Abstract
The latency-associated nuclear antigen (LANA) of Kaposi's sarcoma-associated herpesvirus functions as an origin-binding protein (OBP) and transcriptional regulator. LANA binds the terminal repeats via the C-terminal DNA-binding domain (DBD) to support latent DNA replication. To date, the structure of LANA has not been solved. Sequence alignments among OBPs of gammaherpesviruses have revealed that the C terminus of LANA is structurally related to EBNA1, the OBP of Epstein–Barr virus. Based on secondary structure predictions for LANADBD and published structures of EBNA1DBD, this study used bioinformatics tools to model a putative structure for LANADBD bound to DNA. To validate the predicted model, 38 mutants targeting the most conserved motifs, namely three α-helices and a conserved proline loop, were constructed and functionally tested. In agreement with data for EBNA1, residues in helices 1 and 2 mainly contributed to sequence-specific DNA binding and replication activity, whilst mutations in helix 3 affected replication activity and multimer formation. Additionally, several mutants were isolated with discordant phenotypes, which may aid further studies into LANA function. In summary, these data suggest that the secondary and tertiary structures of LANA and EBNA1 DBDs are conserved and are critical for (i) sequence-specific DNA binding, (ii) multimer formation, (iii) LANA-dependent transcriptional repression, and (iv) DNA replication.
INTRODUCTION
Kaposi's sarcoma-associated herpesvirus (KSHV; human herpesvirus 8) is a DNA tumour virus associated with Kaposi's sarcoma, primary effusion lymphomas and a plasmablastic variety of multicentric Castleman's disease (Cesarman et al., 1995; Chang et al., 1994; Soulier et al., 1995). The latency-associated nuclear antigen (LANA), encoded by ORF73, interacts with multiple cellular proteins to affect various signal transduction pathways (Gao et al., 1996; Kedes et al., 1996). LANA also functions as an origin-binding protein (OBP) by binding to the viral latent origin, and recruits the host cellular replication machinery to ensure replication of viral episomes during S phase. Additionally, LANA tethers viral genomes to mitotic chromosomes via its N-terminal chromosome-binding motif, thereby contributing to episomal maintenance (Ballestas & Kaye, 2001; Ballestas et al., 1999; Barbera et al., 2006; Cotter & Robertson, 1999; Garber et al., 2002; Hu et al., 2002; You et al., 2006).
The C-terminal LANA DNA-binding domain (LANADBD, aa 775–1003; Garber et al., 2001) binds cooperatively to LANA binding sites 1 and 2 (LBS1/2) within viral terminal repeats (TRs) for replication of TR-containing plasmids (Garber et al., 2001, 2002; Hu et al., 2002). LANA predominantly forms dimers, and the dimerization domain has been mapped to the LANADBD (Schwam et al., 2000), which also has partial replication activity (Hu et al., 2002). LANA and EBNA1, the OBP of Epstein–Barr virus (EBV), are functional homologues with respect to DNA binding and supporting DNA replication by recruitment of cellular origin recognition complex proteins. Both proteins form dimers in solution and bind to two sites within their respective origins of replication in a cooperative manner (Lim et al., 2002; Schepers et al., 2001; Stedman et al., 2004; Verma et al., 2006).
Neither the structure of full-length LANA nor its DNA-binding domain (DBD) has been determined to date. In contrast, crystal structures of EBNA1 in the presence and absence of DNA (Bochkarev et al., 1995, 1996) and E2, the OBP of human papillomavirus (HPV) (Hegde et al., 1992), have been solved. Although EBNA1 and E2 share very limited primary sequence homology and are encoded by different classes of DNA tumour virus, their DBDs revealed a common core domain structure. The core domain consists of a series of interspersed β-sheets, which form a β-barrel within the dimer interface, a proline loop, which interacts with cellular proteins, and three α-helices, which make direct or indirect contacts to DNA and stabilize higher-order multimers (Bochkarev et al., 1995, 1996; Ceccarelli & Frappier, 2000). To gain insights into the possible structure of the LANADBD in the absence of a crystal structure, we performed detailed sequence alignments among the LANADBDs of different rhadinoviruses and performed bioinformatics-based modelling to predict a potential structure. We investigated our model by mutational analysis and by functional testing of mutants targeting residues most conserved between different LANADBDs and EBNA1DBD.
RESULTS
High evolutionary conservation of LANADBDs in gammaherpesviruses and bioinformatics-based predicted structure of KSHV LANADBD
Grundhoff & Ganem (2003) first noted a limited secondary structure homology between the C termini of LANA and EBNA1. Furthermore, sequencing LANA from a retroperitoneal fibromatosis-associated herpesvirus variant from Macaca nemestrina (RFHVMn) and Macaca nemestrina rhadinovirus 2 (MneRV2) revealed that the C-terminal amino acids of their LANAs showed the strongest sequence conservation (Burnside et al., 2006). To analyse these homologies further, we performed amino acid alignment among the LANADBDs of KSHV, RFHVMn and rhesus monkey rhadinovirus (RRV) and the EBV EBNA1DBD using the bioinformatics programs praline, 3d-pssm and T-Coffee (Fig. 1 and Table 1). This analysis revealed that KSHV LANADBD had greater than 50 % similarity to the DBDs of RFHVMn and RRV. Although EBNA1DBD had less than 16 % overall amino acid sequence identity to LANADBDs (Table 1), there was significant structural similarity such as the presence of three α-helices, as noted previously (Grundhoff & Ganem, 2003). In addition, we found a proline-rich loop motif that was conserved between KSHV LANADBD (930PHPGPDQSP938) and EBV EBNA1DBD (545PGPGPQPGP553) (Fig. 1b), which is important for the protein–protein interactions of EBNA1 (Bochkarev et al., 1996). We also noted that, among KSHV, RFHVMn and RRV, residues within the α-helices were more highly conserved than the surrounding residues (Fig. 1a). Based on these observations, we performed bioinformatics modelling to predict the KSHV LANADBD structure, a common approach for related proteins for which crystals cannot easily be obtained (Hantz et al., 2009; Hass et al., 2008; Purta et al., 2005).
Fig. 1.
Sequence alignments of LANADBDs of gammaherpesviruses and EBNA1DBD reveals structural conservation. (a) Multiple alignments of amino acid sequences among LANADBDs of KSHV, RFHVMn and RRV using the praline and T-Coffee programs. Conserved amino acids among the OBPs are labelled in bold above the sequences. (b) Binary amino acid alignments between EBV EBNA1DBD and the LANADBDs of KSHV, RFHVMn and RRV using the 3d-pssm program. Conserved helices among the proteins are shown as shaded dark grey boxes. Proline loops are indicated in italic within light grey boxes. Numbers in parentheses refer to corresponding amino acid numbers from BC-1 KSHV LANA (Kelley-Clarke et al., 2007). Short dashes indicate missing amino acids; + and − indicate similarity or no similarity between amino acids.
Table 1.
