INTRODUCTION
Structured abstracts contain distinct labeled sections (e.g., “RESULTS”). The MEDLINE/PubMed database incorporates English-language abstracts that appear in the journals that the US National Library of Medicine (NLM) indexes. If English-language structured abstracts appear in a journal that is indexed, the labels in these abstracts usually appear in all uppercase letters, generally followed by a colon, in MEDLINE/PubMed citations [1].
Several years after formats for more informative abstracts were proposed [2–5], NLM studied the structured abstracts that appeared in MEDLINE from 1989–1991 as an initial step in exploring their utility in enhancing bibliographic retrieval [6]. This early study showed that structured abstracts were an emerging, but rapidly growing phenomenon; that MEDLINE records with structured abstracts tended to have more access points (Medical Subject Headings [MeSH] terms and text words) than MEDLINE records as a whole; and that there was significant variation in the structured abstract formats that different journals prescribed.
Implementation of structured abstracts by biomedical journals has been examined on a small scale in the clinical medicine domain [7, 8], but no large-scale examination across all of MEDLINE has occurred since the first exploratory study by NLM. Hence, the objective of this study was to conduct a retrospective cohort study to measure and characterize the growth in structured abstracts in MEDLINE since 1991, with a view, again, toward exploring their utility in enhancing information display and retrieval.
METHODS
Data source
NLM produces annual, static versions of the MEDLINE data (MEDLINE/PubMed Baseline Repository) that are freely available for use by any researcher [9]. The 2007 MEDLINE baseline dataset [10] was used in this study. Fifteen years of MEDLINE records (7,163,494 records) with record completion dates from November 25, 1991, to November 14, 2006, were extracted from this baseline dataset. Two types of records that had been added to MEDLINE/PubMed during the relevant time period were then excluded from the final research dataset because they were not comparable to records used in the 1989–1991 study: (1) PubMed records that had not received MEDLINE indexing and (2) recently digitized records for pre-1966 Index Medicus citations. Then, all records without abstracts were excluded, leaving a MEDLINE subset of 5,483,473 records. Because the NLM indexing year ended in mid-November, records for articles with 2006 publication dates that were processed by NLM after November 14, 2006, were not part of this subset.
Algorithm for identification of structured abstracts
The 5,483,473 MEDLINE records with abstracts were used to develop and validate a new algorithm for identifying structured abstracts:
Step 1
The algorithm used in the 1989–1991 study was applied to the 1992–2006 set of MEDLINE records with abstracts. However, the twenty-seven labels used in the earlier study were known to be insufficient to retrieve all the variations in structured abstract formats that had appeared since 1991. An additional set of sixty-six structured abstract labels had been identified based on label analyses done on some smaller MEDLINE subsets, information provided by a few experts in the field, and serendipitous discoveries of labels in abstracts in individual MEDLINE citations.
Step 2
To identify a more complete set of labels for use in isolating structured abstracts, a new label discovery algorithm was developed and validated. The discovery algorithm examined each abstract and identified all uppercase strings that (1) appeared in the first character word position of the abstracts, (2) were followed by either a colon or period (e.g., “PRACTICES AND PATIENTS:”), and (3) occurred in an abstract that also contained at least 1 of the following strings: “RESULT:,” “RESULTS:,” “CONCLUSION:,” or “CONCLUSIONS:.” An additional 458 labels, not present in the original list of 27 or in the supplemental list of 66, were identified by this algorithm.
Step 3
Knowledge gained from the discovery algorithm and random samples of records showed that a more comprehensive identification of structured abstracts could be achieved through the use of a set of labels that included slight variations, such as mixed-case, spelling variants, plurals, and punctuation variations. A set of 1,335 labels, including 784 minor variants of previously identified labels, was extracted for use in identifying the structured abstracts to be analyzed in this study.
