Abstract
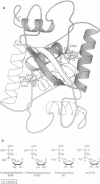
We have studied quantitatively the effect of the corepressor, S-adenosylmethionine (SAM), on the interaction between the E. coli methionine repressor, MetJ, and an idealised operator fragment, by recording measurements of surface plasmon resonance using a BIAcore instrument. We have recorded kinetic binding data in the presence of SAM, which carries a net positive charge, and two corepressor analogues, adenosylornithine (AO) and aza-SAM, which differ in the location of the atom carrying the positive charge. Our data support the hypothesis that the effect of the corepressor is electrostatic in origin. The difference in electrostatic interaction energy between the SAM- and AO-repressor-operator complexes of approximately 3.5 kJ/mol calculated from the known three-dimensional structure is within the range of our experimentally determined values of 2.8-4.3 kJ/mol. These results illustrate the potential of SPR measurements for studying protein-nucleic acid interactions.
Full text
PDF
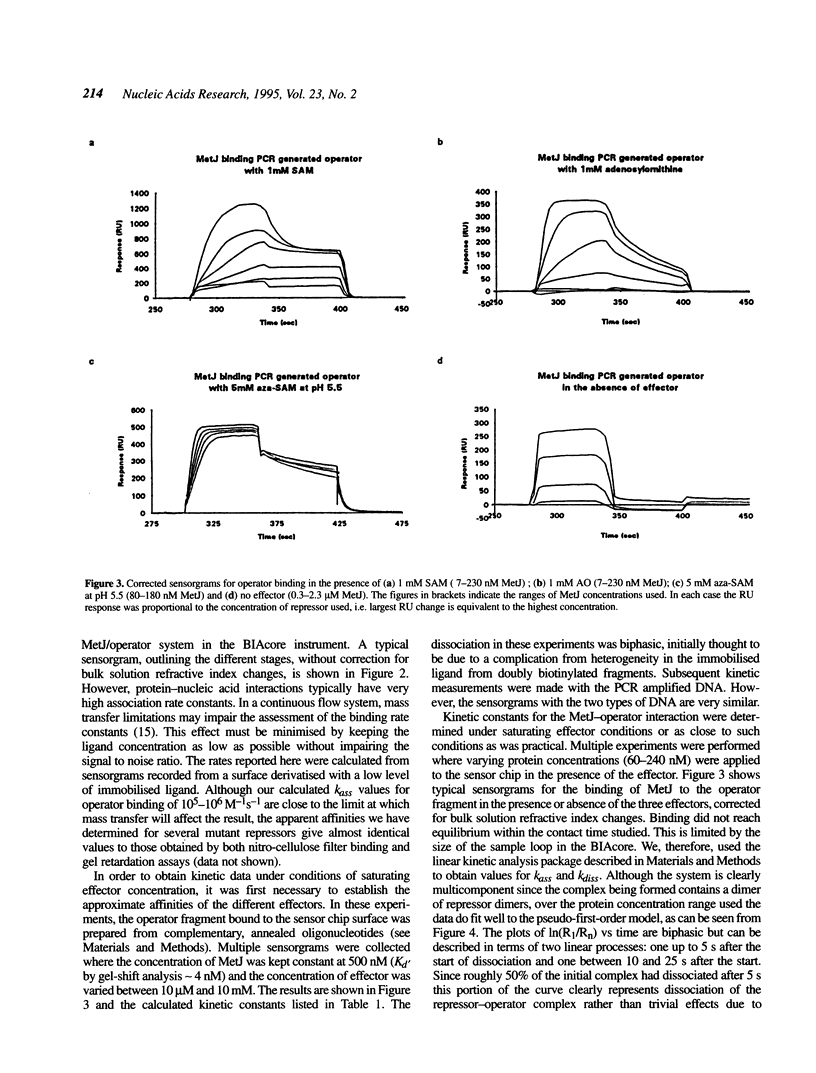
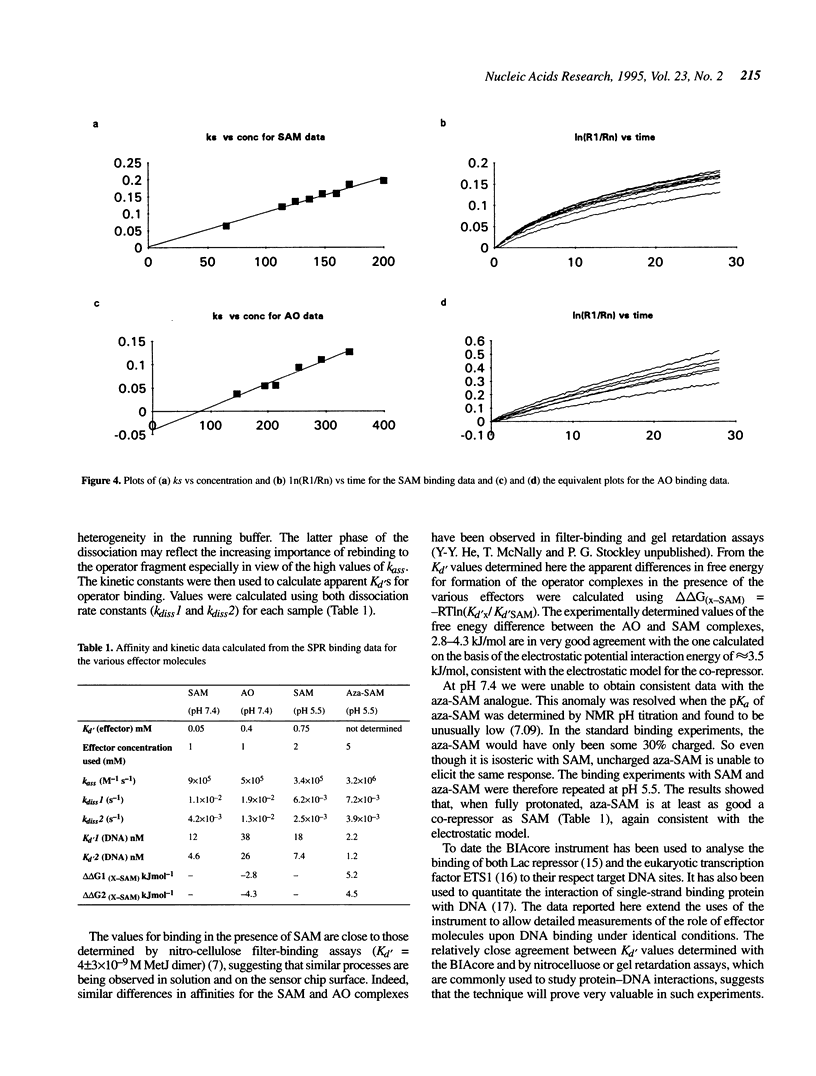

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bondeson K., Frostell-Karlsson A., Fägerstam L., Magnusson G. Lactose repressor-operator DNA interactions: kinetic analysis by a surface plasmon resonance biosensor. Anal Biochem. 1993 Oct;214(1):245–251. doi: 10.1006/abio.1993.1484. [DOI] [PubMed] [Google Scholar]
- Davidson B. E., Saint Girons I. The Escherichia coli regulatory protein MetJ binds to a tandemly repeated 8 bp palindrome. Mol Microbiol. 1989 Nov;3(11):1639–1648. doi: 10.1111/j.1365-2958.1989.tb00149.x. [DOI] [PubMed] [Google Scholar]
- Fisher R. J., Fivash M., Casas-Finet J., Erickson J. W., Kondoh A., Bladen S. V., Fisher C., Watson D. K., Papas T. Real-time DNA binding measurements of the ETS1 recombinant oncoproteins reveal significant kinetic differences between the p42 and p51 isoforms. Protein Sci. 1994 Feb;3(2):257–266. doi: 10.1002/pro.5560030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y. Y., McNally T., Manfield I., Navratil O., Old I. G., Phillips S. E., Saint-Girons I., Stockley P. G. Probing met repressor-operator recognition in solution. Nature. 1992 Oct 1;359(6394):431–433. doi: 10.1038/359431a0. [DOI] [PubMed] [Google Scholar]