Similarity and identity of the C termini among gammaherpesvirus OBPs
| Amino acid similarity (identity) (%) | ||||
|---|---|---|---|---|
| KSHV | RRV | RFHVMn | EBV | |
| KSHV | 100 | – | – | – |
| RRV | 53 (30) | 100 | – | – |
| RFHVMn | 54 (40) | 46 (26) | 100 | – |
| EBV | 53 (14) | 43 (16) | 30 (13) | 100 |
KSHV LANADBD residues 868–960 were modelled with the 3d-jigsaw modelling tool (Bates et al., 2001) using the EBNA1DBD structure (PDB accession no. 1B3T) as template (Bochkarev et al., 1996). The LANADBD residues 929–939 did not have defined coordinates after 3d-jigsaw analysis and were modelled using ModLoop (Fiser & Sali, 2003). Despite the relatively low residue homology, the structure for a LANADBD monomer was very similar to chain A of EBNA1DBD. The root mean square deviation (RMSD) was 0.85 Å between the EBNA1DBD structure and the predicted LANADBD model, suggesting close similarity.
To predict the multimer structure of LANADBD, the program m-zdock (Pierce et al., 2005) was run using the LANADBD homology model to perform a full search of possible homodimeric interfaces. The output models from m-zdock were then filtered based on similarity to the EBNA1DBD dimer interfaces, the ability to fit double-stranded DNA and the score of the model from the program zrank (Pierce et al., 2007). We next selected two m-zdock models for the LANADBD dimer using these criteria, which were joined to construct a tetramer (Fig. 2a). The RMSD for the LANADBD dimer versus the two EBNA1DBD chains was 2 Å.
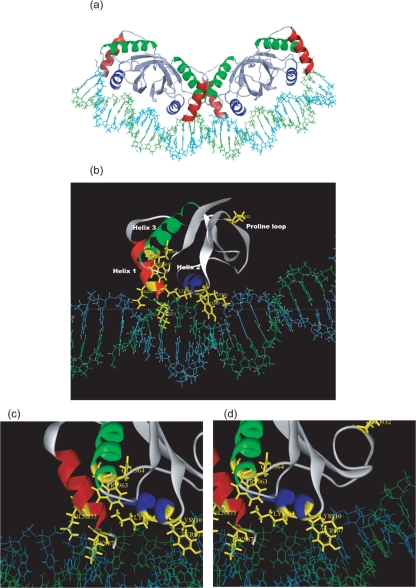
Fig. 2.
Computational model of the LANADBD and multimer structure bound to DNA. The figure shows the LANADBD model with the specific DNA-binding site predicted by the m-zdock program based on alignment with the structure of EBNA1DBD. (a) The tetramer formed by combining two dimers bound to their respective LBS1/2 (DNA helix in light blue and green). The β-barrel bundle is made of four β-strands from each monomer at the dimer interface. (b) Each monomer is composed of four β-strands and three helices (helix 1 in red, helix 2 in blue and helix 3 in green). (c, d) Crucial amino acids for DNA contact or dimerization are shown in yellow: 871K and 875Q for helix 1, 963W and 964E for helix 3 (c) and 907Y, 910K and 911K for helix 2 (d). The monomer pictures were generated using ViewerLite 4.2 (Accelrys).
We have shown previously that LANA binds to LBS1/2 within the TRs, which are spaced by 21–22 nt (Garber et al., 2002), and Wong & Wilson (2005) demonstrated that LANA occupying both sites induces a bend of about 11 °. Whilst the sequence composition between EBNA1-binding sites (AT-rich) and LANA-binding sites (GC-rich) is very different, both the spacing and the induced DNA bending are conserved features. Accordingly, the DNA conformation was initially taken from the structure of EBNA1DBD bound to DNA (Bochkarev et al., 1996) and fitted to the two dimers in the LANADBD tetramer. The linking DNA between the two dimer-binding sites was extended from the existing DNA strands. The Rosetta program (Havranek et al., 2004) was then used to restore the DNA sequence to the LBS1/2 sequences and repack the LANA side chains accordingly.
The resulting model for the LANADBD tetramer bound to DNA (Fig. 2a) shared the defining β-barrel core domain structure with both EBNA1DBD and E2DBD. The dimer of LANADBD was composed of eight antiparallel β-strands within their core domains, and flanking domains including helices 1 (Fig. 2a in red) (Bochkarev et al., 1995), which were positioned at the outside of each monomer towards the dimer interfaces. The β-barrel formation was composed of four β-strands from each monomer, and the β-strands were connected by crossover of the two α-helices (within each core domain) (Fig. 2a in blue and green) on the outside of each barrel. Hence, our model incorporated all known data on the LANADBD–DNA interaction (Garber et al., 2002; Wong & Wilson, 2005), and suggested similar secondary and quaternary structures for LANADBD and EBNA1DBD.
Mutagenesis of KSHV LANADBD and expression of mutant proteins
To test the LANADBD model, we performed a detailed mutational analysis by targeting conserved residues in the three α-helices and the proline loop. A total of 38 single, double or triple alanine substitution mutants were generated by site-directed mutagenesis. Wild-type (wt) and mutant proteins were expressed using the modified vaccinia virus Ankara (MVA)/T7 RNA polymerase expression system in CV-1 cells as described previously (Garber et al., 2001, 2002). Briefly, constructs containing T7 promoter were transfected into MVA/T7-infected cells. The cells were harvested 36 h post-transfection and the proteins were enriched by affinity purification. Expression levels for all mutant proteins were monitored by Western blotting (see Supplementary Fig. S1, available in JGV Online).
Evaluation of wt and mutant KSHV LANADBDs for DNA binding by electrophoretic mobility shift assay (EMSA)
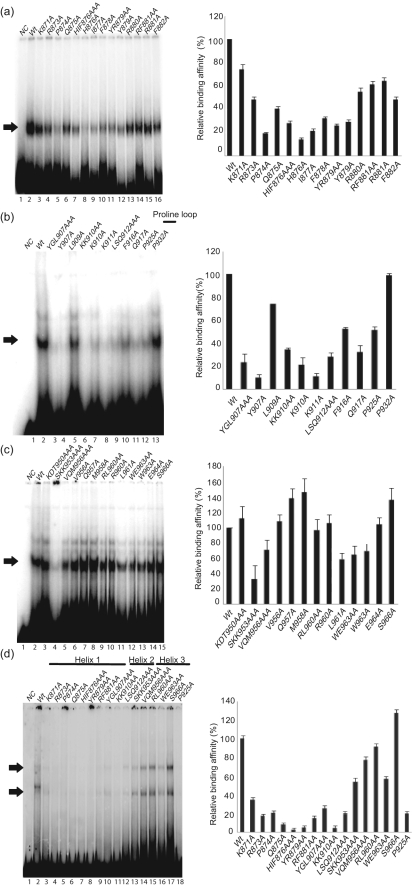
We reported previously that LANADBD binds to its high-affinity binding site (LBS1) with a Kd of 1.51±0.16 nM (Garber et al., 2002). To determine the effect of mutations on DNA binding, equal amounts of wt and mutant LANADBD proteins were incubated with radiolabelled probes containing either LBS1 or LBS1/2 (Fig. 3). After electrophoresis, the gels were dried and signals were quantified by phosphoimaging. Representative autoradiographs from three independent experiments are shown.
