Abstract
O-Methyltransferases, which catalyze the production of small molecules in plants, play a crucial role in determining biosynthetic pathways in secondary metabolism because of their strict substrate specificity. Using three O-methyltransferase (OMT) cDNAs that are involved in berberine biosynthesis, we investigated the structure that was essential for this substrate specificity and the possibility of creating a chimeric enzyme with novel substrate specificity. Since each OMT has a relatively well-conserved C-terminal putative S-adenosyl-L-methionine-binding domain, we first exchanged the N-terminal halves of different OMTs. Among the 6 combinations that we tested for creating chimeric OMTs, 5 constructs produced detectable amounts of recombinant proteins, and only one of these with an N-terminal half of 6-OMT and a C-terminal half of 4′-OMT (64′-OMT) showed methylation activity with isoquinoline alkaloids as a substrate. Further enzymological analysis of 64′-OMT reaction product indicated that 64′-OMT retained the regio-specificity of 6-OMT. Further examination of the N-terminal region of 64′-OMT showed that about 90 amino acid residues in the N-terminal half were critical for reaction specificity. The creation of OMTs with novel reactivity is discussed.
Keywords: Coptisjaponica, isoquinoline alkaloid, O-methyltransferase, chimeric enzyme
Introduction
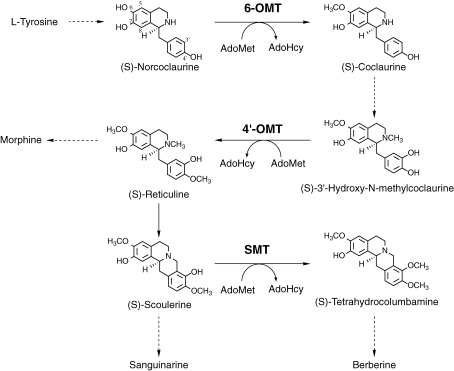
Methyltransferases are crucial for directing intermediates to specific biosynthetic pathways in plant secondary metabolism.1) We have been studying the O-methyltransferases (OMTs) in isoquinoline alkaloid biosynthesis, since these alkaloids include pharmaceutically important alkaloids such as analgesic morphinan alkaloids (e.g. morphine), anti-microbial berberine alkaloids (e.g. berberine), and anti-microbial benzophenanthridine alkaloids (e.g. sanguinarine). In the biosynthetic pathway of berberine, three OMTs [S-adenosyl-L-methionine:norcoclaurine 6-O-methyltransferase (6-OMT);2–4) S-adenosyl-L-methionine:3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase (4′-OMT)4,5) and S-adenosyl-L-methionine:scoulerine 9-O-methyltransferase (SMT)6–8)] have been isolated and characterized (Fig. 1 ). Each OMT shows strict substrate specificity, despite the structural similarities of the various substrates. Isolation of the cDNA of OMT from cultured Coptis japonica cells4) and successful expression with other biosynthetic genes in Escherichia coli clearly indicated that the reaction specificities of OMTs determine the biosynthetic pathway of reticuline, an important intermediate in berberine biosynthesis.9)
Figure 1.
Schematic biosynthetic pathway for a variety of isoquinoline alkaloids. 6-OMT, S-adenosyl-L-methionine:norcoclaurine 6-O-methyltransferase; 4′-OMT, S-adenosyl-L-methionine:3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase; SMT, S-adenosyl-L-methionine:scoulerine 9-O-methyltransferase; AdoMet, S-adenosyl-L-methionine; AdoHcy, S-adenosyl-L-homocysteine.
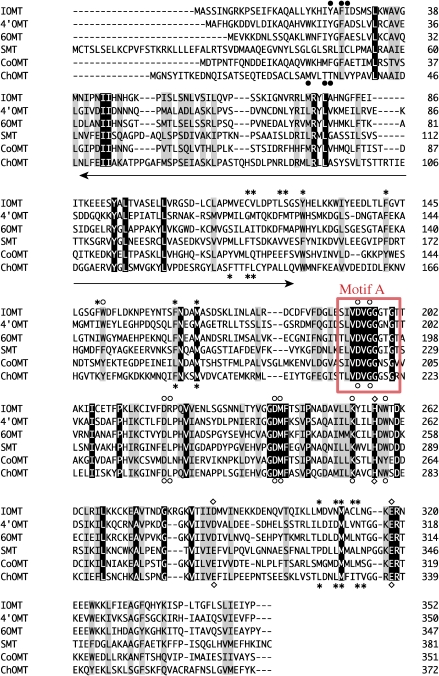
Comparisons with the sequences of plant OMTs have shown that while the C-terminal ends of these OMTs are highly conserved for putative S-adenosyl-L-methionine (AdoMet)-binding,10) the N-terminal ends are more divergent, even among these related OMTs (Fig. 2 ). Recent reports on the three-dimensional structures of several plant OMTs have provided further evidence that the conserved C-terminal domains are actually involved in AdoMet binding.11,12) Furthermore, catalytic residues are also conserved in the C-terminal region. Whereas the substrate-binding residues are dispersed in both the N- and C-terminals, the C-terminal domain is more conserved. These findings suggest that the N-terminal half plays an important role in substrate-binding and that modification of the N-terminal domain may change the substrate specificities of the enzymes.
Figure 2.