This study ultimately defined a structured abstract as one that contained at least 3 labels that represented 3 distinct concepts (e.g., “INTRODUCTION,” “METHODS,” “CONCLUSIONS”), of which 1 must be considered an ending concept (such as “RESULTS” or “CONCLUSIONS”) from the list of 1,335 labels [11]. This algorithm found 4.6% more structured abstracts than were identified by the 27 labels used in the 1989–1991 study.
Final structured abstract research dataset
This structured abstract definition was applied to the records in the MEDLINE subset, identifying 938,772 records. To confirm that this new structured abstract subset contained only valid structured abstracts, a random sample of 671 records (99% confidence interval; margin of error 5 for a universe size of 938,772 [12]) was generated and examined. All 671 abstracts conformed to the study's definition of a structured abstract.
Manual categorization of structured abstract labels
The frequency of occurrence of each unique label in the structured abstract subset was computed. Unique labels considered variants of the same concept were then grouped under the most frequently occurring label for that concept, called the metaterm, and the frequencies of all the variants were combined to arrive at an overall total for that concept. The resulting list of metaterms was examined to determine if the metaterms could be logically grouped into a small number of higher-level categories (by small group consensus), such as “METHODS,” as a potential aid to data mining and retrieval applications.
RESULTS
Growth in structured abstracts in biomedical journals
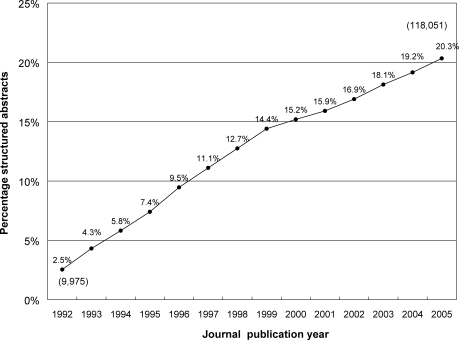
The percentage of new MEDLINE records containing structured abstracts rose from 2.5% for 1992 to 20.3% for 2005 (Figure 1). Because the number of articles indexed each year was also increasing throughout this period, the absolute number of structured abstracts increased even more substantially, from 9,975 (1992) to 118,051 (2005, the last publication year with complete data in the research dataset), more than 1,000.0%.
Figure 1.
Percentage* of structured abstracts per publication year
Characteristics of MEDLINE records with structured abstracts
MEDLINE records with structured abstracts were larger in size than MEDLINE as a whole (7,163,494 records), and they had more access points (Table 1, online only). The average size of a structured abstract record was 758 bytes bigger than the average size of a MEDLINE record. Abstracts from the structured abstract set (938,772 records) had an average of about 57 more words than abstracts from the entire MEDLINE set (5,483,473 records). MEDLINE records with structured abstracts contained an average of 4 labels, but the structured abstract labels alone were not responsible for the larger record sizes. The average title of an article with a structured abstract contained about 2 more words than the average title in MEDLINE records as a whole. Records with structured abstracts had an average of 2.4 more MeSH terms and 1.3 more authors than records in MEDLINE as a whole.
Analysis of structured abstract labels
The majority of the 1,335 structured abstract labels identified in the study refer to 1 or more of only 100 concepts. Each of these 100 concepts was assigned its most frequently occurring name as a metaterm. Table 2 (online only) presents the metaterm “PROBLEM” and its 32 label variants as an example.
