This study shows that mTOR signaling plays important roles in regulating aging of the RPE.
Abstract
Purpose.
Mammalian target of rapamycin (mTOR)-mediated pathways play central roles in regulating aging. The purpose of the present study was to characterize the mTOR cascade in human retinal pigment epithelial (RPE) cells and to investigate its potential roles in controlling RPE senescence.
Methods.
Expression of major components of the mTOR signaling networks was evaluated by Western blot analyses. Formations of the two signaling complexes of mTOR, mTORC1, and mTORC2 were determined by coimmunoprecipitation. The activation of mTORC1 was monitored by measuring the phosphorylation status of the downstream substrate protein S6. Senescence of the cultured human RPE cells was assessed by measuring both the senescence associated-β-galactosidase (SA-β-Gal) activity and the expression level of p16, a cyclin-dependent kinase inhibitor.
Results.
Human RPE cells contained functional mTORC1 and mTORC2 signaling complexes. The assembly and activity of mTORC1 were regulated by upstream nutrient and growth factor signals. The sensitivity of mTORC1 to extracellular nutrient stimuli increased in RPE cells that had developed in vitro senescence. Suppression of the mTORC1 by rapamycin prevented the appearance of senescence markers in the RPE.
Conclusions.
The mTOR pathway presented age-associated changes in human RPE cells, and downregulation of mTORC1 could delay the aging process of the RPE.
Advanced age is a major risk factor for a number of vision-threatening eye diseases, including cataract, glaucoma, diabetic retinopathy, and age-related macular degeneration (AMD). Although the mechanistic link between aging and AMD remains elusive, age-dependent degeneration of the RPE is a hallmark pathologic change of AMD, especially in the atrophic form.1–3 The RPE is part of the blood-retina barrier and is responsible for nutrient support and metabolism.4 With aging, the RPE monolayer loses cells, whereas the remaining cells are subjected to increased metabolic demand.5,6 Decreased mitochondrial function and compromised antioxidant capacity are evident in the aged RPE.7 The production of neurotrophic and angiogenic factors secreted by the RPE shows age-related changes.8,9 Recently, it has been shown that the RPE/choroid complex becomes immunoreactive with aging10; this may be associated with chronic inflammation and abnormal immune responses under certain genetically susceptible backgrounds such as complement factor H mutation. The molecular mechanisms underlying these age-related changes are largely unknown.
The process of aging is determined by a genetically programmed molecular clock and is subjected to regulation by multiple pathways in which TOR-mediated signaling is one of the central players.11,12 TOR is a large serine/threonine protein kinase that integrates upstream signals from nutrients, growth factors, and energy levels to control cell growth and proliferation.13,14 In mammals, mTOR resides in two distinct signaling complexes (mTORC): mTORC1 (mTOR/Raptor/GβL) and mTORC2 (mTOR/Rictor/GβL/SIN1). The rapamycin-sensitive mTORC1 regulates translation and protein synthesis largely through the downstream effector proteins S6K and 4E-BP1.15 The phosphorylation status of these two proteins and substrate S6, further downstream, are commonly used as indicators of mTORC1 activity. In contrast to mTORC1, mTORC2 is resistant to short-term exposure to rapamycin16 and is a principal kinase responsible for the phosphorylation of Akt at Ser473.17–19 Negative regulation of aging by TOR signaling has been firmly established in various invertebrate models.12 Very recently, it has been shown that knocking out S6K increases life span in mice.20 Consistently, reduced mTOR/S6K activity has been observed in mice with extended longevity.21 However, whether mTOR regulates tissue-specific degeneration of the retina has not been well studied.
In the present study, we have performed an initial characterization of the mTOR pathway in both immortalized ARPE-19 cells and human RPE cells isolated from donor eye tissues. Our results demonstrate that the RPE cells contain mTOR complexes that can be activated by various upstream stimuli. Cultured primary RPE cells entered replicative senescence after serial passages, and downregulation of mTORC1 signaling relieved senescence-associated phenotypical changes. Furthermore, we found that the response of mTORC1 to nutrient signal was upregulated in the aged RPE cells. Our data indicate that changes in the mTOR-related signaling network will likely contribute to degeneration of the RPE.