Fig. 3.
DNA-binding activity of LANADBD mutants. Purified LANADBD wt and mutant proteins were incubated with radiolabelled LBS1 or LBS1/2 as described previously (Garber et al., 2001). The DNA-binding affinity is represented as the percentage for mutants compared with wt LANADBD, which was set to 100 %. In each assay, all mutants were tested for DNA-binding activity with LBS1 (a–c) or LBS1/2 (d). EMSA results are shown for helix 1 mutants (a), helix 2 mutants (b), helix 3 mutants (c) and adapted mutants from each helix (d). Arrows indicate specific protein–DNA complexes. NC, Probe alone as a negative control; wt, wt LANADBD. Results on graphs are shown as means±sd from three independent experiments.
Most mutants in helix 1 significantly reduced the binding affinity to both LBS1 and LBS1/2 (Fig. 3a, d). In particular, P874A and H876A reduced the DNA-binding affinity to less than 20 % of that of wt (Fig. 3a, lanes 5 and 8). Helix 1 (871K–882F) contains the polar residues 871K, 873R and 879Y, which are highly conserved residues among KSHV, RFHVMn and RRV (Fig. 1a). These residues potentially contact DNA either directly or indirectly by stabilizing the secondary structure of the N-terminal domain of LANADBD. From the structure of EBNA1DBD, polar residue 477K within helix 1 and residues 461K–469R within the N terminus have been shown to contact DNA directly (Bochkarev et al., 1996). In agreement with the binding data, the predicted structure (Fig. 2) suggested that the N-terminal residues of helix 1 (871K, 873R and 875Q) are located in close approximation to the DNA (Fig. 2b, c). For EBNA1DBD, residues within helix 2, which was originally termed the DNA recognition helix (Bochkarev et al., 1995), also contribute to DNA binding. The recognition helices of all HPV E2 proteins contain several highly conserved residues in a consensus motif (338LXXLRY343), which is also conserved in EBNA1DBD (517LYNLRR522) (Fujita et al., 2001). Within LANADBD, 906PYGLKK911 in helix 2 has a similar surface charge to EBNA1DBD helix 2 (Fig. 1). Moreover, in the model, 907Y and 910KK911, like 518Y and 521RR522 of EBNA1DBD, were predicted to be in close contact with the DNA (Fig. 2d). Indeed, all mutants in helix 2, except L909A, showed dramatically reduced DNA-binding affinities to both LBS1 and LBS1/2 (Fig. 3b, d).
For EBNA1DBD, it was shown that the proline loop (545PGPGPQPGP553) between helix 2 and the β-barrel bundle contributes to DNA binding as well as to protein–protein interactions with cellular transcription factors (Bochkarev et al., 1995, 1996). Mutant P932A in the centre of the proline loop (930PHPGPDQSP938) of LANADBD did not reduce DNA-binding affinity (Fig. 3b, lane 13); however, P925A located inside the β-barrel bundle reduced binding affinity by about 50 % (Fig. 3b, lane 12).
Helix 3 (950K–966S) followed the proline loop and continued towards the inside of the β-barrel through an extended strand. In contrast to mutants in helices 1 and 2, helix 3 mutants, except for SKK953AAA, L961A, W963A and WE963AA, did not show significant changes in DNA-binding affinity (Fig. 3c, d and Table 2). SKK953AAA in helix 3 may change folding by interrupting hydrogen bonds with basic residues of helix 1. Thus, these helix 3 residues contribute towards stabilizing protein–DNA interactions and, in contrast to residues within helices 1 and 2, are not directly involved in DNA binding.
Table 2.
Summary of LANADBD mutants and their relative activities in DNA binding, dimerization, replication and transcription repression
nt, Not tested.
| Position | Mutant | EMSA* | IP† | RA‡ | Repression assay§ | |
|---|---|---|---|---|---|---|
| LBS1 | LBS1/2 | |||||
| Helix 1 | K871A | 74 (±4.8) | 35 (±2.2) | 74 | nt | 79 (±8) |
| R873A | 48 (±2.9) | 18 (±1.6) | 135 | nt | 103 (±12) | |
| P874A | 19 (±1.3) | 21 (±2.1) | 98 | nt | 77 (±13) | |
| Q875A | 40 (±2.5) | 8 (±2.02) | 57 | nt | 87 (±11) | |
| HIF876AAA | 28 (±1.9) | 3 (±1.85) | 127 | − | 41 (±7) | |
| H876A | 14 (±1.6) | nt | nt | nt | 66 (±5) | |
| I877A | 21 (±2.4) | nt | nt | nt | 65 (±6) | |
| F878A | 32 (±1.8) | nt | nt | nt | – | |
| YR879AA | 26 (±1.5) | 5 (±2.11) | 73 | − | 67 (±8) | |
| Y879A | 29 (±2.2) | nt | nt | nt | 75 (±4) | |
| R880A | 55 (±3.4) | nt | nt | nt | 7.4 (±0.6) | |
| RF881AA | 61 (±3.5) | 15 (±2.54) | 31 | nt | nt | |
| R881A | 64 (±3.6) | nt | nt | nt | nt | |
| F882A | 48 (±2.6) | nt | nt | nt | nt | |
| Helix 2 | YGL907AAA | 23 (±7.8) | 25 (±3.01) | 73 | − | – |
| Y907A | 10 (±2.8) | nt | nt | nt | 71 (±6) | |
| L909A | 74 (±0) | nt | nt | nt | nt | |
| KK910AA | 34 (±2.1) | 5 (±2.29) | 210 | − | nt | |
| K910A | 21 (±6.5) | nt | nt | nt | nt | |
| K911A | 11 (±2.8) | nt | nt | nt | nt | |
| LSQ912AAA | 28 (±3.6) | 19 (±2.97) | 248 | − | 50 (±3) | |
| F916A | 52 (±2.1) | nt | nt | nt | nt | |
| Q917A | 32 (±6.3) | nt | nt | nt | nt | |
| P925A | 51 (±3.5) | 20 (±2.3) | 200 | nt | 72 (±7) | |
| Proline loop | P932A | 99 (±2.1) | nt | nt | nt | 83 (±6) |
| Helix 3 | KDT950AAA | 112 (±16) | nt | 74 | nt | 93 (±9) |
| SKK953AAA | 33 (±18) | 54 (±3.82) | 64 | nt | 65 (±11) | |
| VQM956AAA | 71 (±13) | 77 (±4.1) | 88 | nt | nt | |
| V956A | 108 (±9) | nt | nt | nt | nt | |
| Q957A | 138 (±13) | nt | 147 | nt | 86 (±5) | |
| M958A | 146 (±18) | nt | nt | 94 (±2) | ||
| RL960AA | 97 (±14) | 91 (±3.87) | 104 | nt | nt | |
| R960A | 106 (±11) | nt | nt | nt | nt | |
| L961A | 59 (±7.8) | nt | nt | nt | 69 (±6) | |
| WE963AA | 65 (±11.5) | 56 (±3.33) | 29 | − | 0.2 (±4) | |
| W963A | 70 (±12) | nt | nt | nt | 64 (±4) | |
| E964A | 104 (±9.1) | nt | nt | nt | 94 (±7) | |
| S966A | 136 (±16) | 127 (±4.6) | 188 | +/− | 82 (±13) | |
*Detection levels by EMSA in the presence of a single or double DNA-binding site. Numbers indicate the percentage of relative binding affinity compared with that of wt.
