetal bovine serum (Hyclone) and penicillin/streptomycin (Sigma-Aldrich) in a humidified atmosphere with 5% CO2 at 37°C. Transient transfection of expression plasmids was performed using FuGENE6 (Roche) according to the manufacturer's instructions. DSBs were generated in the cell nuclei of transfected cells by irradiation with a laser microbeam at 365 nm, and the behavior of YFP-XLF proteins in living cells was monitored by time-lapse imaging as described previously [23].
2.5. Immunoprecipitation
FLAG-XLF expression plasmids were introduced into human 293T cells using the FuGENE6 reagent (Roche). The transfected cells were collected 48 h after transfection and lysed in a lysis buffer containing 20 mM Tris-Cl (pH 7.5), 150 mM NaCl, 5 mM EDTA, 1% Igepal and a protease inhibitor cocktail (Complete, Roche). The lysates were cleared by centrifugation at 20,000 × g for 10 min and subsequently incubated with anti-FLAG monoclonal antibody (M2, Sigma-Aldrich) conjugated with protein G agarose beads at 4°C for 3 h. Antigen-antibody complexes were collected by brief centrifugation and washed three times with lysis buffer. Immunoprecipitated proteins were eluted by boiling in SDS-sample buffer and analyzed by SDS-polyacrylamide gel electrophoresis followed by Western blotting using anti-FLAG M2 , anti-Ku80 and anti-XRCC4 antibodies as described previously [15].
3. Results
3.1. XLF binds to the heterodimeric domain of Ku
Ku is a heterodimer of Ku70 and Ku80, containing three structural domains that include a heterodimeric domain, Ku70 CTD, and Ku80 CTD (Fig. 1A) [4–6]. We tested whether CTDs are required for the recruitment of XLF to DSBs. We transiently co-transfected an expression plasmid for YFP-XLF with an expression plasmid for either full-length or C-terminally deleted Ku70 into Ku70-deficient cells. Under our experimental conditions, the amount of the exogenously expressed full-length Ku70 protein was comparable to that of C-terminally deleted Ku70 protein, and the amounts of YFP-XLF were equal in three transfected cells (Supplementary Fig. 1A). Using these transfected cells, DSBs were generated in a defined area of the nucleus by irradiation with a laser microbeam. The behavior of YFP-XLF immediately after laser microirradiation was monitored by a live cell imaging technique. When the expression plasmid for YFP-XLF was cotransfected with a vacant vector into Ku70-deficient cells, YFP-XLF failed to accumulate at laser-induced DSBs (Fig. 1B), consistent with our previous observation that Ku is essential for the recruitment of XLF to DSBs. When YFP-XLF was coexpressed with full-length Ku70 in Ku70-deficient cells, YFP-XLF rapidly accumulated at DSBs. When Ku70 without its CTD was coexpressed with YFP-XLF, we observed similar rapid accumulation of YFP-XLF at DSBs (Fig. 1B and C), indicating that the Ku70 CTD is nonessential for the recruitment of XLF to DSBs in vivo.
Fig. 1. Ku CTDs are nonessential for the recruitment of XLF to DSBs in vivo.