Materials and Methods
Cell Culture and Treatment
Cultures of human adult retinal pigment epithelial (haRPE) cells were established as described previously.22 Primary fetal RPE cells (hfRPE) derived from human fetal eyes (Advanced Bioscience Resources, Alameda, CA) were prepared according to published procedures.23 ARPE-19 cells were obtained from American Type Culture Collection (ATCC, Manassas, VA). Cells were cultured in Dulbecco's modified Eagle's/F12 medium supplemented with 10% fetal bovine serum (Sigma-Aldrich, St. Louis, MO) in a humidified CO2 incubator at 37°C.
Replicative senescence was introduced by repeatedly passaging cells at a 1:4 ratio. Population doubling level (PDL) at each passage was calculated using the formula PDL = 3.32 × log (total viable cells at harvest/total cells at seed), and the cumulative PDL (CPD) was used to determine the age of the cells.24 Under our experimental conditions, hfRPE cells had a CPD ranging from 34 to 42, depending on the individual line. Cell senescence was monitored by their morphologic changes and SA-β-Gal staining.
For amino acid starvation and repletion, cells were serum starved for 16 hours and then transferred to phosphate-buffered saline (PBS) containing 1 mg/mL d-glucose for an additional 90 minutes. An amino acid stock mixture in the form of 50× the standard concentration (Sigma-Aldrich) was added back to the medium to the final concentration of 1× and incubated for 30 minutes.25,26 For serum or insulin stimulation, cells were serum starved in DMEM for 16 hours. Cells were then incubated with either 10% FBS for 90 minutes or with 100 nM insulin for 30 minutes. Rapamycin (Sigma-Aldrich) was used at a final concentration of 2 nM unless otherwise indicated.
Western Blot Analysis and Antibodies
Cells were rinsed with ice-cold PBS twice and harvested with a cell scraper. After centrifugation, the pellet was lysed in modified RIPA buffer (50 mM Tris-HCl [pH 7.4], 1% NP-40, 150 mM NaCl, 1 mM EDTA, 1 mM Na3VO4, 1 mM NaF) supplemented with protease inhibitor cocktail (Sigma-Aldrich). Samples were resolved on SDS-PAGE and transferred to nitrocellulose membranes. Membranes were probed with specific antibodies, and signals were detected by an infrared imaging system (Odyssey; LI-COR, Lincoln, NE). Anti-p16 was purchased from BD Biosciences (San Jose, CA), and all other antibodies were from Cell Signaling Technology (Danvers, MA).
Coimmunoprecipitation
Coimmunoprecipitation was carried out according to Kim et al.27 Briefly, cells were washed with ice-cold PBS twice and lysed in buffer containing 40 mM HEPES (pH 7.4), 120 mM NaCl, 1 mM EDTA, 10 mM pyrophosphate, 10 mM glycerophosphate, 50 mM NaF, 1.5 mM Na3VO4, and 0.3% CHAPS and were supplemented with protease inhibitor cocktail. The lysates were precleared with protein G agarose for 30 minutes. Precleared lysates were incubated with either anti-mTOR antibody or control IgG for 3 hours at 4°C, followed by incubation with protein G agarose beads for 1 hour. Beads were washed four times with lysis buffer and resuspended in SDS sample buffer. Immunoprecipitates were resolved on SDS-PAGE and analyzed by immunoblotting.
Measurement of SA-β-Gal Activity
SA-β-Gal activity was evaluated using a staining kit (Cell Signaling) according to the manufacturer's instructions. Briefly, cells were fixed in 1× fixative solution for 15 minutes at room temperature. After two washes with PBS, cells were incubated overnight at 37°C in 1× staining solution containing isopropyl β-D-1-thiogalactopyranoside as the substrate. Thereafter, cells were washed with PBS once, and the number of SA-β-Gal–positive cells (blue staining) was monitored under bright-field illumination.