†IP, Detection levels by immunoprecipitation. Numbers indicate the percentage of dimerization activity compared with that of wt.
‡RA, Replication activity. –, No activity; +/−, reduced activity compared with that of LANADBD.
§Transcription repression activity. Numbers indicate the percentage of transcriptional repression activity compared with that of wt.
Evaluation of multimerization of KSHV LANADBD by co-immunoprecipitation assays
Schwam et al. (2000) first demonstrated that LANADBD in solution and in the absence of TR DNA exists predominantly as a homodimer. To analyse dimerization of a subset of mutants with reduced DNA-binding activities, we performed co-immunoprecipitation assays. Flag-tagged wt or mutant LANADBDs were tested for their ability to interact with haemagglutinin (HA)-tagged wt LANADBD. LANADBD complexes were immunoprecipitated by anti-Flag beads and separated by SDS-PAGE. The amount of wt LANADBD precipitated was detected and quantified by Western blotting using anti-HA antibody. The dimerization activity for each mutant is reported as the percentage relative to that of HA- and Flag-tagged wt LANADBD, which was set to 100 %.
Mutants HIF876AAA and YR879AA in helix 1, which showed drastically reduced DNA-binding affinities, did dimerize at a level comparable to wt (Fig. 4a, lanes 5–8). RF881AA and Q875A reduced dimerization only (Fig. 4b, lanes 5 and 6, and Table 2), further suggesting that most helix 1 residues contribute directly to DNA binding but not to dimer formation.
Fig. 4.

Co-immunoprecipitation assays with alanine substitution mutants. The dimerization ability of Flag-tagged wt or mutant LANADBDs with HA-tagged wt LANADBD was tested. Dimerization activity for each mutant was normalized based on the expression level of Flag-tagged wt or mutant LANADBD proteins. L, Cell lysate, IP; immunoprecipitated samples; Wt (N), HA-tagged wt only as a negative control; Wt (P), Flag-tagged and HA-tagged wt as a positive control.
Similarly, except for YGL907AAA, which showed a moderate decrease (73 %) in dimerization (Fig. 4b, lanes 7 and 8), helix 2 mutants had largely unaltered or increased dimerization activities compared with wt (Fig. 4c, lanes 5–8). This result was expected, as helix 2 of EBNA1DBD and presumably LANADBD function as a DNA recognition domain. In addition, P925A within the β-barrel connected to the proline loop did not affect dimerization (Table 2).
Within helix 3, several mutants had reduced dimerization (Fig. 4d–f). Dimerization for WE963AA and SKK953AAA was reduced to 29 and 64 %, respectively (Fig. 4f, lanes 7 and 8, and Table 2). Within the EBNA1DBD, the corresponding mutants in helix 3 showed the loss of both dimerization and DNA replication activities (Bochkarev et al., 1996).
Analysis of DNA replication activity of wt and mutant KSHV LANADBDs
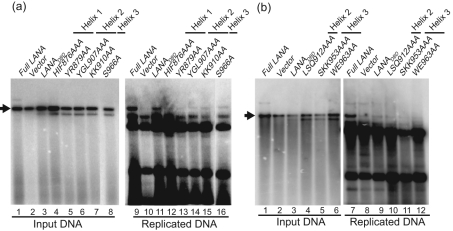
Mutagenesis of the LBS1/2 showed that the replication efficiency of TR-containing plasmids is dependent on the LBS1 (Garber et al., 2002). To test the inverse, we chose a subset of mutants with reduced DNA binding or dimerization and performed transient replication assays as described previously (Garber et al., 2002; Hu et al., 2002). Briefly, a plasmid containing four copies of the TR was co-transfected with plasmid expressing wt or mutant LANADBD into 293 cells. Replicating DNA was extracted and analysed by Southern blotting after DpnI digestion. As described previously, LANADBD replicated with about 20 % efficiency compared with full-length LANA (compare Fig. 5a, lanes 9–11, and Fig. 5b, lanes 7–9) (Hu et al., 2002).
Fig. 5.
Analysis of DNA replication mediated by alanine substitution mutants using a short-term replication assay. LANADBD-expressing constructs were co-transfected with pPuro/4TR into 293 cells; 10 % of the extracted DNA (Hirt, 1967) was digested with HindIII as input (a, lanes 1–8, and b, lanes 1–6) and the remaining DNA was double digested with HindIII and DpnI (a, lanes 9–16, and b, lanes 7–12). The DNA was detected by Southern blotting with a radiolabelled 4TR probe. Full-length LANA was transfected as a positive control. The arrow indicates the position of full-length pPuro-4TR.
The mutants with reduced binding affinity in helix 1 (HIF876AAA and YR879AA), helix 2 (YGL907AAA, KK910AA and LSQ912AAA) and helix 3 (SKK953AAA) did not replicate to detectable levels (Fig. 5a, lanes 12–15, and b, lanes 10 and 11). Furthermore, WE963AA in helix 3, which strongly reduced dimerization, was also inactive in the replication assay (Fig. 5b, lane 12). In contrast, S966A, which had no phenotype in either binding or dimerization, showed residual replication activity (Fig. 5a, lane 16). These data further confirmed that LANA dimerization and high-affinity binding to the TR are required for replication. Interestingly, whilst the proline loop mutant P932A bound to DNA and dimerized like the wt, it did not support replication (Table 2 and data not shown). For EBNA1DBD, it has been shown that the proline loop contributes to spatial orientation of helices 1 and 2 and interacts with cellular proteins (Bochkarev et al., 1995, 1996; Ceccarelli & Frappier, 2000).