Amino acid sequence alignment of Coptis and flavonoid OMTs. The substrate-binding residues (asterisk), AdoMet-binding residues (open circle), catalytic residues (square) and substrate-binding from the partner monomer (solid circle) are shown for S-adenosyl-L-methionine:isoflavone O-methyltransferase (IOMT) and S-adenosyl-L-methionine:chalcone O-methyltransferase (ChOMT).11) Amino acids conserved in all of the sequences are highlighted, and similar ones are shaded. The arrow indicates the 90-amino acid region that is believed to be important for alkaloid recognition. Motif A is boxed in red.
To examine this possibility, we constructed all possible expression vectors for chimeric OMTs from 6-OMT, 4′-OMT and SMT, and then expressed them in E. coli and characterized their reaction specificities. Whereas many constructs produced soluble protein, only one construct with an N-terminal half of 6-OMT and a C-terminal half of 4′-OMT showed methylation activity, and the results indicated that the N-terminal domain could determine the reaction specificity. Further examination of the N-terminal domain of chimeric enzyme showed that the 90 amino acids of the N-terminus were important for substrate recognition. A more detailed analysis of the chimeric enzyme with several substrates indicated that the chimeric enzyme had a unique reaction specificity that was distinct from those of the original 6-OMT and 4′-OMT. We discuss the creation of OMTs with novel reaction specificity in isoquinoline alkaloid biosynthesis.
Materials and methods
Chemicals.
(R,S)-Norlaudanosoline and (R,S)-laudanosoline were obtained from Sigma Aldrich. (R)- and (S)-Coclaurine were gifts from Drs. N. Nagakura and K. Iwasa of Kobe Women’s College of Pharmacy. The other alkaloids used were gifts from Mitsui Petrochemical Industries, Ltd.
Construction of expression vectors for chimeric OMTs with different N- and C-terminal halves.
Expression vectors for chimeric OMTs that consisted of N- and C-terminal halves from different OMTs were constructed without extra peptides derived from the vector sequence of pET-21d (Novagen). First, each OMT cDNA was subcloned into pBluescript II SK-4,8) and this construct was used for PCR amplification. The N-terminals of 4′-OMT, 6-OMT and SMT were amplified by PCR with the following forward and reverse primer pairs: 5′-GAGAATTAACATGTCTTTCCATGG-3′ (4′-ATG primer) and 5′-TCCACCGACGTCAACAAGTGAGTC-3′ (4′motif A RV primer) for 4′-OMT, 5′-CAGAATCATAGACATGTTAGTGAAGAAGAAGG-3′ (6-ATG primer) and 5′-CCACCACCGACGTCCACAAGTGTAG-3′ (6-motif A RV primer) for 6-OMT, and 5′-ACCAACCTATACATGTGCACTAGC-3′ (S-ATG primer) and 5′-GCGGATCCCCACCACCGACGTCAACCAACTC-3′ (S-motif A RV primer) for SMT. The forward primers were designed to introduce an Afl III site (ACPuPyGT) at the ATG start codon, and reverse primers were designed to introduce an Aat II site (GACGTC) and a BamH I site (GGATCC) with no modification of the amino acid sequence. Underlined bases were modified to introduce restriction sites. The C-terminal halves of 4′-OMT, 6-OMT and SMT were amplified with the M13 primer and the following respective forward primers: 5′-TCACTTGTTGACGTCGGTGGAGG-3′ (4′-motif A FW primer) for 4′-OMT, 5′-CACTTGTGGACGTCGGTGGTGG-3′ (6-motif A FW primer) for 6-OMT and 5′-CGGGATCCGTTGGTTGACGTCGGTGGTGG-3′ (S-motif A FW primer) for SMT. These primers were also designed to introduce Aat II and BamH I sites with no change in the amino acid sequence.
Chimeric constructs with the N-terminal half of 6-OMT and the C-terminal half of 4′-OMT (64′-OMT) or with the N-terminal half of 4′-OMT and the C-terminal half of 6-OMT (4′6-OMT) were produced as follows. PCR-amplified fragments for the N- and C-terminals were digested respectively with Afl III and Aat II, and with Aat II and Xho I, while pET-21d was digested with Nco I and Xho I. The digested N- and C-terminal fragments were mixed with pET-21d and ligated.
The N- and C-terminal fragments of SMT were subcloned into pET-21d vector using Nco I/BamH I and BamH I/Xho I sites, to produce pET-SN and pET-SC, respectively. Chimeric constructs containing the SMT C-terminus with the N-terminal half of 4′- or 6-OMT (i.e., 4′S- and 6S-OMT) were constructed in the Nco I/Aat II site of pET-SC by inserting PCR-amplified N-terminal fragments that were digested with Afl III and Aat II. Similarly, chimeric constructs (S4′- or S6-OMT) that contained the N-terminal half of SMT and the C-terminal half of 4′- or 6-OMT were constructed at the Aat II/Xho I site of pET-SN. All constructs were sequenced to confirm that there was no unexpected sequence modification during construction.
DNA shuffling between the 4′- and 6-OMT N-terminal halves.