Alternatively, the activity of SA-β-Gal was quantitatively measured using a fluorogenic substrate 4-methylumbelliferyl-β-D-galactopyranoside (MUG). SA-β-gal can convert 4-MUG into 4-methylumbelliferone (4-MU), which has a bright blue fluorescence.28 To perform the assay, cells were lysed in lysis buffer (5 mM CHAPS, 40 mM citric acid, 40 mM sodium phosphate, pH 6.0, supplemented with protease inhibitor cocktails). The lysate was incubated with equal volumes of 2× reaction buffer (40 mM citric acid, 40 mM sodium phosphate, 300 mM NaCl, 10 mM β-mercaptoethanol, and 4 mM MgCl2, pH 6.0; freshly added 4-MUG at the final concentration of 1.7 mM) for 30 minutes at 37°C. The reaction was stopped by the addition of 10× volume of 0.4 M sodium carbonate. Fluorescence was measured using a plate reader (Spectramax M2; Molecular Devices, Sunnyvale, CA) with excitation 360 nm and emission 465 nm. The activity of SA-β-Gal was calculated using a 4-MU standard curve and normalized by the number of cells in each reaction.
Statistical Analysis
Results from experiments measuring SA-β-Gal activity and S6 phosphorylation were repeated at least three times and were averaged and expressed as mean ± SEM. Statistical analyses were performed with commercially available software (InStat3; GraphPad Software, Inc., San Diego, CA) using Student's t-tests.
Results
Functional mTOR Signaling Network in Cultured Human RPE Cells
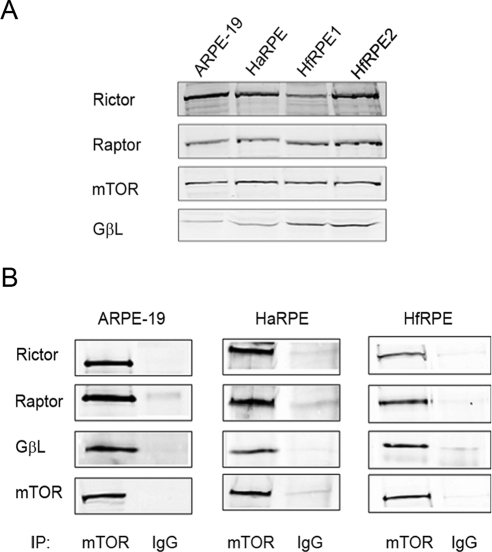
Because of their critical roles in regulating cell growth, the mTOR-associated signaling pathways have been studied extensively in cancer cells. However, few studies have been performed in nontransformed normal cells. We explored this signaling network in the RPE using an immortalized cell line, ARPE-19, as well as in primary cultures of both human adult (haRPE) and fetal (hfRPE) RPE cells. Western blot analyses showed that the key components of the mTOR signaling complexes are expressed in all three types of the RPE cells investigated (Fig. 1A). To detect the presence of mTORC1 and mTORC2 complexes in human RPE cells, coimmunoprecipitation with a specific antibody against mTOR was performed. As shown in Figure 1B, both Raptor and Rictor were detected in the mTOR immunoprecipitates, indicating the existence of both signaling complexes. GβL, the common component in both complexes, was also coimmunoprecipitated with mTOR.
Figure 1.
Expression of mTOR signaling complexes in human RPE. (A) Cell lysates were prepared from an immortalized cell line (ARPE-19) and from cultured primary haRPE and hfRPE cells. Components of mTOR signaling complexes were detected by specific antibodies. (B) Coimmunoprecipitation was performed using either anti–mTOR antibody or control IgG. The complex formation with Rictor, Raptor, and GβL was analyzed by immunoblotting. Data presented are representative of three separate experiments.