Transcriptional repressor activity of wt and mutant LANADBDs
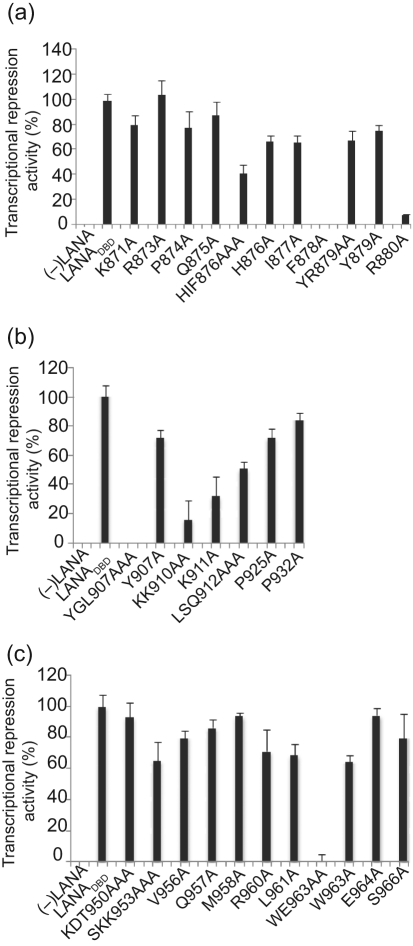
We and others have previously shown that the TR sequences have enhancer activity, which can be repressed by LANA. Furthermore, LANADBD alone is sufficient for repression (Garber et al., 2001). To test mutants for repressor activity, plasmids encoding wt or mutant LANADBD were co-transfected with pGL3/7TR reporter plasmid into 293 cells. Cell lysates were assayed for luciferase activity and normalized as described previously (Renne et al., 2001). The data for mutants in all three helices is shown as the percentage repression activity compared with that of wt, which was set to 100 % (Fig. 6).
Fig. 6.
Analysis of the activity of LANA-dependent transcriptional repression by alanine substitution. Graphical representation of data from luciferase reporter assays. pGL3/7TR luciferase reporter and wt or mutant LANADBD plasmid were co-transfected. RLU values were normalized to total protein concentration as described previously (Renne et al., 2001). The percentage of suppression activity was compared with that of LANADBD wt, which was set to 100 %. Results are shown as means±sd from three independent experiments. (−)LANA, Negative control with no LANADBD.
Within helix 1, eight out of 11 mutants had only moderately decreased repression activity of between 80 and 65 % compared with wt. Repression activity of HIF876AAA and F878A was decreased to 41 % and to less than 1 % of wt, which was concordant with strongly reduced binding activity (Fig. 6a and Table 2). Interestingly, R880A showed only 7 % repression activity despite its DNA binding activity only being reduced to 55 % (Fig. 6a and Table 2).
Within helix 2, four out of five tested mutants showed significantly decreased repression activity, the exception being Y907A. These were mostly concordant with either loss of or a strong reduction in DNA binding (Table 2). These data further confirmed that residues in helix 2 significantly contribute to DNA recognition and binding. Interestingly, the DNA binding of Y907A was strongly reduced, although it displayed 71 % repressor activity (Fig. 6b).
In agreement with the DNA-binding data, most mutants in helix 3, including the proline loop, had a modest or no effect on transcriptional repression (Fig. 6b, c). However, WE963AA completely abolished repressor activity. Interestingly, although WE963AA had only modestly reduced DNA-binding activity (Table 2), it had strongly reduced homodimer formation, suggesting that these residues may interact with helix 1 to stabilize the homodimer or contribute to interactions with cellular proteins conveying transcriptional repression.
In summary, these data showed that, for most of the mutants, DNA binding and transcriptional repression were similarly affected. However, we observed some mutants, notably R880A and WE963AA, that could bind to the TR but did not repress transcription, and one mutant, Y907A, that bound poorly to the TR but still repressed transcription.
DISCUSSION
Many mechanistic details on the role of LANA in transcriptional regulation, latent DNA replication, tethering of viral episomes to host chromatin and interaction with multiple host cellular proteins have been reported (reviewed by Lieberman et al., 2007; Verma et al., 2007). In contrast, with the exception of a small 23 aa peptide in the N-terminal histone H2A-binding domain (Barbera et al., 2006), no structural data is available on LANA. We have expressed LANADBD protein using vaccinia virus, baculovirus and Escherichia coli systems, but have not yielded concentrations of soluble protein amenable to crystallization. A further complicating factor is that all published DNA-binding assays have been performed in the presence of BSA, substitution for which will be crucial to solve the LANADBD structure in the presence of its cognate binding site (Ballestas & Kaye, 2001; Cotter & Robertson, 1999; Garber et al., 2002).
In the meantime, we performed bioinformatics modelling based on the observed sequence homology between the DBDs of KSHV, RRV and RFHVMn LANA and the DBD of EBNA1 to predict a structure for KSHV LANADBD. We note that the X-ray structures of the EBNA1DBD and E2DBD core domains, which show no discernible sequence homology, superimpose almost perfectly (Bochkarev et al., 1996; reviewed by Grossman & Laimins, 1996; Hegde et al., 1992; Liang et al., 1996). In contrast, the DBDs of LANA and EBNA1 showed 14 % identity and 53 % similarity (Table 1) and the highest conservation was within motifs that are crucial for the overall core domain structure (Fig. 1b) (Grundhoff & Ganem, 2003). As a result, the predicted model (Fig. 2) indicated a high degree of structural relatedness.
To functionally validate this model, we targeted the three α-helices and the proline loop, which showed the highest conservation (Fig. 1b) and for which phenotypes have been described for EBNA1DBD. We identified residues in all three α-helices that are crucial for DNA binding (helices 1 and 2) or multimerization (helix 3). Both efficient DNA binding and dimerization are crucial for LANA's ability to support replication of the TR-containing plasmids. The functional data for all mutants is summarized in Table 2. The key observations were that charged residues within a conserved motif in helix 2 (906PYGLKK911) were crucial for DNA binding (Fig. 2d). Helix 1 of LANA DBD also contributed to binding, presumably through direct interactions with DNA (Fig. 3a, d). These data are in agreement with those for EBNA1DBD where helices 1 and 2 both significantly contribute to DNA binding. Interestingly, within the crystal structure, helix 1 of EBNA1DBD was located much closer to the DNA than helix 2. However, biochemical data by Cruickshank et al. (2000) clearly demonstrated that helix 2 is also critical for DNA binding. To explain the difference between the crystal structure of EBNA1DBD bound to DNA and the biochemical data, it was suggested that EBNA1 binds to DNA via a two-step mechanism: sequence-specific binding is initiated by helix 2 followed by interactions of helix 1 residues. The observation that LANA residues from both helices contribute to binding activity points to a conserved DNA-binding mechanism for EBNA1 and the rhadinovirus LANA proteins, which has also been suggested for the HPV E2 protein (reviewed by de Prat-Gay et al., 2008; Hegde et al., 1992; Liang et al., 1996).
Most mutations in helices 1 and 2 reduced transcriptional repressor activity as well as reducing DNA binding (Table 2). These data are consistent with the previous observation that high and low affinities of LBS1/2 determine DNA binding and replication (Garber et al., 2002). In contrast, most mutants in helix 3 had only moderate effects on transcriptional suppression; however, mutant WE963AA displayed greatly reduced repression but only moderately reduced DNA binding (Fig. 3c and Table 2), indicating a role in protein–protein interactions that conveys LANA-dependent repression.