To characterize the sequence that was essential for reaction specificity in the N-terminal halves of 6-OMT and 4′-OMT, these N-terminals were shuffled using PCR.13) pET-4′OMT4) and pET-64′OMT were first digested with Xba I and Xho I and each OMT insert was purified by gel electrophoresis and extracted from agarose gel. The 4′-OMT fragment was then digested with Sau3AI, and the 64′-OMT fragment was digested with Sau3A I and Nco I. The resultant DNAs (200 ng each) were mixed and randomly assembled by PCR without primers. PCR was carried out for 30 cycles under the following conditions with Tbr EXT DNA polymerase (Finnzymes, Espoo, Finland): denaturation at 94 ℃ for 30 sec; annealing at 55 ℃ for 30 sec; extension at 72 ℃ for 30 sec; and a final extension at 72 ℃ for 5 min. The PCR products were then amplified using Tbr EXT DNA polymerase with the following 4 primer pairs: (i) 4′-ATG and 6-motif A RV, (ii) 6-ATG and 4′-motif A RV, (iii) 4′-ATG and 4′ -motif A RV, and (iv) 6-ATG and 6-motif A RV primer. The thermal cycle program was 30 cycles of 94 ℃ for 30 sec, 52 ℃ for 40 sec and 72 ℃ for 1 min, and then 72 ℃ for 5 min of final extension. Each PCR product was digested with Afl III and Aat II, and ligated into the Nco I/Aat II site of pET-4′C that contained the C-terminal half of 4′-OMT in pET-21d. pET-4′C was constructed from pET-SC after being digested with Aat II and Xho I, and ligation of the C-terminal half of 4′-OMT into the resultant vector.
Heterologous expression of the chimeric OMTs in Escherichia coli.
The expression vectors for the chimeric OMTs were introduced into E. coli BL21 (DE3). The expression of chimeric OMTs was induced with 1 mM isopropylthiogalactoside. After incubation at 16 ℃ for 24 h, cells were harvested and sonicated in extraction buffer (0.1 M Tris-HCl, pH 7.5, containing 10% glycerol and 20 mM 2-mercaptoethanol). After centrifugation at 12,000g for 10 min, the supernatant was desalted through an NAP-5 column (Amersham Biosciences) and used to measure OMT activity toward the alkaloid substrates.
Assay of O-methylation activity.
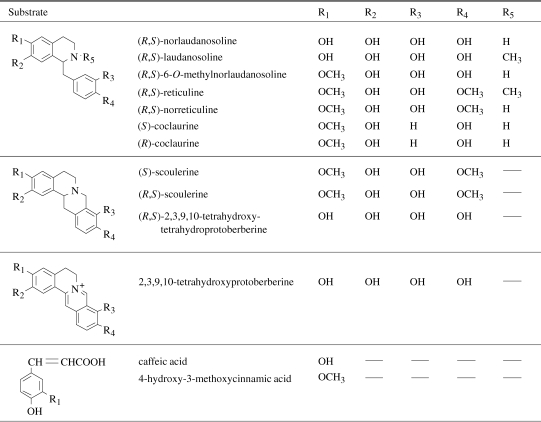
OMT activities were detected by HPLC and liquid chromatography-mass spectrography (LC/MS). The isoquinoline alkaloids and chemicals tested as methyl-group acceptors are listed in Table 1 . The standard OMT reaction mixture consisted of 0.3 M Tris-HCl (pH 7.5), 25 mM sodium ascorbate, 1 mM methyl-group acceptor, 1 mM AdoMet, and crude enzyme preparation (ca. 100 µg protein). The assay mixture was incubated at 30 ℃ for 1 h, after which the reaction was terminated by the addition of methanol. After protein precipitation, the reaction product was analyzed by reversed-phase HPLC (mobile phase, 20% methanol/H2O containing 1% acetic acid; column, LiChrospher 100 RP-18(e) (4 × 250 mm; MERCK); flow rate, 0.5 ml/min; detection, absorbance measurement at 280 nm). Mass ion chromatograms were obtained with an LC/MS-2000 (Shimadzu).
Table 1.
The isoquinoline alkaloids and chemicals tested as a methyl-group acceptor
 |
To quantify the enzymatic activity of chimeric OMTs, transfer of the 3H-labeled methyl group of S-adenosyl-L-[methyl-3H]methionine (0.5 MBq/µmol) (PerkinElmer) to the product was measured as described elsewhere.3)
Purification of chimeric OMTs from E. coli lysate.
64′- and 4′6-OMT were purified from 600 ml cultures of E. coli as described elsewhere.4) In brief, proteins were extracted in buffer (100 mM Tris-HCl, pH 7.5, containing 20 mM 2-mercaptoethanol and 10% glycerol) and purified with Q-Sepharose Fast Flow (Amersham Biosciences) and a Bio-Gel HTP column (Bio-Rad).
Rat liver catechol O-methyltransferase for the preparation of 6- or 7-O-methylated isoquinoline alkaloid.
Catechol OMT (COMT) was obtained from rat liver to prepare (R,S)-6- or 7-O-methylnorlaudanosoline as a standard as described by Meyerson et al.14) The reaction products were used for LC/MS analysis.
Other methods.
The subunit molecular mass of the enzyme was determined by SDS-PAGE (10% polyacrylamide), and the molecular mass of the native enzyme was determined by gel filtration chromatography through a Superose 12 column (Amersham Biosciences) in fast protein liquid chromatography. Protein concentrations were determined according to Bradford15) with bovine serum albumin as a standard. Chimeric OMTs were analyzed by immunoblotting with polyclonal antibodies that had been raised in rabbit against purified 4′-OMT or SMT.4,7)
Results
Expression of chimeric OMTs with N- and C-terminals derived from different OMTs in E. coli.