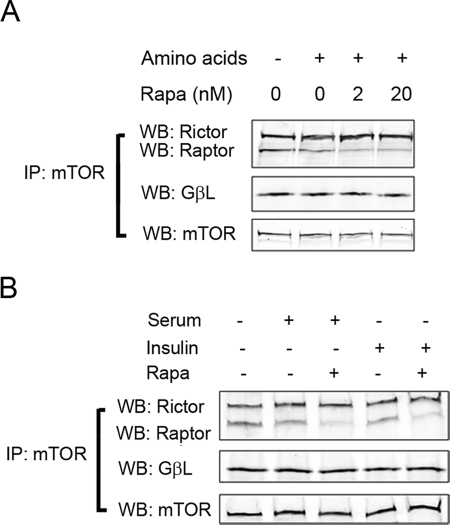
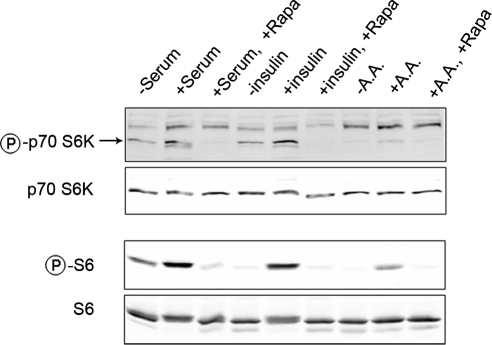
To further characterize the responses of mTOR to upstream stimuli, we measured the formation of mTORC1 in haRPE cells exposed to nutrients and growth factors (Fig. 2). Similar to previous studies in other cell types,27,29 upstream signals caused a moderate decline in the interaction between Raptor and mTOR, but not the binding of GβL to mTOR. The destabilization of the mTOR-Raptor interaction may indicate the occurrence of conformational changes that are required for exposing substrates to the kinase domain mTOR.30 The data also showed that mTORC1, but not mTORC2, was sensitive to pretreatment of rapamycin, which disrupted the formation of mTORC1 at a concentration as low as 2 nM (Fig. 2A). The phosphorylation status of p70 S6K is an indicator of mTORC1 activity. Incubation with growth factors and amino acids resulted in the phosphorylation of p70 S6K and its further downstream substrate S6 in a rapamycin-sensitive manner (Fig. 3), verifying the involvement of mTORC1. Nutrient and growth factor signals stimulated mTOR signaling in both hfRPE and ARPE-19 cells; the latter were slightly less responsive to upstream stimuli (data not shown). Thus, cultured human RPE cells have functional mTOR signaling complexes that can respond to upstream stimuli from diverse environmental factors.
Figure 2.
hRPE cells contained rapamycin-sensitive mTORC1 whose formation could be regulated by nutrients and growth factors. haRPE were deprived of amino acids for 60 minutes and restimulated with amino acids for 10 minutes with or without preincubation of rapamycin at the indicated concentrations (A) or serum-starved for 16 hours and then stimulated with 20% FBS for 30 minutes or 100 nM insulin for 10 minutes in the presence or absence of 2 nM rapamycin (B). Cells were lysed, and coimmunoprecipitation was performed with an antibody against mTOR. Levels of Rictor, Raptor, and GβL in the pellets were analyzed by Western blot analysis. Data presented are representative of two separate experiments.
Figure 3.
mTORC1-mediated signaling responded to extracellular stimuli in a rapamycin-sensitive manner. haRPE cells were treated with FBS, insulin, or amino acids (AA). The phosphorylation status of p70 S6K and S6 was determined with phosphorylation-specific antibodies. Data presented here are representative of two separate experiments.
mTORC1 Signaling in RPE Senescence
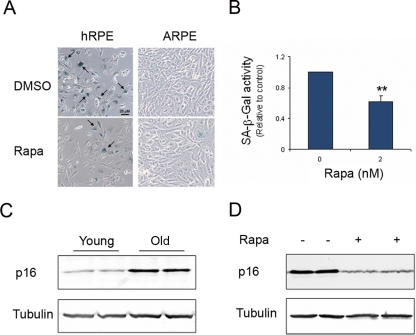
Normal cells have limited capacity for replication in culture and enter senescence after a certain number of in vitro passages.31 Consistent with previous studies,32 in primary RPE cells with increasing passage numbers, an increased percentage of cells displayed positive staining for SA-β-Gal, the most accepted marker of senescence,33 and more than 90% of cells presented SA-β-Gal staining at the highest CPD (Fig. 4A). Increased SA-β-Gal was not observed in the immortalized cell line, ARPE-19. To seek an independent marker of cellular aging, we further measured the protein level of a CDK inhibitor, p16Ink4a. The accumulation of p16 in aged cells is another marker of senescence and has been demonstrated in various types of cells, including stem cells.34,35 As shown in Figure 4C, increased expression of p16 was evident in aged hfRPE cells.