These 100 metaterms covered 99.9% of more than 4 million structured abstract label occurrences identified in the study. The 100 metaterms were then manually grouped into 5 higher-level categories, called metacategories:
“BACKGROUND”: 38 metaterms (for 403,301 or 10.0% of label occurrences)
“OBJECTIVE”: 7 metaterms (for 657,146 or 16.2% of label occurrences)
“METHODS”: 33 metaterms (for 1,142,491 or 28.2% of label occurrences)
“RESULTS”: 8 metaterms (for 906,521 or 22.4% of label occurrences)
“CONCLUSIONS”: 14 metaterms (for 942,329 or 23.2% of label occurrences)
DISCUSSION
Both the number of individual records with structured abstracts and the number of journals publishing structured abstracts has increased steadily since 1991. Structured abstracts appeared in 20.3% of the 580,583 articles indexed for MEDLINE in 2005, the last full year covered by this study, and the upward trend is continuing (23.0% in 2008). Records with structured abstracts represented 13.1% (938,772/7,163,494) of 1992–2006 MEDLINE as a whole. This is a dramatic increase from 1989–1991, when structured abstracts appeared in only 0.4% (3,873/924,748) of MEDLINE records. A total of 3,166 journal titles contributed structured abstracts from 1992–2006, in comparison to only 78 journals in 1989–1991. More than 1,000 journals have continuously published structured abstracts (starting in one year and continuing through 2005) in contrast to only 10 journals in 1989–1991.
The 1989–1991 study had some indication that the additional MeSH terms assigned to records with structured abstracts were the result of more indexing of patient demographic characteristics, such as sex or age groups. The current study confirms this. For example, in both studies, roughly 60% of records with structured abstracts were assigned the MeSH term “Female,” in comparison to about 30% of MEDLINE records as a whole. Similar substantial differences occurred for the MeSH terms “Male,” “Adult,” and “Middle Age.”
CURRENT STATUS AND FUTURE DIRECTIONS
Now that structured abstracts appear in a sizeable fraction of MEDLINE records (nearly a quarter of all abstracts added to MEDLINE in a year), NLM is exploring their utility in enhancing display, assisting the indexing process, and improving information retrieval and discovery. NLM has made two changes as a direct result of this updated research on the occurrence of structured abstracts in MEDLINE/PubMed citations:
the PubMed abstract display changed in August 2010 to show the labels in bold with each section starting on a new line [13] and
the 2011 document type definition (DTD) for the MEDLINE citation <http://www.nlm.nih.gov/databases/dtd/nlmmedlinecitationset_110101.dtd> has been enhanced so that each section of a structured abstract is encoded in extensible markup language (XML), separating the label from the text and recording the metacategory map so that data recipients do not have to translate the multitude of possible labels into the metacategories.
The first change improves the readability of structured abstracts in PubMed. The second change makes it possible to improve even further on the readability; for example, a technique such as color-coded labels for the metacategory to which they map could be employed to make it faster for readers to scan for the section of most interest to the search at hand. The second change also makes it possible for NLM to conduct experiments to determine the utility of features such as:
a PubMed limit for citations having structured abstracts;
new search tags that restrict retrieval to the terms occurring in certain sections of an abstract, for example, the “OBJECTIVE” or the “RESULTS”;
modifications to the Related Citations algorithm [14] in PubMed to weight words from the various sections differently (for example, decreasing the weight for the “BACKGROUND” section, while increasing the weight for the “OBJECTIVE” section); and
modifications to the NLM Medical Text Indexer [15] software application that parses journal article titles and abstracts to suggest possible MeSH terms for the human indexer to select; for example, the “OBJECTIVE” and “RESULTS” sections could be targeted for increased weighting in the suggestion of terms.
Licensees of NLM MEDLINE data will be able to conduct their own experiments and implement new functionality for users of their applications of the data, too, now that NLM has coded the structured abstracts for the 2011 DTD.
Work is also underway to identify new labels that have appeared since 2006 and to map them to the five metacategories. Detailed information about NLM structured abstract research is available on the Structured Abstracts in MEDLINE site <http://structuredabstracts.nlm.nih.gov>.
Electronic Content
Acknowledgments
The authors are grateful to former NLM associate fellow, Amy McNeely, bookshare librarian, Benetech Initiative, Palo Alto, California, for her careful analysis that did the initial mapping of the structured abstract segment labels found in MEDLINE records into metaterms and metacategories and for her assistance in validating the structured abstract algorithm.