We constructed chimeric OMT vectors to examine the roles that the N- and C-terminal halves play in substrate- and reaction-specificities. Chimeric OMTs were connected at the middle of motif A, since motif A was the most conserved region in OMTs.10) Each N- and C-terminal half was amplified by PCR to introduce a common restriction site for ligation and chimeric OMTs were produced without any changes in the amino acid sequences of the connection. Hereafter, chimeric OMTs with the N-terminal half of 6-OMT and the C-terminal half of 4′-OMT or with the N-terminal half of SMT and the C-terminal half of 6-OMT, and so on, are referred to as 64′-OMT, S6-OMT, and so on.
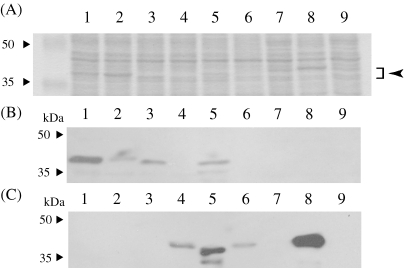
Recombinant OMTs were expressed in E. coli and the expression was monitored with anti-4′-OMT or anti-SMT polyclonal antibodies as well as Coomassie Brilliant Blue R250 (CBB)-staining after SDS-PAGE. Immunoblot analysis with anti-4′OMT and anti-SMT polyclonal antibodies indicated that the recombinant chimeric OMTs were successfully expressed in soluble form, except for 6S-OMT (Fig. 3 ), whereas CBB staining indicated that only some chimeric OMTs were accumulated at detectable levels. Immunoblot analysis also indicated that anti-4′-OMT antibodies could detect the N-terminal half of 4′-OMT more effectively, whereas anti-SMT antibodies detected both the N- and C-terminals.
Figure 3.
Expression of chimeric OMTs with N- and C-terminals derived from different OMTs. (A) The crude E. coli lysate (10 µg protein) expressing the chimeric OMTs was separated by SDS-PAGE and stained with Coomassie Brilliant Blue R250 (CBB). The molecular mass of the chimeric OMTs is ca. 40 kDa (indicated by arrow). (B) Immunoblot analysis with anti-4′-OMT polyclonal antibodies. (C) Immunoblot analysis with anti-SMT polyclonal antibodies. M, molecular mass markers; lane 1, pET-4′OMT; lane 2, pET-64′OMT; lane 3, pET-4′6OMT; lane 4, pET-S4′OMT; lane 5, pET-4′SOMT; lane 6, pET-S6OMT; lane 7, pET-6SOMT; lane 8, pET-SMT; lane 9, pET-21d (vector control).
Since the expression levels of chimeric OMTs differed, we tried to measure the OMT activities for various substrates (Table 1). The OMT assays with 3H-labeled AdoMet clearly showed that only cell lysate of recombinant E. coli with pET-64′OMT had methylation activity for norlaudanosoline (data not shown). The other chimeric enzymes showed no methylation activity. Since the expression level varied, we partially purified 4′6-OMT and measured its OMT activity. However, no activity was detected for 4′6-OMT. This result suggested that many chimeric OMTs might produce an inactive form, and only limited combinations allowed the production of a functional form, even though the C-terminal halves of 4′- and 6-OMT were highly conserved.
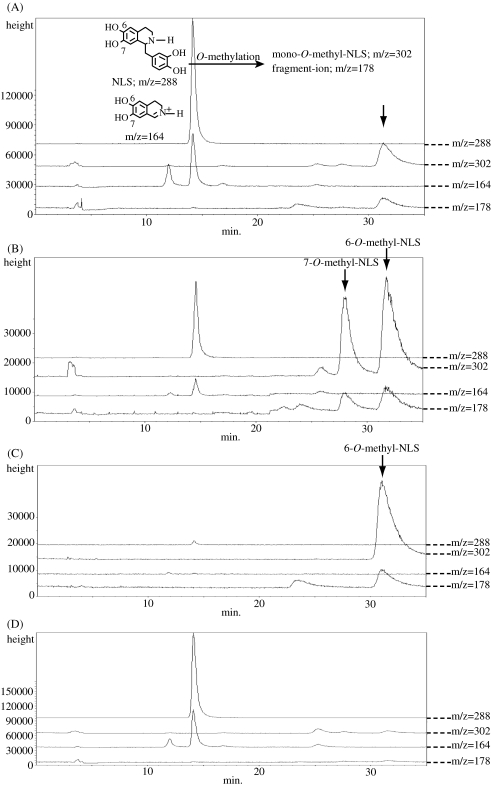
Since 64′-OMT was a newly constructed chimeric enzyme, we carefully examined the reaction product. HPLC and liquid chromatography-mass spectrography (LC/MS) analyses indicated that 64′-OMT product was mono-methylated at the A-ring of norlaudanosoline, but these analyses did not identify the position of methylation, i.e. 6- or 7-OH. Since rat-liver catechol O-methyltransferase (COMT) has been shown to methylate norlaudanosoline to produce both 6- and 7-O-methylated products,14) we compared the 64′-OMT product with 6-O-mono-methylated and 7-O-mono-methylated norlaudanosoline prepared with rat-liver COMT. LC/MS analysis of the 64′-OMT and rat-liver COMT products and authentic 6-O-methylnorlaudanosoline clearly indicated that 64′-OMT methylated the 6-OH group of norlaudanosoline (Fig. 4 ), which showed that the N-terminal half of 6-OMT determined the regio-specificity of 64′-OMT.