These data strongly suggest functional homology between all three α-helices and the proline loop of KSHV LANADBD and EBNA1DBD. In addition, this analysis yielded at least one mutant in each helix and in the proline loop that showed discordance in phenotype with regard to DNA binding, homodimer formation, transcriptional repression or DNA replication. Within helix 1, R880A bound to the TR but had almost no repressor activity. Conversely, Y907A in helix 2 significantly reduced DNA binding but still repressed transcription, and WE963AA in helix 3 had only moderately reduced binding but completely lost repression activity. Finally, proline loop mutant P932A had no defect in either binding or dimerization, but did not support DNA replication. These mutants will be useful for further mechanistic studies on LANA function and some may function as dominant-negative proteins, which have not been described to date for LANA.
Previously, two studies have performed mutational analysis of the LANA C-terminal domain. First, Wong & Wilson (2005) introduced a limited set of mutations and analysed their effect on DNA binding and found that binding to DNA induced 5 ° bending or greater for LBS1 and about 11 ° for occupation on LBS1/2; furthermore, mutations preventing bending also greatly affected DNA binding of LANA. We observed similar results for mutants SKK953AAA and WE963AA in helix 3, confirming that changes in DNA bending do contribute to decreased DNA binding and replication activity (Wong & Wilson, 2005). Additionally, Kelley-Clarke et al. (2007) performed an unbiased mutational analysis across LANADBD by introducing triple alanine substitutions to define residues important for binding to the TR and attachment to host chromatin.
With respect to the importance of helix 2 for DNA recognition, our data are in agreement with both previous studies and add further details by identifying several residues whose mutation alone eliminates DNA binding. In particular, 909L, 910K, 911K and 917Q partly overlap with the conserved LXXLRY motif present in the core domains of EBNA1 and many HPV E2 proteins (Fujita et al., 2001).
With respect to helices 1 and 3, we identified several residues that contribute to DNA binding but were not identified previously (Kelley-Clarke et al., 2007). Specifically, HIF876AAA, YR879AA and all corresponding single amino acid substitutions showed drastically reduced DNA binding (Figs 3 and 4 and Table 2). In agreement with our observation, the corresponding EBNA1DBD residues are also important for DNA binding and bending, either by contacting the DNA directly or by stabilizing the N-terminal domain of DBD (Bochkarev et al., 1996). No significant changes in DNA binding were observed within helix 3 mutants. However, RL960AA, which was previously shown not to bind to DNA (Kelley-Clarke et al., 2007), bound to LBS1 or LBS1/2 with wt activity levels (Fig. 4) and also formed dimers. Observed differences between the two studies may in part be due to differences in protein expression and purification method utilized.
In summary, our data suggest that LANADBD has a high degree of structural conservation with EBNA1DBD, which is critical for sequence-specific DNA binding, multimer formation, protein–protein interactions required for its DNA replication activity and LANA-dependent transcriptional repression.
METHODS
Amino acid alignment of gammaherpesviruses LANADBDs of different primate species and EBNA1.
The sequences of KSHV LANADBD (aa 775–1003; NCBI Protein accession no. AAK50002), the reference BC-1 KSHV LANA (aa 934–1162; NCBI Protein accession no. AAC55944), EBV EBNA1DBD (aa 461–641; NCBI Protein accession no. P03211), RFHVMn LANADBD (aa 849–1071; NCBI Protein accession no. ABH07414) and RRV LANADBD (aa 251–448; NCBI Protein accession no. AAF60071) were binarily and multiply aligned using 3d-pssm version 2.6.0 (http://www.sbg.bio.ic.ac.uk/servers/3dpssm/index.html), praline (http://www.ibi.vu.nl/programs; reviewed by Pirovano & Heringa, 2010) and T-Coffee version 7.71 (http://www.tcoffee.org/; Notredame et al., 2000).
Computational prediction of the LANADBD multimer structure.
The m-zdock program (http://zlab.bu.edu/m-zdock) was used to predict putative LANADBD dimer and tetramer complexes. m-zdock is a specially developed algorithm for predicting the structure of multimers based on the structure of unbound (or partially bound) monomers (Pierce et al., 2005). The predicted tetramer of LANADBD bound to LBS1/2 was modelled based on solved structures of EBNA1DBD (Bochkarev et al., 1996).
Plasmid constructs.
pcDNA 3.1 Flag-LANADBD has been described previously (Garber et al., 2001). Fragments containing LBS1 or LBS1/2 used as EMSA probes were produced by XhoI/XbaI digestion from pAG31 containing LBS1 and pAG43 containing LBS1/2, respectively, as described previously (Garber et al., 2002).
pPuro/4TR, used for the short-term replication assay, was constructed by cloning four TR units from pCRII/4TR (Garber et al., 2002; Hu et al., 2002) into a pPur vector (BD Biosciences). PGL3/7TR, which contains seven TR units, was constructed from pAG9 (Garber et al., 2001) and used as a reporter for LANA-dependent transcriptional repression assays.
Alanine substitution mutagenesis.
A PCR-based QuikChange Site-directed Mutagenesis kit (Stratagene) was used to generate alanine substitution mutants in LANADBD, as recommended by the manufacturer. Primers containing the desired alanine substitution were designed using the web-based program Primer X (http://bioinformatics.org/primerx) (see Supplementary Table S1, available in JGV Online). All constructs were confirmed by sequencing (Davis Sequencing Co.).
Cell lines.
CV-1 cells, African green monkey fibroblasts, 293 cells, human embryonic kidney cells, were obtained from ATCC. Cell monolayers were maintained in Dulbecco's modified Eagle's medium supplemented with 10 % fetal bovine serum, 2 mM l-glutamine, 5 U penicillin ml−1 and 5 μg streptomycin ml−1 (all from Mediatech) at 37 °C under a 5 % CO2 atmosphere.
Expression of wild-type and mutant LANADBD proteins with the MVA/T7 expression system.
Wild-type and mutant LANADBD proteins were produced by using an MVA/T7 expression system (Moss et al., 1990). Briefly, highly confluent CV-1 cells in 10 cm plates were infected with MVA/T7 virus as described previously (Garber et al., 2001; Moss et al., 1990) and transfected at 3 h post-infection using a slightly modified calcium phosphate methods (Sambrook & Russell, 2001). Cells were harvested at 36–40 h post-transfection. His-tagged wt or mutant LANADBD proteins were purified using Ni2+/Tris(carboxymethyl)ethylenediamine columns (Active Motif). Protein concentrations were determined by BCA assays (Bio-Rad) and protein expression levels were determined by Western blot analysis using anti-Flag M2 antibody (Sigma-Aldrich).
EMSA.
For probe labelling, fragments containing LBS1 or LBS1/2 were labelled using T4 polynucleotide kinase (NEB) in the presence of [γ-32P]ATP (Amersham Biosciences) following the manufacturer's instructions. EMSAs were performed as described previously (Garber et al., 2001). Captured protein–DNA complex signals on the phosphor screen were analysed using a Typhoon 9410 phosphorimager system (Amersham Bioscience).