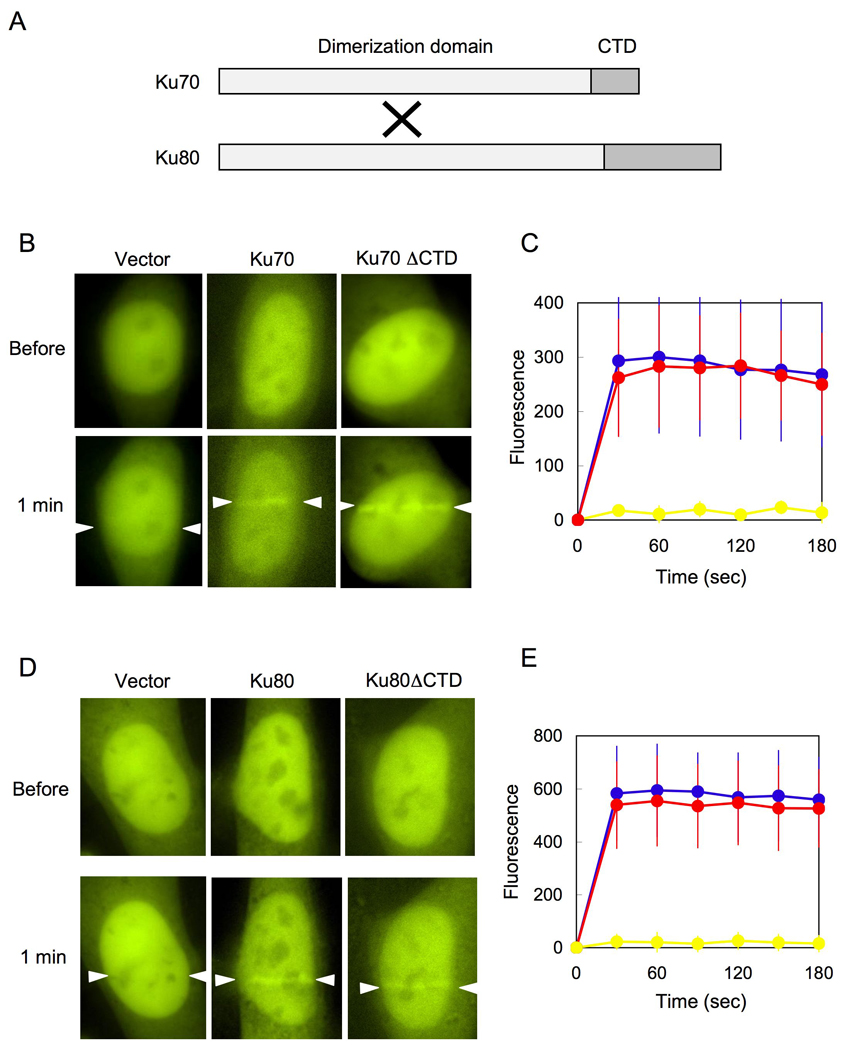
(A) Domains of human Ku70 and Ku80. Each protein consists of a dimerization domain and a CTD. Ku70 and Ku80 form a heterodimer that is composed of three distinct structural moieties, namely a heterodimeric domain, Ku70 CTD and Ku80 CTD.
(B) The Ku70 CTD is nonessential for the accumulation of YFP-XLF at DSBs. An expression plasmid for YFP-XLF was cotransfected with expression plasmids for full-length Ku70 (Ku70) or Ku70 lacking CTD (Ku70ΔCTD) into Ku70-deficient cells. The nuclei of the transfected cells were irradiated with a laser microbeam (indicated by white arrows). Microscopy images of YFP-XLF were taken before and after laser microirradiation.
(C) A time course of accumulation of YFP-XLF at laser-induced DSBs in Ku70-deficient cells transfected with a vacant vector (yellow), expression plasmids for full-length Ku70 (blue) and Ku70ΔCTD (red). Images were obtained at 30 sec intervals for 180 sec, and fluorescence intensities at the damage sites were quantified. Mean values of the fluorescence intensities with standard deviation were calculated from ten independent measurements.
(D) The Ku80 CTD is nonessential for the accumulation of YFP-XLF at DSBs. An expression plasmid for YFP-XLF was cotransfected with expression plasmids for full-length Ku80 (Ku80) or Ku80 lacking CTD (Ku80ΔCTD) into Ku80-deficient cells. Accumulation of YFP-XLF at DSBs was examined as described in (B).
(E) A time course of accumulation of YFP-XLF at laser-induced DSBs in Ku80-deficient cells transfected with a vacant vector (yellow), expression plasmids for full-length Ku80 (blue) and Ku80ΔCTD (red). Images were obtained at 30 sec intervals for 180 sec, and fluorescence intensities at the damage sites were quantified. Mean values of the fluorescence intensities with standard deviation were calculated from ten independent measurements.