Figure 4.
LC/MS analysis of the 64′-OMT reaction product (A), rat-liver COMT reaction product (B), authentic 6-O-methylnorlaudanosoline (C) and control reaction (pET-21d) (D). Arrows indicate the respective reaction products. NLS, norlaudanosoline.
Purification and characterization of 64′-OMT.
Since 64′-OMT was successfully expressed and showed 6-OMT activity, we tried to compare other enzymological properties of 64′-OMT with those of 6- and 4′-OMT. We first highly purified 64′-OMT from E. coli to determine its enzymological properties. The subunit molecular mass of 64′-OMT was about 40 kDa (SDS-PAGE) and the molecular mass of active 64′-OMT was estimated to be about 63 kDa by gel filtration chromatography, indicating that 64′-OMT also formed a dimer like 4′- and 6-OMT. Enzyme assays at various pH values indicated that the optimum pH of 64′-OMT for the methylation of norlaudanosoline was about pH 7.6. This optimal pH of 64′-OMT was lower than that of 6-OMT (pH 9.0)3) but similar to that of 4′-OMT (pH 8.0).4)
64′-OMT did not require divalent cations for activity, as with 4′- and 6-OMT. The addition of divalent cations at 5 mM inhibited 64′-OMT activity more severely than for 4′-OMT and 6-OMT; Fe2+, Cu2+, Co2+, Zn2+, Ni2+ and Mn2+ by 99 %, 98 %, 100 %, 100 %, 100 % and 64 %, respectively. Other cations (Ca2+, Mg2+) had no effect on 64′-OMT activity. Interestingly, the enzymatic activity of 64′-OMT was inhibited by SH-reagents (5 mM), i.e., p-chloromercuribenzenesulfonate (100%) and iodoacetamide (23%), whereas those of 4′- and 6-OMT were not inhibited.3,4)
We also determined the substrate specificity of 64′-OMT using the incorporation of radioactivity from S-adenosyl-L-[methyl-3H]methionine into the products as an index. 64′-OMT methylated (R,S)-norlaudanosoline (relative activity 100%) and (R,S)-laudanosoline (7.8%), whereas no significant methylation was found for the other substrates, as indicated with crude E. coli lysate. This substrate specificity resembled that of 6-OMT, but 64′-OMT showed a greater preference for norlaudanosoline.
The substrate-saturation kinetics of the purified 64′-OMT for (R,S)-norlaudanosoline and AdoMet were the typical Michaelis–Menten-type. Therefore, the kinetic parameters were estimated from double-reciprocal plots of the initial velocity versus the substrate concentration. By varying the concentrations of norlaudanosoline and [3H]AdoMet in the range of 1 mM–30 µM, we could calculate a set of apparent Km and Vmax values and we replotted them to determine the real Km and Vmax. The respective Km values of 64′-OMT for (R,S)-norlaudanosoline and AdoMet were 109 and 187 µM, with a Vmax of 1.5 nkat/mg protein. These Km values were much smaller than those of 4′-OMT. The pattern of the primary reciprocal plots was representative of a sequential substrate-binding mechanism (data not shown). 4′-OMT has been reported to follow an ordered Bi–Bi mechanism,4) while 6-OMT follows a ping-pong Bi–Bi mechanism.3)
S-Adenosyl-L-homocysteine (AdoHcy) inhibited 64′-OMT activity as well as those of 4′- and 6-OMT. AdoHcy inhibition was noncompetitive with norlaudanosoline and competitive with AdoMet. The respective Ki values for AdoHcy versus norlaudanosoline and AdoMet were 168 and 70 µM. The Ki values of AdoHcy for 4′- and 6-OMT were respectively reported to be 43 and 180 µM for methyl-group acceptor (6-O-methylnorlaudanosoline for 4′-OMT and norlaudanosoline for 6-OMT) and 27 µM and 2.1 mM for AdoMet.3,4)
DNA shuffling of the N-terminal halves of 4′- and 6-OMT and their chimeric OMTs.
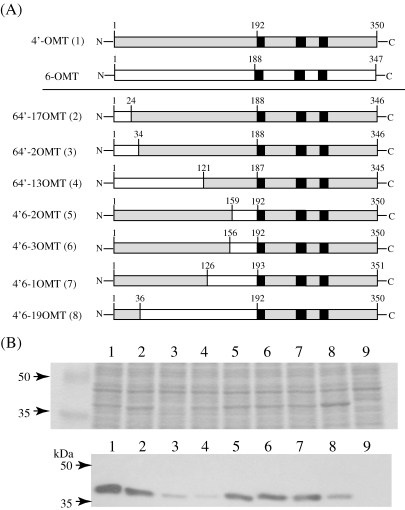
To elucidate which parts of the N-terminal regions of 4′- and 6-OMT are required for substrate recognition, we produced more complicated chimeric constructs using the DNA shuffling method.13) Eventually, 7 clones with proper reading frames were obtained (Fig. 5A ) and their enzymatic activities were analyzed by LC/MS. Immunoblot analysis of E. coli lysate with anti-4′-OMT antibodies and CBB staining showed that these chimeric OMTs were successfully produced, whereas clone 64′-2OMT was less abundant (Fig. 5B). The weak signal in 64′-13OMT also indicated that 4′-OMT antibodies were more sensitive to the N-terminal end of 4′-OMT.