Co-immunoprecipitation.
Plasmids expressing wt and mutant Flag-tagged LANADBD proteins and a plasmid expressing wt HA-tagged LANADBD were co-transfected to evaluate dimer formation. Co-transfected cells were harvested at 36–40 h post-transfection, lysed in lysis buffer and pre-cleared by centrifugation. Lysates were co-immunoprecipitated with anti-Flag M2 beads. LANADBD complexes were separated by SDS-PAGE and the amount of HA-tagged wt LANADBD was detected and quantified by Western blotting using anti-HA antibody. Dimerization activity for each mutant was normalized based on the expression level of Flag-tagged wt or mutant LANADBD proteins.
Short-term DNA replication assays.
Co-transfection of 3×106 293 cells with 8 μg pPuro/4TR plasmid and 2 μg wt or mutant LANADBD expression plasmids was carried out using TransIT-293 Transfection Reagent (Mirus). Transfection efficiency was monitored using pcDNA3/LacZ. Short-term DNA replication assays were performed as described previously (Hu et al., 2002). Captured signals on the phosphor screen were analysed using a Typhoon 9410 phosphorimager system.
Luciferase reporter assays.
For transcriptional repression assays, 20 ng pGL3/7TR luciferase plasmid as a reporter and 380 ng wt or mutant plasmid as an effector were co-transfected into 3×105 293 cells using TransIT-293 Transfection Reagent. To monitor transfection efficiency, pMaxGFP plasmid was co-transfected with these plasmids and transfection efficiency was over 90 %. Relative light units (RLUs) were measured at 48 h post-transfection. Protein concentrations were determined by BCA assay, and RLU values were normalized to the protein concentration. This was based on previous observations that LANA modulates a wide range of promoters (Renne et al., 2001). Reporter gene activity values represented the mean of several independent transfections performed in triplicate (means±sd).
Supplementary Material
Acknowledgments
We thank Dr Robert McKenna and Dr David C. Bloom for helpful advice and critical reading of this manuscript. This work was supported by grants R01 CA88763 and R01 CA119917 from the NIH National Cancer Institute to R. R.
Footnotes
A supplementary figure and table are available with the online version of this paper.
References
- Ballestas, M. E. & Kaye, K. M. (2001). Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J Virol 75, 3250–3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballestas, M. E., Chatis, P. A. & Kaye, K. M. (1999). Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284, 641–644. [DOI] [PubMed] [Google Scholar]
- Barbera, A. J., Chodaparambil, J. V., Kelley-Clarke, B., Joukov, V., Walter, J. C., Luger, K. & Kaye, K. M. (2006). The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science 311, 856–861. [DOI] [PubMed] [Google Scholar]
- Bates, P. A., Kelley, L. A., MacCallum, R. M. & Sternberg, M. J. (2001). Enhancement of protein modeling by human intervention in applying the automatic programs 3d-jigsaw and 3d-pssm. Proteins (Suppl. 5), 39–46. [DOI] [PubMed]
- Bochkarev, A., Barwell, J. A., Pfuetzner, R. A., Furey, W., Jr, Edwards, A. M. & Frappier, L. (1995). Crystal structure of the DNA-binding domain of the Epstein–Barr virus origin-binding protein EBNA 1. Cell 83, 39–46. [DOI] [PubMed] [Google Scholar]
- Bochkarev, A., Barwell, J. A., Pfuetzner, R. A., Bochkareva, E., Frappier, L. & Edwards, A. M. (1996). Crystal structure of the DNA-binding domain of the Epstein–Barr virus origin-binding protein, EBNA1, bound to DNA. Cell 84, 791–800. [DOI] [PubMed] [Google Scholar]
- Burnside, K. L., Ryan, J. T., Bielefeldt-Ohmann, H., Bruce, A. G., Thouless, M. E., Tsai, C. & Rose, T. M. (2006). RFHVMn ORF73 is structurally related to the KSHV ORF73 latency-associated nuclear antigen (LANA) and is expressed in retroperitoneal fibromatosis (RF) tumor cells. Virology 354, 103–115. [DOI] [PubMed] [Google Scholar]
- Ceccarelli, D. F. & Frappier, L. (2000). Functional analyses of the EBNA1 origin DNA binding protein of Epstein–Barr virus. J Virol 74, 4939–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesarman, E., Chang, Y., Moore, P. S., Said, J. W. & Knowles, D. M. (1995). Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med 332, 1186–1191. [DOI] [PubMed] [Google Scholar]
- Chang, Y., Cesarman, E., Pessin, M. S., Lee, F., Culpepper, J., Knowles, D. M. & Moore, P. S. (1994). Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266, 1865–1869. [DOI] [PubMed] [Google Scholar]
- Cotter, M. A. & Robertson, E. S. (1999). The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264, 254–264. [DOI] [PubMed] [Google Scholar]
- Cruickshank, J., Shire, K., Davidson, A. R., Edwards, A. M. & Frappier, L. (2000). Two domains of the Epstein–Barr virus origin DNA-binding protein, EBNA1, orchestrate sequence-specific DNA binding. J Biol Chem 275, 22273–22277. [DOI] [PubMed] [Google Scholar]
- de Prat-Gay, G., Gaston, K. & Cicero, D. O. (2008). The papillomavirus E2 DNA binding domain. Front Biosci 13, 6006–6021. [DOI] [PubMed] [Google Scholar]
- Fiser, A. & Sali, A. (2003). ModLoop: automated modeling of loops in protein structures. Bioinformatics 19, 2500–2501. [DOI] [PubMed] [Google Scholar]
- Fujita, T., Ikeda, M., Kusano, S., Yamazaki, M., Ito, S., Obayashi, M. & Yanagi, K. (2001). Amino acid substitution analyses of the DNA contact region, two amphipathic α-helices and a recognition-helix-like helix outside the dimeric β-barrel of Epstein–Barr virus nuclear antigen 1. Intervirology 44, 271–282. [DOI] [PubMed] [Google Scholar]
- Gao, S. J., Kingsley, L., Li, M., Zheng, W., Parravicini, C., Ziegler, J., Newton, R., Rinaldo, C. R., Saah, A. & other authors (1996). KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi's sarcoma. Nat Med 2, 925–928. [DOI] [PubMed] [Google Scholar]
- Garber, A. C., Shu, M. A., Hu, J. & Renne, R. (2001). DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J Virol 75, 7882–7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber, A. C., Hu, J. & Renne, R. (2002). Latency-associated nuclear antigen (LANA) cooperatively binds to two sites within the terminal repeat, and both sites contribute to the ability of LANA to suppress transcription and to facilitate DNA replication. J Biol Chem 277, 27401–27411. [DOI] [PubMed] [Google Scholar]
- Grossman, S. R. & Laimins, L. A. (1996). EBNA1 and E2: a new paradigm for origin-binding proteins? Trends Microbiol 4, 87–89. [DOI] [PubMed] [Google Scholar]