Next, we tested the requirement of the Ku80 CTD for XLF recruitment to DSBs. We cotransfected the YFP-XLF expression plasmid with a vacant vector, an expression plasmid for full-length Ku80, or a plasmid for Ku80 without its CTD into Ku80-deficient cells. As shown in Supplementary Fig. 1B, the expression of full-length Ku80 was equivalent to that of Ku80 without CTD, and the amounts of YFP-XLF in the transfected cells were roughly equal. We used these cells for live cell imaging and observed rapid accumulation of YFP-XLF at laser-induced DSBs in Ku80-deficient cells transiently expressing either full-length or C-terminally deleted Ku80, but not in cells cotransfected with a vacant vector (Fig. 1D and E). Taken together, these results indicate that the CTDs of both Ku70 and Ku80 are nonessential for the recruitment of XLF to DSBs and suggest that the central heterodimeric domain of Ku is sufficient for the DSB recognition of XLF in vivo.
We further confirmed these observations in vivo by EMSA to examine the direct interaction of XLF with Ku lacking both Ku70- and Ku80-CTDs. We prepared purified Ku heterodimer consisting only of the heterodimeric domain by co-expressing each subunit lacking its CTD in insect cells. Purified Ku and XLF proteins were mixed with a 32P-labeled 65-bp DNA probe, and the complex formation between Ku and XLF on DNA was analyzed by EMSA. As reported previously, XLF has the unique feature of not associating with a short DNA such as a 65-bp DNA fragment (Fig. 2 lane 2) [15, 24]. When Ku and the probe were mixed, we observed two shift bands, representing a DNA bound to one Ku molecule and a DNA bound to two Ku molecules (Fig. 2 lane 3). The inclusion of XLF in the Ku-DNA mixture resulted in the appearance of an additional supershift band (Fig. 2 lane 4), which we have previously demonstrated to be a complex consisting of Ku, XLF and DNA [15]. When the same reactions were performed using C-terminally deleted Ku, we observed a similar pattern of two Ku-DNA bands (Fig. 2 lane 5) with an additional supershift band (Fig. 2 lane 6). When BSA was substituted for XLF, no supershift bands were observed (Fig. 2 lane 7). This further supports the idea that XLF binds to the heterodimeric domain of Ku.