Figure 5.
(A) Schematic drawing of chimeric OMTs obtained by DNA shuffling. The polypeptides derived from 4′-OMT are shaded, and sequences derived from 6-OMT are indicated by an open bar. Figures indicate the number of residues counted from the 1st Met. Solid squares indicate the conserved sequence motifs of plant AdoMet-dependent methyltransferases.10) Figures in parentheses indicate the lane number in panel (B). (B) Expression of chimeric OMTs obtained by DNA shuffling. Upper panel: The crude E. coli lysate (10 µg protein) expressing the chimeric OMTs was separated by SDS-PAGE (10% acrylamide) and stained with CBB. The molecular mass of the chimeric OMTs is ca. 40 kDa. Lower panel: Immunoblot analysis with anti-4′-OMT polyclonal antibodies. M, molecular mass markers; lane 1, pET-4′OMT; lane 2, pET-64′-17OMT; lane 3, pET-64′-2OMT; lane 4, pET-64′-13OMT; lane 5, pET-4′6-2OMT; lane 6, pET-4′6-3OMT; lane 7, pET-4′6-1OMT; lane 8, pET-4′6-19OMT; lane 9, pET-21d (vector control).
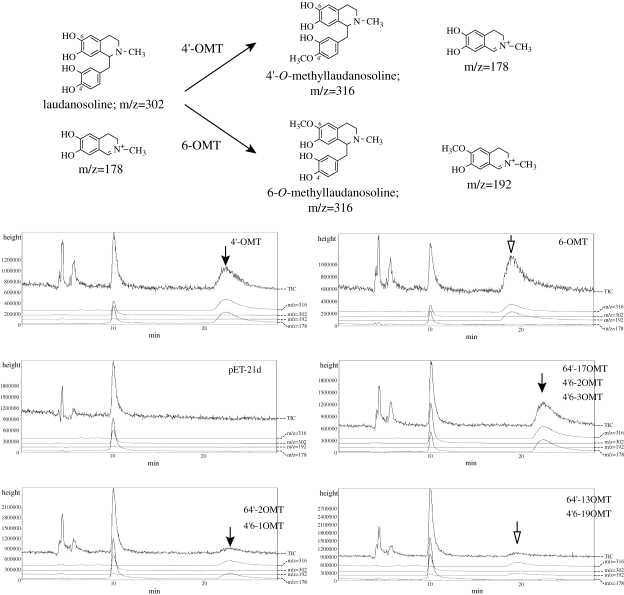
Enzyme activity was measured with [3H]AdoMet using laudanosoline and norlaudanosoline as substrates. While all constructs showed detectable activity, the activities differed (data not shown). We used LC/MS analysis to determine the regio-specificity of chimeric OMTs, since 4′- and 6-O-methylation of laudanosoline showed different fragmentation patterns; i.e., the 4′-OMT-type reaction produces a fragment of m/z = 178 and the 6-OMT-type gives a fragment of m/z = 192 (Fig. 6 ). The chimeric OMTs, i.e., 64′-17, 64′-2, 4′6-2, 4′6-3 and 4′6-1 OMTs, produced 4′-OMT-type products, whereas 64′-13 and 4′6-19 OMTs produced 6-OMT-type products. These results indicated that the region between the 34th and 125th amino acids is crucial for the determination of regio-specificity, whereas some other region might be involved in substrate recognition.
Figure 6.
LC/MS analysis of the chimeric OMT reaction products using laudanosoline as a substrate. Upper panel; OMT reactions and their predicted ions in LC/MS analysis. Lower panels; Mass ion chromatogram of the reaction products. Product peaks with the same retention time and fragmentation ion as the 4′-OMT product (22 min, fragment of m/z = 178) are indicated by solid arrows, and those with the same parameters as 6-OMT (19 min, m/z = 192) are shown by open arrows.
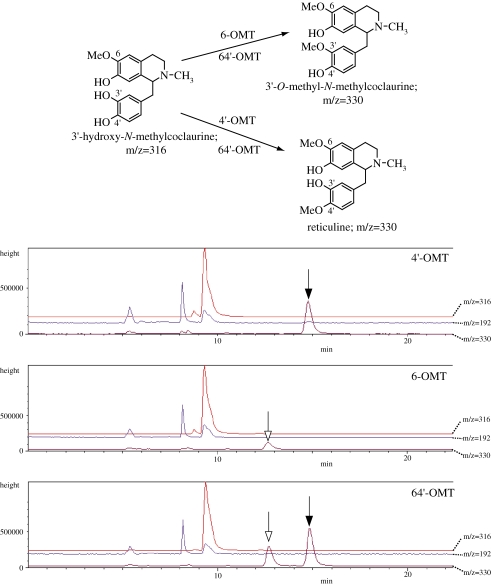
The regio-specificities of chimeric enzymes were further characterized with another substrate, i.e., 3′-hydroxy-N-methylcoclaurine (Fig. 7 ). As expected, 4′-OMT and 4′-OMT-type chimeric enzymes (64′-17, 64′-2, 4′6-1, 4′6-2, 4′6-3 OMTs) showed 4′-O-methylation activity to produce reticuline. Unexpectedly, however, 6-OMT showed a weak but evident peak that was distinct from that of the 4′-O-methylation product. This novel peak was speculated to be a 3′-O-methylation product from the fragment signal of m/z 192 (data not shown). Interestingly, other 6-OMT-type enzymes, 64′-OMT and 4′6-19 OMT, showed both 3′-O-methylation and 4′-O-methylation activities, whereas another 6-OMT-type chimeric enzyme, 64′-13OMT, showed only 4′-O-methylation activity.