Figure 4.
Rapamycin delayed the appearance of senescence markers in human RPE. (A) Cells were cultured in the presence or absence of 2 nM rapamycin (Rapa) for 6 days. SA-β-Gal (arrow) was stained with the colorimetric substrate 5-bromo-4-chloro-3-indolyl β-D-galactopyranoside. (B) Alternatively, the activity of SA-β-Gal was quantitated using a fluorogenic substrate, 4-MUG. Data represented are the averages of three separate experiments (mean ± SE). Significant differences from vehicle treatment was determined by Student's t-test and indicated as **P < 0.01. The accumulation of p16 protein was measured as a second marker of senescence in hfRPE cells (C). Expression of p16 was barely detected in cells at early passage (CPD ∼12) but largely increased in cells at late passage (CPD ∼32). Tubulin was used as a loading control. Rapamycin treatment prevented the accumulation of p16 associated with aging (D). Data presented are representative of two separate experiments and are reproducible in different cell lines of hfRPE.
TOR signaling is one of the central players in the regulation of aging. To determine whether mTOR signaling regulates senescence of the RPE, we cultured primary RPE cells in the presence or absence of the mTORC1 inhibitor rapamycin. At a concentration as low as 2 nM, rapamycin reduced the percentage of cells with positive staining of SA-β-Gal and the overall enzymatic activity (Figs. 4A, 4B). Increasing concentrations of rapamycin did not further decrease SA-β-gal activity (data not shown). Decreased SA-β-Gal activity by rapamycin was observed in both haRPE and hfRPE cells, but not in APRE-19 cells. Consistently, p16 accumulation was reversed by the treatment of rapamycin (Fig. 4D).
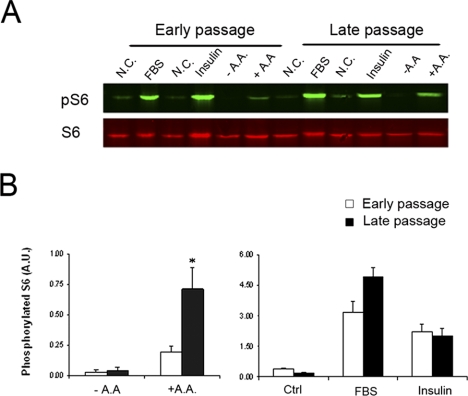
Given the importance of mTOR in controlling organism aging and cellular senescence, we compared mTORC1 signaling between young and aged RPE cells to known stimuli. Cells at early passages (CPD 12∼18) and late passages (CPD 32∼36) were exposed to nutrient and growth factors, and the activation of downstream effector S6 was measured (Fig. 5). All stimuli could activate mTORC1 signaling in hfRPE cells at early and late passages; however, the extent of activation showed age- and stimulus-dependent differences. In aged RPE cells, S6 phosphorylation had a statistically significant increase in response to amino acid signal. Response of mTORC1 signaling to serum also had a tendency to increase in old cells; however, no statistical significance was achieved. In contrast, there was no difference in the sensitivity of mTORC1 in response to insulin.
Figure 5.
Increased sensitivity of mTORC1 in response to extracellular nutrient signal in senescent hfRPE cells. (A) hfRPE cells at both early and late passage were exposed to FBS, insulin, or AA. Activation of mTORC1 was determined by phosphorylation status of S6. (B) Quantification of the Western blot analysis data. Phosphorylation status of S6 was calculated by intensity of phosphorylated S6 divided by intensity of total S6. Data represented are the averages of four separate experiments (mean ± SE). Significant differences from early passage were determined by Student's t-test and indicated as *P < 0.05.