This research is supported by the Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Footnotes
REFERENCES
- 1.US National Library of Medicine. Structured abstracts [Internet] Bethesda, MD: The Library; 20 May 2008. [rev. 9 Aug 2010; cited 9 Aug 2010]. < http://www.nlm.nih.gov/bsd/policy/structured_abstracts.html>. [Google Scholar]
- 2.Ad Hoc Working Group for Critical Appraisal of the Medical Literature. A proposal for more informative abstracts of clinical articles. Ann Intern Med. 1987 Apr;106(4):598–604. [PubMed] [Google Scholar]
- 3.Haynes R.B, Mulrow C.D, Huth E.J, Altman D.G, Gardner M.J. More informative abstracts revisited. Ann Intern Med. 1990 Jul 1;113(1):69–76. doi: 10.7326/0003-4819-113-1-69. [DOI] [PubMed] [Google Scholar]
- 4.Mulrow C.D, Thacker S.B, Pugh J.A. A proposal for more informative abstracts of review articles. Ann Intern Med. 1988 Apr;108(4):613–5. doi: 10.7326/0003-4819-108-4-613. [DOI] [PubMed] [Google Scholar]
- 5.Hayward R.S, Wilson M.C, Tunis S.R, Bass E.B, Rubin H.R, Haynes R.B. More informative abstracts of articles describing clinical practice guidelines. Ann Intern Med. 1993 May 1;118(9):731–7. doi: 10.7326/0003-4819-118-9-199305010-00012. [DOI] [PubMed] [Google Scholar]
- 6.Harbourt A.M, Knecht L.S, Humphreys B.L. Structured abstracts in MEDLINE, 1989–1991. Bull Med Libr Assoc. 1995 Apr;83(2):190–5. [PMC free article] [PubMed] [Google Scholar]
- 7.Nakayama T, Hirai N, Yamazaki S, Naito M. Adoption of structured abstracts by general medical journals and format for a structured abstract. J Med Libr Assoc. 2005 Apr;93(2):237–42. [PMC free article] [PubMed] [Google Scholar]
- 8.Kulkarni H. Structured abstracts: still more. Ann Intern Med. 1996 Apr 1;124(7):695–6. doi: 10.7326/0003-4819-124-7-199604010-00021. [DOI] [PubMed] [Google Scholar]
- 9.US National Library of Medicine. MEDLINE®/PubMed® baseline repository (MBR) [Internet] Bethesda, MD: The Library; [rev. 24 May 2010; cited 3 Aug 2010]. < http://mbr.nlm.nih.gov>. [Google Scholar]
- 10.US National Library of Medicine. 2007 MEDLINE®/PubMed® baseline: 16,120,074 citations found [Internet] Bethesda, MD: The Library; 4 Jan 2007. [cited 3 Aug 2010]. < http://www.nlm.nih.gov/archive//20090811/bsd/licensee/2007_stats/2007_LO.html>. [Google Scholar]
- 11.US National Library of Medicine. Structured abstracts in MEDLINE® [Internet] Bethesda, MD: The Library; 18 May 2010. [rev. 4 Jun 2010; cited 3 Aug 2010]. < http://structuredabstracts.nlm.nih.gov>. [Google Scholar]
- 12.RaoSoft. Raosoft® sample size calculator [Internet] Raosoft. 2004. [cited 3 Aug 2010]. < http://www.raosoft.com/samplesize.html>.
- 13.Ripple A.M, Mork J.G, Knecht L.S. Structured abstracts: a new look for the PubMed® abstract display. NLM Tech Bull. 2010 Jul–Aug. p. e11.
- 14.US National Library of Medicine. PubMed help: computation of related citations [Internet] Bethesda, MD: The Library; [rev. 28 Jun 2010; cited 11 Aug 2010]. < http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=helppubmed&part=pubmedhelp#pubmedhelp.Computation_of_Related_Citati>. [Google Scholar]
- 15.Aronson A.R, Mork J.G, Gay C.W, Humphrey S.M, Rogers W.J. The NLM indexing initiative's medical text indexer. Stud Health Technol Inform. 2004;107(pt 1):268–72. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.