Figure 7.
LC/MS analysis of the chimeric OMT reaction products using 3′-hydroxy-N-methylcoclaurine as a substrate. Upper panel; OMT reactions and their predicted ions in LC/MS analysis. Lower panels; Mass ion chromatogram of the reaction products. Product peaks with the same retention time as the 4′-OMT product (15 min, m/z = 330) are indicated by solid arrows, and those considered to be 3-OMT product (ca.13 min, m/z = 330) are shown by open arrows.
Discussion
We have described here the successful production of chimeric OMTs that play a role in isoquinoline alkaloid biosynthesis. Among 6 chimeric OMTs with N- and C-terminal halves derived from different Coptis OMTs, 5 constructs produced detectable amounts of soluble proteins. However, only 64′-OMT showed enzyme activity with known isoquinoline alkaloid substrates. One of the reasons for this functional complementation of the N-terminus of 6-OMT and the C-terminus of 4′-OMT may be the relatively high sequence similarity of 6- and 4′-OMT, whereas it is not clear why 4′6-OMT did not show enzyme activity. Even though their high sequence similarity, especially of the C-terminal halves, would provide a basis for exchange of the N-terminal half, both the N- and C-terminal halves contributed to substrate-binding.11,12) Such interaction between the N- and C-terminal halves might limit their functional reconstitution. Notably, 4′-OMT-specific antibodies could distinguish the N-terminal halves of 4′-OMT and 6-OMT.4)
Since 64′-OMT showed 6-O-methylation activity for norlaudanosoline, the N-terminal half would contribute to the substrate recognition and regio-specificity of Coptis OMT. When the substrate specificities of 64′- and 6-OMT were compared, however, 64′-OMT exhibited a stronger preference for norlaudanosoline than 6-OMT. Moreover, 64′-OMT was sensitive to SH reagent, while 4′-OMT and 6-OMT were not. These differences between 64′-OMT and ‘native’ OMTs can provide a useful basis for understanding the function of the residues in this activity.
Interestingly, when another substrate, 3′-hydroxy N-methylcoclaurine was reacted with a 6-OMT-type chimeric enzyme, i.e., 64′-OMT, 64′-OMT showed 4′-OMT activity for 3′-hydroxy N-methylcoclaurine as well as the 3′-O-methylation activity found for authentic 6-OMT (Fig. 7). Even more interestingly, another 6-OMT-type chimeric enzyme, 64′-13OMT, showed only 4′-O-methylation activity, while this activity was much weaker. These changes in reaction specificity with different substrates may provide a clue to understanding the molecular diversification of OMTs.
Since 4′- and 6-OMT showed a high degree of similarity to IOMT (Fig. 2), the three-dimensional structures of Coptis OMTs were simulated with 3D-PSSM (http://www.sbg.bio.ic.ac.uk/~3dpssm/) and SWISS-MODEL (http://swissmodel.expasy.org//SWISS-MODEL.html) based on the structure of IOMT. Comparison of these structures with the architecture of IOMT dimer indicated that the N-terminal halves of Coptis OMTs form a domain and closely interact with the partner monomer (data not shown). This finding implies that the substrate-recognition regions of Coptis OMTs that were identified in this work may be important for the formation of a substrate-binding pocket with the partner monomer, and indicates that the N-terminus of 6-OMT and the C-terminus of 4′-OMT interact with each other in 64′-OMT dimer. This interaction may also be responsible for the sensitivity of 64′-OMT to SH-reagent. Residue Cys-105 of 6-OMT is predicted to be located at the intersection of the subunits in 64′-OMT dimer. This residue might be involved in the sensitivity to this reagent in chimeric OMT. We speculated that this cysteine residue might be outside of an active site in native OMTs, but might face inward in 64′-OMT dimer. Thus, substrate binding in 64′-OMT might be sterically hindered by modification with SH-reagent. Our site-directed mutation of Cys-105 to Ala in 64′-OMT also suggested that this residue plays an important role in 64′-OMT since the mutant C105A showed no enzyme activity.
Conclusion
A chimeric 64′-OMT with an N-terminal half of 6-OMT and a C-terminal half of 4′-OMT was produced in E. coli. Whereas this 64′-OMT showed 6-O-methylation activity for norlaudanosoline and laudanosoline, it showed 4′-O-methylation activity and 3′-O-methylation activity when 3′-hydroxy-N-methylcoclaurine was used as a substrate. This chimeric enzyme construction may be useful for the diversification of isoquinoline alkaloid biosynthesis.