Discussion
The mTOR-mediated signaling pathway is a central player in controlling cell growth and metabolism in response to a variety of signals. Recent studies have indicated that the signaling network plays key roles in regulating organism aging and in cell senescence. Although the TOR kinase is conserved in all eukaryotes, several lines of evidence have suggested that mTOR signaling pathways may have distinct features in different cell types and tissues. For example, the relative amounts of mTORC1 and mTORC2 are variable in different cancer cell lines.16,18 The sensitivity of mTORC1 in response to stimuli depends on tissue context.36 Therefore, it is important to characterize the tissue-specific regulatory mechanisms by mTOR in aging and age-related degeneration.
In the present study, we characterized the mTOR pathway in cultured human RPE cells. Using Western blot analyses and coimmunoprecipitation approaches, we verified the existence of mTORC1 and mTORC2 complexes in the RPE. Nutrient and growth factor signals activated mTORC1 signaling, as shown by the phosphorylation of downstream effector S6 (Fig. 3). The prototypical inhibitor rapamycin dissociated mTORC1 and inhibited the activation of downstream events (Figs. 2, 3). The functional mTOR network in the RPE will link various environmental factors to critical cellular functions, such as mRNA translation and protein synthesis.
We further compared the mTOR signaling between young and old fetal RPE cells. Results showed that mTORC1 underwent age-related changes in terms of sensitivity to upstream stimuli (Fig. 3). However, the alteration was not a general phenomenon for all stimuli because it was only detected in response to amino acid, but not growth factors. Such a difference may be explained by the complexity of mTOR signaling. Upstream signals convey signals to mTOR by way of different mediators.37,38 Growth factors rely on PI3K/Akt to transduce signal through negative regulator TSC1/TSC2 to mTOR, though amino acids relay signals to mTORC1 by way of the Ras-related small GTP-binding protein Rag.38–40 The amino acid-specific alteration of mTORC1 signaling with increased age suggests that age-related changes may occur upstream of mTOR in RPE cells.
Negative regulatory roles of mTOR signaling in determining the life spans of mammals have been postulated. Recently, it has been shown that by reducing mTORC1 signaling, either by knocking out the downstream effector S6K or by using the inhibitor rapamycin, extends the life span of mice.20,41 In the present study, we demonstrated that inhibiting mTORC1 by rapamycin could prevent replicative senescence in cultured RPE cells that were neither transformed nor immortalized. Rapamycin at 2 nM effectively inhibited both SA-β-Gal activity and p16 upregulation (Fig. 4), two well-established markers of senescence. In addition, we observed that serially passaged RPE cells gradually lost RPE markers such as RPE65 and cellular retinaldehyde-binding protein (CRALBP). Although similar findings have been reported,42 future studies will be needed to determine whether decreases of these marker proteins are present in aged RPE cells in vivo or whether they are merely associated events of in vitro cell culture.
A recent study using four mouse models of retinitis pigmentosa showed that cone degeneration after rod cell death was associated with nutrient deprivation.43 Applying rapamycin to the drinking water of wild-type mice caused the downregulation of red/green opsin, though insulin treatment, which activated mTORC1, could delay cone cell loss. The underlying mechanisms by which mTOR signaling controls aging and cell death remain to be elucidated but will likely involve the process of autophagy. Autophagy is a conserved lysosomal pathway to remove long-lived proteins and cytoplasmic organelles, and (m)TOR signaling is one of major upstream regulators negatively controlling the process.44,45 Under conditions of high-level stress, such as progressive photoreceptor degeneration, autophagy results in cell death, and activating mTOR will confer protection. On the other hand, emerging evidence suggests that aging is associated with decreased autophagy, and knocking out of genes in the autophagy machinery accelerates the aging rate and decreases the life span of invertebrates.46–50 Furthermore, extending life spans by knocking down TOR signaling requires the presence of intact autophagy machinery,48 suggesting that the regulation of aging by mTOR occurs at least partially through the regulation of autophagy. These findings are consistent with our observation that inhibiting mTOR can delay the aging process of the RPE.