- Grundhoff, A. & Ganem, D. (2003). The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus permits replication of terminal repeat-containing plasmids. J Virol 77, 2779–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantz, S., Couvreux, A., Champier, G., Trapes, L., Cotin, S., Denis, F., Bouaziz, S. & Alain, S. (2009). Conserved domains and structure prediction of human cytomegalovirus UL27 protein. Antivir Ther 14, 663–672. [PubMed] [Google Scholar]
- Hass, M., Lelke, M., Busch, C., Becker-Ziaja, B. & Gunther, S. (2008). Mutational evidence for a structural model of the Lassa virus RNA polymerase domain and identification of two residues, Gly1394 and Asp1395, that are critical for transcription but not replication of the genome. J Virol 82, 10207–10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havranek, J. J., Duarte, C. M. & Baker, D. (2004). A simple physical model for the prediction and design of protein–DNA interactions. J Mol Biol 344, 59–70. [DOI] [PubMed] [Google Scholar]
- Hegde, R. S., Grossman, S. R., Laimins, L. A. & Sigler, P. B. (1992). Crystal structure at 1.7 Å of the bovine papillomavirus-1 E2 DNA-binding domain bound to its DNA target. Nature 359, 505–512. [DOI] [PubMed] [Google Scholar]
- Hirt, B. (1967). Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol 26, 365–369. [DOI] [PubMed] [Google Scholar]
- Hu, J., Garber, A. C. & Renne, R. (2002). The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus supports latent DNA replication in dividing cells. J Virol 76, 11677–11687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedes, D. H., Operskalski, E., Busch, M., Kohn, R., Flood, J. & Ganem, D. (1996). The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat Med 2, 918–924. (erratum: Nat Med (1996). 2, 1041) [DOI] [PubMed] [Google Scholar]
- Kelley-Clarke, B., Ballestas, M. E., Srinivasan, V., Barbera, A. J., Komatsu, T., Harris, T. A., Kazanjian, M. & Kaye, K. M. (2007). Determination of Kaposi's sarcoma-associated herpesvirus C-terminal latency-associated nuclear antigen residues mediating chromosome association and DNA binding. J Virol 81, 4348–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, H., Petros, A. M., Meadows, R. P., Yoon, H. S., Egan, D. A., Walter, K., Holzman, T. F., Robins, T. & Fesik, S. W. (1996). Solution structure of the DNA-binding domain of a human papillomavirus E2 protein: evidence for flexible DNA-binding regions. Biochemistry 35, 2095–2103. [DOI] [PubMed] [Google Scholar]
- Lieberman, P. M., Hu, J. & Renne, R. (2007). Gammaherpesvirus maintenance and replication during latency. In Human Herpesvirus: Biology. Therapy and Immunoprophylaxis, pp. 379–402. Edited by A. Arvin, G. Campadelli-Fiume, E. Mocarski, P. S. Moore, B. Roizman, R. Whitley and K. Yamanishi. New York: Cambridge University Press.
- Lim, C., Sohn, H., Lee, D., Gwack, Y. & Choe, J. (2002). Functional dissection of latency-associated nuclear antigen 1 of Kaposi's sarcoma-associated herpesvirus involved in latent DNA replication and transcription of terminal repeats of the viral genome. J Virol 76, 10320–10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss, B., Elroy-Stein, O., Mizukami, T., Alexander, W. A. & Fuerst, T. R. (1990). Product review. New mammalian expression vectors. Nature 348, 91–92. [DOI] [PubMed] [Google Scholar]
- Notredame, C., Higgins, D. G. & Heringa, J. (2000). T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302, 205–217. [DOI] [PubMed] [Google Scholar]
- Pierce, B., Tong, W. & Weng, Z. (2005). m-zdock: a grid-based approach for Cn symmetric multimer docking. Bioinformatics 21, 1472–1478. [DOI] [PubMed] [Google Scholar]
- Pierce, B., Tong, W. & Weng, Z. (2007). zrank: reranking protein docking predictions with an optimized energy function. Proteins 67, 1078–1086. [DOI] [PubMed] [Google Scholar]
- Pirovano, W. & Heringa, J. (2010). Protein secondary structure prediction. Methods Mol Biol 609, 327–348. [DOI] [PubMed] [Google Scholar]
- Purta, E., van Vliet, F., Tricot, C., De Bie, L. G., Feder, M., Skowronek, K., Droogmans, L. & Bujnicki, J. M. (2005). Sequence–structure–function relationships of a tRNA (m7G46) methyltransferase studied by homology modeling and site-directed mutagenesis. Proteins 59, 482–488. [DOI] [PubMed] [Google Scholar]
- Renne, R., Barry, C., Dittmer, D., Compitello, N., Brown, P. O. & Ganem, D. (2001). Modulation of cellular and viral gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J Virol 75, 458–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. & Russell, D. W. (eds) (2001). Molecular Cloning: a Laboratory Manual, 3rd edn. New York: Cold Spring Harbor Laboratory Press.
- Schepers, A., Ritzi, M., Bousset, K., Kremmer, E., Yates, J. L., Harwood, J., Diffley, J. F. & Hammerschmidt, W. (2001). Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein–Barr virus. EMBO J 20, 4588–4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwam, D. R., Luciano, R. L., Mahajan, S. S., Wong, L. & Wilson, A. C. (2000). Carboxy terminus of human herpesvirus 8 latency-associated nuclear antigen mediates dimerization, transcriptional repression, and targeting to nuclear bodies. J Virol 74, 8532–8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulier, J., Grollet, L., Oksenhendler, E., Cacoub, P., Cazals-Hatem, D., Babinet, P., d'Agay, M. F., Clauvel, J. P., Raphael, M. & Degos, L. (1995). Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86, 1276–1280. [PubMed] [Google Scholar]
- Stedman, W., Deng, Z., Lu, F. & Lieberman, P. M. (2004). ORC, MCM, and histone hyperacetylation at the Kaposi's sarcoma-associated herpesvirus latent replication origin. J Virol 78, 12566–12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma, S. C., Choudhuri, T., Kaul, R. & Robertson, E. S. (2006). Latency-associated nuclear antigen (LANA) of Kaposi's sarcoma-associated herpesvirus interacts with origin recognition complexes at the LANA binding sequence within the terminal repeats. J Virol 80, 2243–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma, S. C., Lan, K. & Robertson, E. (2007). Structure and function of latency-associated nuclear antigen. Curr Top Microbiol Immunol 312, 101–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, L. Y. & Wilson, A. C. (2005). Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen induces a strong bend on binding to terminal repeat DNA. J Virol 79, 13829–13836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You, J., Srinivasan, V., Denis, G. V., Harrington, W. J., Jr, Ballestas, M. E., Kaye, K. M. & Howley, P. M. (2006). Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with bromodomain protein Brd4 on host mitotic chromosomes. J Virol 80, 8909–8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.