Acknowledgments
We thank Drs. N. Nagakura and K. Iwasa and Mitsui Petrochemical Industries Ltd. for their generous gifts of the alkaloids. We also appreciate Dr. Kawata for his kind gift of rat liver. This research was supported in part by a Research for the Future Program Grant (JSPS-RFTF00L01607) and a Research Grant A (JSPS-21248013) from the Japan Society for the Promotion of Science (to F. S.), and by a fellowship from the Japan Society for the Promotion of Science (to T. M.).
Abbreviations
- 4′-OMT
S-adenosyl-L-methionine:3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase
- 6-OMT
S-adenosyl-L-methionine:norcoclaurine 6-O-methyltransferase
- AdoMet
S-adenosyl-L-methionine
- CBB
Coomassie Brilliant Blue
- ChOMT
S-adenosyl-L-methionine:chalcone O-methyltransferase
- AdoHcy
S-adenosyl-L-homocysteine
- CNMT
S-adenosyl-L-methionine:coclaurine N-methyltransferase
- HPLC
high performance liquid chromatography
- IOMT
S-adenosyl-L-methionine:isoflavone O-methyltransferase
- LC/MS
liquid chromatography-mass spectrography
- OMT
O-methyltransferase
- SMT
S-adenosyl-L-methionine:scoulerine 9-O-methyltransferase.
References
- 1).Poulton, J.E. (1981) Transmethylation and demethylation reactions in the metabolism of secondary plant products. In The Biochemistry of Plants. Vol. 7, Secondary Plant Products (ed. Conn, E.E.). Academic Press, New York, pp. 667–723. [Google Scholar]
- 2).Rueffer M., Nagakura N., Zenk M.H. (1983) Partial purification and properties of S-adenosylmethionine: (R),(S)-norlaudanosolin-6-O-methyltransferase from Argemone platyceras cell cultures. Planta Med. 49, 131–137 [DOI] [PubMed] [Google Scholar]
- 3).Sato F., Tsujita T., Katagiri Y., Yoshida S., Yamada Y. (1994) Purification and characterization of S-adenosyl-L-methionine: norcoclaurine 6-O-methyltransferase from cultured Coptis japonica cells. Eur. J. Biochem. 225, 125–131 [DOI] [PubMed] [Google Scholar]
- 4).Morishige T., Tsujita T., Yamada Y., Sato F. (2000) Molecular characterization of the S-adenosyl-L-methionine: 3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase involved in isoquinoline alkaloid biosynthesis in Coptis japonica. J. Biol. Chem. 275, 23398–23405 [DOI] [PubMed] [Google Scholar]
- 5).Frenzel T., Zenk M.H. (1990) S-Adenosyl-L-methionine: 3′-hydroxy-N-methyl-(S)-coclaurine-4′-O-methyl transferase, a regio- and stereoselective enzyme of the (S)-reticuline pathway. Phytochemistry 29, 3505–3511 [Google Scholar]
- 6).Muemmler S., Rueffer M., Zenk M.H. (1985) S-Adenosyl-L-methionine: (S)-scoulerine 9-O-methyltransferase, a highly stereo- and regio-specific enzyme in tetrahydroprotoberberine biosynthesis. Plant Cell Rep. 4, 36–39 [DOI] [PubMed] [Google Scholar]
- 7).Fujiwara H., Takeshita N., Terano Y., Fitchen J.H., Tsujita T., Katagiri Y., et al. (1993) Expression of (S)-scoulerine 9-O-methyltransferase in Coptis japonica plants. Phytochemistry 34, 949–954 [Google Scholar]
- 8).Takeshita N., Fujiwara H., Mimura H., Fitchen J.H., Yamada Y., Sato F. (1995) Molecular cloning and characterization of S-adenosyl-L-methionine:scoulerine-9-O-methyltransferase from cultured cells of Coptis japonica. Plant Cell Physiol. 36, 29–36 [PubMed] [Google Scholar]
- 9).Minami H., Kim J.-S., Ikezawa N., Takemura T., Katayama T., Kumagai H., et al. (2008) Microbial production of plant benzylisoquinoline alkaloids. Proc. Natl. Acad. Sci. USA 105, 7393–7398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Joshi C.P., Chiang V.L. (1998) Conserved sequence motifs in plant S-adenosyl-L-methionine-dependent methyltransferases. Plant Mol. Biol. 37, 663–674 [DOI] [PubMed] [Google Scholar]
- 11).Zubieta C., He X.Z., Dixon R.A., Noel J.P. (2001) Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases. Nat. Struct. Biol. 8, 271–279 [DOI] [PubMed] [Google Scholar]
- 12).Zubieta C., Kota P., Ferrer J.L., Dixon R.A., Noel J.P. (2002) Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase. Plant Cell 14, 1265–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13).Stemmer W.P.C. (1994) DNA shuffling by random fragmentation and reassembly: In vitro recombination for molecular evolution. Proc. Natl. Acad. Sci. USA 91, 10747–10751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14).Meyerson L.R., Cashaw J.L., McMurtrey K.D., Davis V.E. (1979) Stereoselective enzymatic O-methylation of tetrahydropapaveroline and tetrahydroxyberbine alkaloids. Biochem. Pharmacol. 28, 1745–1752 [DOI] [PubMed] [Google Scholar]
- 15).Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]