Different approaches can be used to drive cultured RPE cells into the senescent stage. All have certain advantages and limitations. A commonly used approach is to expose cells to mild to high-dose oxidative stress. RPE cells, including the ARPE-19 cell line, can rapidly enter premature senescence after oxidative injury.51,52 However, the oxidants may affect the signaling transduction pathways independently of senescence and may select clones of cells that are particularly resistant. For instance, at a submillimolar concentration, hydrogen peroxide treatment can activate Akt dose dependently in hRPE cells.53 Peroxide treatment used in premature senescence can also lead to the activation of AMPK, an upstream regulator of mTOR.54 The replicative senescence model can be induced in only primary RPE cells but not in immortalized cell lines, such as ARPE-19, which have an unusual karyotype with deletions and insertions on chromosome 8 and 19.55 It is a gradual change and can be used to monitor passage-dependent changes in senescent phenotype and associated biochemical changes. Although RPE cells do not normally divide under in vivo conditions, the process of replicative senescence is likely controlled by general mechanisms regulating aging of the RPE. It should be noted that the findings of our present study apply to the RPE culture. The in vivo implications of these results should be further assessed in animal models relevant to age and age-related degeneration of the retina and RPE.
Aging is an inevitable biological process and contributes to the pathogenesis of various age-related diseases, including AMD. Manifestations of aging are highly diverse and are dependent on the types of affected cells and tissue. In recent years the mTOR pathway has emerged as one of the central regulatory mechanisms of aging in mammalian cells. AMD is a disease that has been suggested to result at least partially from aging of the RPE. If inhibiting mTOR can slow down the aging process of the RPE, it will become a promising therapeutic strategy to prevent and treat vision loss in elderly people with the atrophic AMD.
Footnotes
Supported by National Institutes of Health Grants EY07892, EY08126, and EY018715; an unrestricted department grant from Research to Prevent Blindness, Inc.; and an Alston Callahan Postdoctoral Scholar Award from International Retinal Research Foundation (YC).
Disclosure: Y. Chen, None; J. Wang, None; J. Cai, None; P. Sternberg, None
References
- 1. Snodderly DM. Evidence for protection against age-related macular degeneration by carotenoids and antioxidant vitamins. Am J Clin Nutr. 1995;62:1448S–1461S [DOI] [PubMed] [Google Scholar]
- 2. Spraul CW, Lang GE, Grossniklaus HE. Morphometric analysis of the choroid, Bruch's membrane, and retinal pigment epithelium in eyes with age-related macular degeneration. Invest Ophthalmol Vis Sci. 1996;37:2724–2735 [PubMed] [Google Scholar]
- 3. Green WR, Key SN., 3rd Senile macular degeneration: a histopathologic study. Trans Am Ophthalmol Soc. 1977;75:180–254 [PMC free article] [PubMed] [Google Scholar]
- 4. Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005;85:845–881 [DOI] [PubMed] [Google Scholar]
- 5. Dorey CK, Wu G, Ebenstein D, Garsd A, Weiter JJ. Cell loss in the aging retina: relationship to lipofuscin accumulation and macular degeneration. Invest Ophthalmol Vis Sci. 1989;30:1691–1699 [PubMed] [Google Scholar]
- 6. Gao H, Hollyfield JG. Aging of the human retina: differential loss of neurons and retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 1992;33:1–17 [PubMed] [Google Scholar]
- 7. Feher J, Kovacs I, Artico M, Cavallotti C, Papale A, Balacco Gabrieli C. Mitochondrial alterations of retinal pigment epithelium in age-related macular degeneration. Neurobiol Aging. 2006;27:983–993 [DOI] [PubMed] [Google Scholar]