Fig. 2. Ku heterodimer without CTDs forms a complex with XLF on DNA.
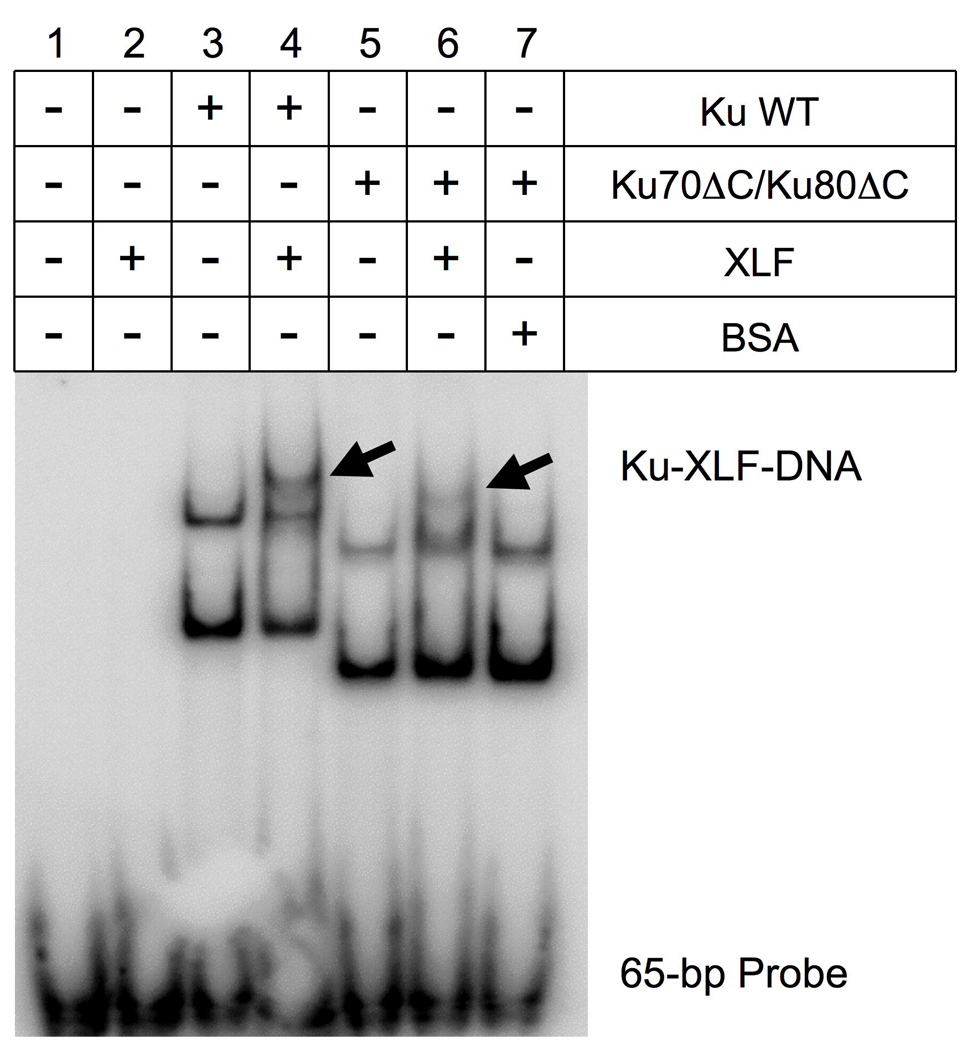
A 32P-labeled 65-bp DNA probe was mixed with purified proteins in the combinations indicated. Complex formation of the proteins and the probe was analyzed by polyacrylamide gel electrophoresis followed by autoradiography. Arrows indicate the bands containing Ku-XLF-DNA complexes.
3.2. The C-terminal region of XLF is essential for DSB recognition in vivo
Next, we analyzed which region of XLF is required for its recruitment to DSBs in vivo. XLF consists of an N-terminal globular domain and a C-terminal helical tail region [18, 19]. The N-terminal domain forms a globular structure, which is the site of XRCC4 interaction [20] (Fig. 3A). Because deletion of the N-terminal domain seems to cause gross structural changes in XLF homodimer and consequently seems to abrogate most physiological functions, we examined only C-terminal deletions (Fig. 3B). Since a consensus sequence of NLS was found in the C-terminal region of XLF, we cloned a series of XLF deletion cDNAs into a plasmid carrying YFP fused with SV40 NLS. We transiently transfected these plasmids into cultured human cells and examined whether the C-terminally deleted YFP-XLF proteins could be recruited to DSBs induced by laser microirradiation. We observed that all C-terminally deleted YFP-XLF proteins examined in this study failed to accumulate at DSBs in the nucleus (Fig. 3B). Remarkably, deletion of 10 amino acids at the C-terminal (XLF aa 1–289) completely abolished the recruitment of XLF to DSBs (Fig. 3C and D), although the amounts of YFP-XLF and YFP-XLF (aa 1–289) in the transefected cells were nearly identical (Supplementary Fig. 1C). This observation indicates that the C-terminal region of XLF plays a critical role in the recruitment of XLF to DSBs in vivo. We found that the C-terminal region of XLF are evolutionarily conserved among vertebrate and mammalian species (Fig. 3E), which further suggests the biological importance of the XLF C-terminal region.
Fig. 3. The end of the C-terminal region is essential for the recruitment of XLF to DSBs in vivo.