- Heyduk T., Lee J. C. Escherichia coli cAMP receptor protein: evidence for three protein conformational states with different promoter binding affinities. Biochemistry. 1989 Aug 22;28(17):6914–6924. doi: 10.1021/bi00443a021. [DOI] [PubMed] [Google Scholar]
- Johnson C. M., Cooper A., Stockley P. G. Differential scanning calorimetry of thermal unfolding of the methionine repressor protein (MetJ) from Escherichia coli. Biochemistry. 1992 Oct 13;31(40):9717–9724. doi: 10.1021/bi00155a027. [DOI] [PubMed] [Google Scholar]
- Karlsson R., Michaelsson A., Mattsson L. Kinetic analysis of monoclonal antibody-antigen interactions with a new biosensor based analytical system. J Immunol Methods. 1991 Dec 15;145(1-2):229–240. doi: 10.1016/0022-1759(91)90331-9. [DOI] [PubMed] [Google Scholar]
- O'Shannessy D. J., Brigham-Burke M., Soneson K. K., Hensley P., Brooks I. Determination of rate and equilibrium binding constants for macromolecular interactions using surface plasmon resonance: use of nonlinear least squares analysis methods. Anal Biochem. 1993 Aug 1;212(2):457–468. doi: 10.1006/abio.1993.1355. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Schevitz R. W., Zhang R. G., Lawson C. L., Joachimiak A., Marmorstein R. Q., Luisi B. F., Sigler P. B. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988 Sep 22;335(6188):321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- Phillips K., Phillips S. E. Electrostatic activation of Escherichia coli methionine repressor. Structure. 1994 Apr 15;2(4):309–316. doi: 10.1016/s0969-2126(00)00032-0. [DOI] [PubMed] [Google Scholar]
- Phillips S. E., Manfield I., Parsons I., Davidson B. E., Rafferty J. B., Somers W. S., Margarita D., Cohen G. N., Saint-Girons I., Stockley P. G. Cooperative tandem binding of met repressor of Escherichia coli. Nature. 1989 Oct 26;341(6244):711–715. doi: 10.1038/341711a0. [DOI] [PubMed] [Google Scholar]
- Rafferty J. B., Somers W. S., Saint-Girons I., Phillips S. E. Three-dimensional crystal structures of Escherichia coli met repressor with and without corepressor. Nature. 1989 Oct 26;341(6244):705–710. doi: 10.1038/341705a0. [DOI] [PubMed] [Google Scholar]