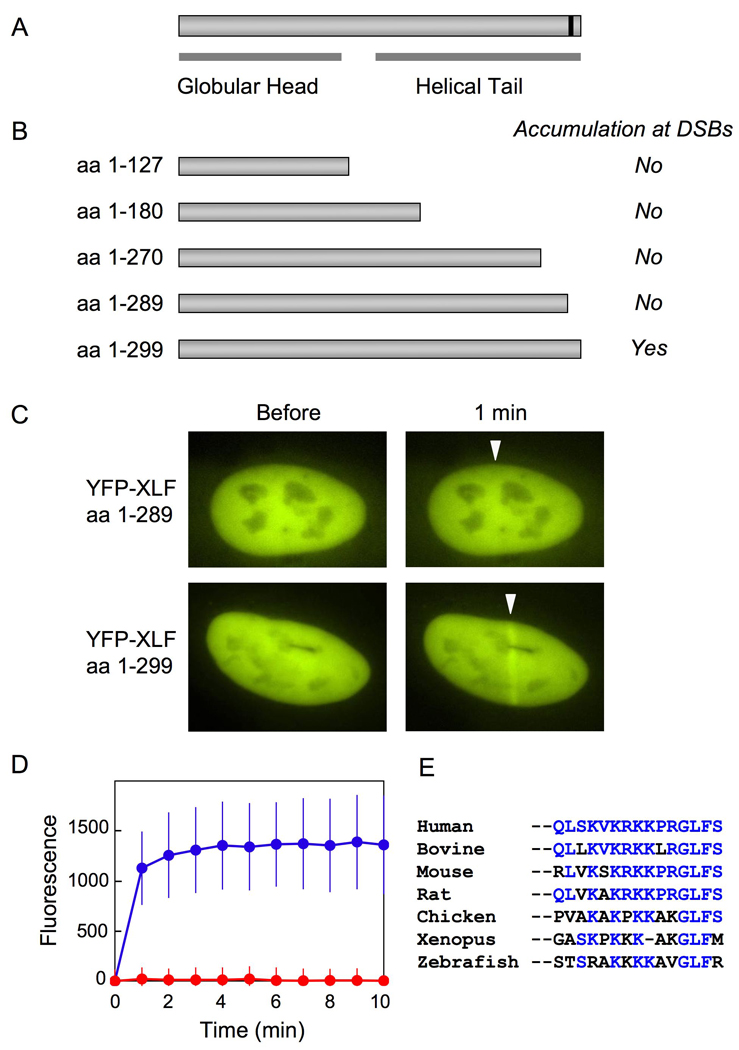
(A) Human XLF consists of an N-terminal globular head domain and a C-terminal helical tail region. A putative NLS at the C-terminal end is marked by a black line.
(B) Full-length and C-terminally deleted XLF proteins that were fused to SV40 NLS, YFP, and a FLAG tag at their N-terminal ends were transiently expressed in U2OS cells. Accumulation of the YFP-XLF deletions at laser-induced DSBs was examined via live cell imaging. The results were summarized on the right.
(C) Typical examples of laser-irradiated nuclei containing full-length YFP-XLF (aa 1–299) and C-terminally deleted YFP-XLF (aa 1–289). White arrows indicate sites of laser microirradiation.
(D) A time course of accumulation of full-length (blue) and C-terminally deleted (aa 1–289, red) YFP-XLF at laser-induced DSBs. Images were obtained at 1 min intervals for 10 min. Mean values of the fluorescence intensities with standard deviation were calculated from ten independent measurements.
(E) Evolutionary conservation of the C-terminal region of XLF. The last 15 amino acid sequences of XLF proteins from seven species are aligned. Amino acid residues identical to those of the human XLF are highlighted in blue.
3.3. The C-terminal region of XLF is required for interaction with Ku
Since Ku is essential for the recruitment of XLF to DSBs [15], we next examined whether the C-terminally deleted XLF associates with Ku. We expressed a series of FLAG-tagged XLF deletions in 293T cells and performed immunoprecipitation analyses. When full-length XLF was transiently expressed in 293T cells and subsequently immunoprecipitated, we detected immunoprecipitated FLAG-XLF along with coprecipitation of endogenous Ku and XRCC4 (Fig. 4 lane 8). However, when the C-terminal deletions of XLF were used for immunoprecipitation, the endogenous Ku was not coprecipitated with any of the XLF deletions (Fig. 4 lanes 6 and 7). This observation, along with imaging analyses of XLF C-terminal deletions, indicates that the XLF C-terminal region is essential for Ku-mediated XLF recruitment to DSBs. Interestingly, although the C-terminal deleted FLAG-XLF proteins contain an intact N-terminal globular domain that is the site of XRCC4 interaction, these deletions resulted in markedly reduced amounts of coprecipitated endogenous XRCC4 (Fig. 4 lanes 6 and 7). This observation suggests that Ku-XLF interaction affects the association of XLF with XRCC4.
Fig. 4. Immunoprecipitation analyses of C-terminally deleted XLF.
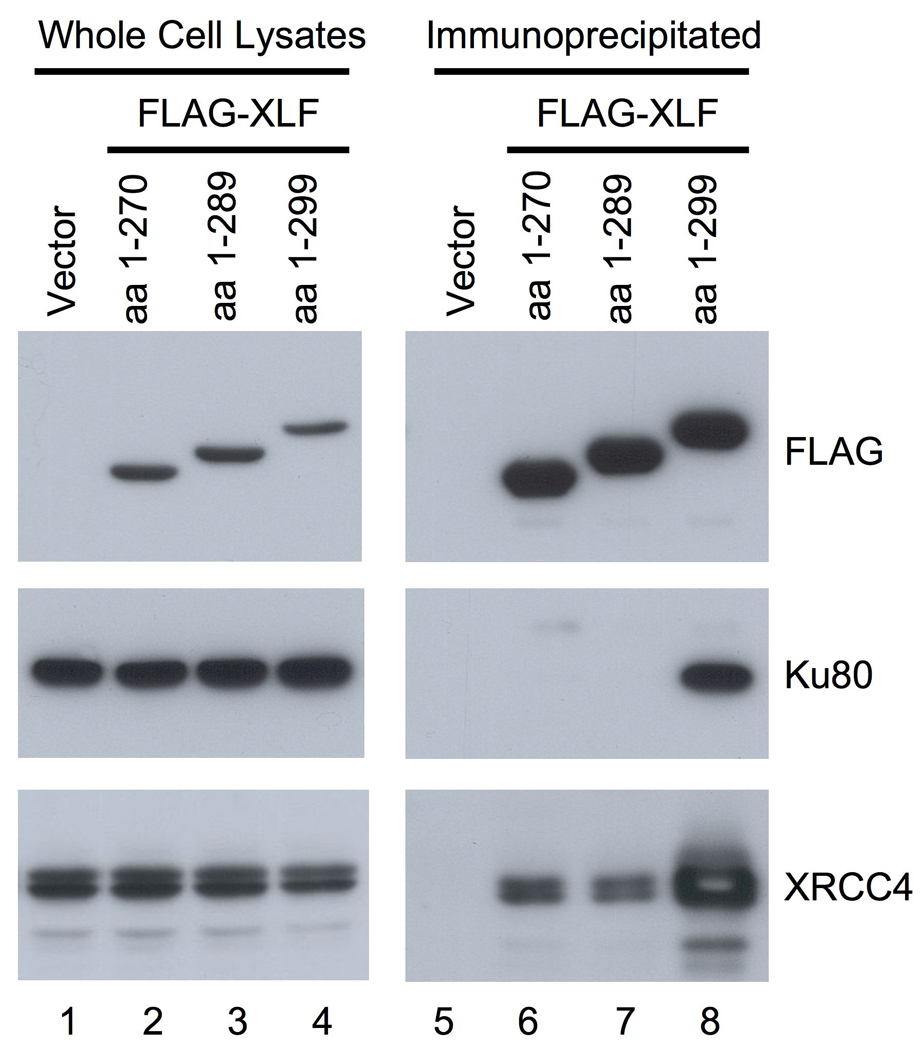
Full-length and C-terminally deleted XLF proteins with SV40 NLS and a FLAG tag fused to their N-terminal ends were transiently expressed in 293T cells. Whole cell lysates were prepared from the transfected cells and subjected to immunoprecipitation using an anti-FLAG antibody. Proteins in the immunoprecipitated samples were examined by Western blotting using antibodies against FLAG, Ku80 and XRCC4.
4. Discussion
In this study, we performed domain analyses of Ku-XLF interaction. We found that the heterodimeric domain of Ku is sufficient for the recruitment of XLF to DSBs in vivo and for the association of Ku with XLF in vitro. Previously, we have demonstrated that highly purified forms of Ku and XLF do not associate with each other and that Ku-XLF interaction occurs only in the presence of DNA. The heterodimeric domain of Ku forms a ring-shaped structure that has extremely high affinity for DNA ends [3, 4]. We speculate that XLF recognizes subtle conformational changes of Ku elicited by DNA-end binding and is recruited to Ku-bound DSBs. The DNA-dependent protein interactions of Ku-XLF [15] and Ku-DNA-PKcs [7] appear to be key steps in the initiation of protein assembly for NHEJ in vivo.
It has been reported that multiple protein-protein interactions are biochemically detectable among the core factors of NHEJ including Ku, DNA-PKcs, XRCC4 and XLF [7, 10, 15, 25]. In our previous study employing live cell imaging of protein dynamics, we observed that the deletion of one NHEJ factor affects the dynamic behavior of other NHEJ factor at DSBs in living cells [15, 16]. Thus, we have proposed that the multiple interactions play critical roles in the assembly of functional NHEJ machinery in vivo [17]. In this study, we observed in immunoprecipitation that the association of XRCC4 with C-terminally truncated XLF is markedly weakened despite the presence of an intact XRCC4-interaction site on the N-terminal globular domain. We interpret this result as an indication that Ku, XRCC4 and XLF stabilize one another at DSBs via multiple protein interactions and that the lack of Ku-XLF interaction leads to a less stable association of this complex with XRCC4. This finding lends further support to our previous model of NHEJ factor assembly [17].
It is currently known that the major function of XLF in NHEJ is stimulation of the ligation activity of XRCC4/DNA-ligase IV [12, 13] and that the N-terminal globular domain of XLF has been defined as the site of XRCC4 binding [20]. Although the functional roles of the C-terminal region of XLF is not yet clear [21], our study suggests that it is required for normal XLF function in NHEJ. Strikingly, a 10-amino acid deletion at the end of the C-terminal region completely abolishes the Ku-XLF interaction and the accumulation of XLF at DSBs, suggesting the critical role of the XLF C-terminal region as the module for the Ku-XLF interaction. Taken together with the evolutionary conservation of the XLF C-terminal region among vertebrate and mammalian species, this result strongly suggests the involvement of the C-terminal region of XLF in regulating NHEJ. Further studies will be performed to complete understanding of the functional role of XLF in NHEJ.
Supplementary Material
Acknowledgements
This work was supported by grants from the National Institute of Health to D.J.C. (CA050519 and CA92584) and by Grants-in-Aid for Scientific Research (C) from MEXT, Japan, Sagawa Foundation for Promotion of Cancer Research, and the Mochida Memorial Foundation for Medical and Pharmaceutical Research to K.Y.
Abbreviations
- CTD
C-terminal domain
- DNA-PKcs
catalytic subunit of DNA-dependent protein kinase
- DSB
DNA double-strand break
- NHEJ
non-homologous end-joining
- NLS
nuclear localization signal
- YFP
yellow fluorescent protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lees-Miller SP, Meek K. Repair of DNA double strand breaks by non-homologous end joining. Biochimie. 2003;85:1161–1173. doi: 10.1016/j.biochi.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 2.Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 3.Downs JA, Jackson SP. A means to a DNA end: the many roles of Ku. Nat Rev Mol Cell Biol. 2004;5:367–378. doi: 10.1038/nrm1367. [DOI] [PubMed] [Google Scholar]
- 4.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Z, Zhu L, Lin D, Chen F, Chen DJ, Chen Y. The three-dimensional structure of the C-terminal DNA-binding domain of human Ku70. J Biol Chem. 2001;276:38231–38236. doi: 10.1074/jbc.M105238200. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Z, Hu W, Cano L, Lee TD, Chen DJ, Chen Y. Solution structure of the C-terminal domain of Ku80 suggests important sites for protein-protein interactions. Structure. 2004;12:495–502. doi: 10.1016/j.str.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 7.Dynan WS, Yoo S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 1998;26:1551–1559. doi: 10.1093/nar/26.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meek K, Gupta S, Ramsden DA, Lees-Miller SP. The DNA-dependent protein kinase: the director at the end. Immunol Rev. 2004;200:132–141. doi: 10.1111/j.0105-2896.2004.00162.x. [DOI] [PubMed] [Google Scholar]
- 9.Critchlow SE, Bowater RP, Jackson SP. Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr Biol. 1997;7:588–598. doi: 10.1016/s0960-9822(06)00258-2. [DOI] [PubMed] [Google Scholar]
- 10.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 11.Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche MC, Sanal O, Plebani A, Stephan JL, Hufnagel M, le Deist F, Fischer A, Durandy A, de Villartay JP, Revy P. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 12.Gu J, Lu H, Tsai AG, Schwarz K, Lieber MR. Single-stranded DNA ligation and XLF-stimulated incompatible DNA end ligation by the XRCC4-DNA ligase IV complex: influence of terminal DNA sequence. Nucleic Acids Res. 2007;35:5755–5762. doi: 10.1093/nar/gkm579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsai CJ, Kim SA, Chu G. Cernunnos/XLF promotes the ligation of mismatched and noncohesive DNA ends. Proc Natl Acad Sci U S A. 2007;104:7851–7856. doi: 10.1073/pnas.0702620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai Y, Kysela B, Hanakahi LA, Manolis K, Riballo E, Stumm M, Harville TO, West SC, Oettinger MA, Jeggo PA. Nonhomologous end joining and V(D)J recombination require an additional factor. Proc Natl Acad Sci U S A. 2003;100:2462–2467. doi: 10.1073/pnas.0437964100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yano K, Morotomi-Yano K, Wang SY, Uematsu N, Lee KJ, Asaithamby A, Weterings E, Chen DJ. Ku recruits XLF to DNA double-strand breaks. EMBO Rep. 2008;9:91–96. doi: 10.1038/sj.embor.7401137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yano K, Chen DJ. Live cell imaging of XLF and XRCC4 reveals a novel view of protein assembly in the non-homologous end-joining pathway. Cell Cycle. 2008;7:1321–1325. doi: 10.4161/cc.7.10.5898. [DOI] [PubMed] [Google Scholar]
- 17.Yano K, Morotomi-Yano K, Adachi N, Akiyama H. Molecular mechanism of protein assembly on DNA double-strand breaks in the non-homologous end-joining pathway. J Radiat Res (Tokyo) 2009;50:97–108. doi: 10.1269/jrr.08119. [DOI] [PubMed] [Google Scholar]
- 18.Andres SN, Modesti M, Tsai CJ, Chu G, Junop MS. Crystal structure of human XLF: a twist in nonhomologous DNA end-joining. Mol Cell. 2007;28:1093–1101. doi: 10.1016/j.molcel.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Chirgadze DY, Bolanos-Garcia VM, Sibanda BL, Davies OR, Ahnesorg P, Jackson SP, Blundell TL. Crystal structure of human XLF/Cernunnos reveals unexpected differences from XRCC4 with implications for NHEJ. EMBO J. 2008;27:290–300. doi: 10.1038/sj.emboj.7601942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malivert L, Ropars V, Nunez M, Drevet P, Miron S, Faure G, Guerois R, Mornon JP, Revy P, Charbonnier JB, Callebaut I, de Villartay JP. Delineation of the Xrcc4-interacting region in the globular head domain of cernunnos/XLF. J Biol Chem. 2010;285:26475–26483. doi: 10.1074/jbc.M110.138156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malivert L, Callebaut I, Rivera-Munoz P, Fischer A, Mornon JP, Revy P, de Villartay JP. The C-terminal domain of Cernunnos/XLF is dispensable for DNA repair in vivo. Mol Cell Biol. 2009;29:1116–1122. doi: 10.1128/MCB.01521-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woodard RL, Lee KJ, Huang J, Dynan WS. Distinct roles for Ku protein in transcriptional reinitiation and DNA repair. J Biol Chem. 2001;276:15423–15433. doi: 10.1074/jbc.M010752200. [DOI] [PubMed] [Google Scholar]
- 23.Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G, Mari PO, van Gent DC, Chen BP, Chen DJ. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J Cell Biol. 2007;177:219–229. doi: 10.1083/jcb.200608077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu H, Pannicke U, Schwarz K, Lieber MR. Length-dependent binding of human XLF to DNA and stimulation of XRCC4: DNA ligase IV activity. J Biol Chem. 2007;282:11155–11162. doi: 10.1074/jbc.M609904200. [DOI] [PubMed] [Google Scholar]
- 25.Costantini S, Woodbine L, Andreoli L, Jeggo PA, Vindigni A. Interaction of the Ku heterodimer with the DNA ligase IV/Xrcc4 complex and its regulation by DNA-PK. DNA Repair (Amst) 2007;6:712–722. doi: 10.1016/j.dnarep.2006.12.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