- 8. Ogawa T, Boylan SA, Oltjen SL, Hjelmeland LM. Changes in the spatial expression of genes with aging in the mouse RPE/choroid. Mol Vis. 2005;11:380–386 [PubMed] [Google Scholar]
- 9. Steinle JJ, Sharma S, Chin VC. Normal aging involves altered expression of growth factors in the rat choroid. J Gerontol A Biol Sci Med Sci. 2008;63:135–140 [DOI] [PubMed] [Google Scholar]
- 10. Chen H, Liu B, Lukas TJ, Neufeld AH. The aged retinal pigment epithelium/choroid: a potential substratum for the pathogenesis of age-related macular degeneration. PLoS ONE. 2008;3:e2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Greer EL, Brunet A. Signaling networks in aging. J Cell Sci. 2008;121:407–412 [DOI] [PubMed] [Google Scholar]
- 12. Martin DE, Hall MN. The expanding TOR signaling network. Curr Opin Cell Biol. 2005;17:158–166 [DOI] [PubMed] [Google Scholar]
- 13. Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer. 2006;6:729–734 [DOI] [PubMed] [Google Scholar]
- 14. Bhaskar PT, Hay N. The two TORCs and Akt. Dev Cell. 2007;12:487–502 [DOI] [PubMed] [Google Scholar]
- 15. Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci U S A. 1998;95:1432–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sarbassov DD, Ali SM, Kim DH, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302 [DOI] [PubMed] [Google Scholar]
- 17. Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101 [DOI] [PubMed] [Google Scholar]
- 18. Frias MA, Thoreen CC, Jaffe JD, et al. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr Biol. 2006;16:1865–1870 [DOI] [PubMed] [Google Scholar]
- 19. Jacinto E, Facchinetti V, Liu D, et al. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137 [DOI] [PubMed] [Google Scholar]
- 20. Selman C, Tullet JM, Wieser D, et al. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science. 2009;326:140–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sharp ZD, Bartke A. Evidence for down-regulation of phosphoinositide 3-kinase/Akt/mammalian target of rapamycin (PI3K/Akt/mTOR)-dependent translation regulatory signaling pathways in Ames dwarf mice. J Gerontol A Biol Sci Med Sci. 2005;60:293–300 [DOI] [PubMed] [Google Scholar]
- 22. Sternberg P, Jr, Davidson PC, Jones DP, Hagen TM, Reed RL, Drews-Botsch C. Protection of retinal pigment epithelium from oxidative injury by glutathione and precursors. Invest Ophthalmol Vis Sci. 1993;34:3661–3668 [PubMed] [Google Scholar]
- 23. Maminishkis A, Chen S, Jalickee S, et al. Confluent monolayers of cultured human fetal retinal pigment epithelium exhibit morphology and physiology of native tissue. Invest Ophthalmol Vis Sci. 2006;47:3612–3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. van der Loo B, Fenton MJ, Erusalimsky JD. Cytochemical detection of a senescence-associated beta-galactosidase in endothelial and smooth muscle cells from human and rabbit blood vessels. Exp Cell Res. 1998;241:309–315 [DOI] [PubMed] [Google Scholar]
- 25. Tee AR, Fingar DC, Manning BD, Kwiatkowski DJ, Cantley LC, Blenis J. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc Natl Acad Sci U S A. 2002;99:13571–13576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J Biol Chem. 2005;280:18717–18727 [DOI] [PubMed] [Google Scholar]
- 27. Kim DH, Sarbassov DD, Ali SM, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175 [DOI] [PubMed] [Google Scholar]
- 28. Gary RK, Kindell SM. Quantitative assay of senescence-associated beta-galactosidase activity in mammalian cell extracts. Anal Biochem. 2005;343:329–334 [DOI] [PubMed] [Google Scholar]
- 29. Acosta-Jaquez HA, Keller JA, Foster KG, et al. Site-specific mTOR phosphorylation promotes mTORC1-mediated signaling and cell growth. Mol Cell Biol. 2009;29:4308–4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Foster KG, Acosta-Jaquez HA, Romeo Y, et al. Regulation of mTOR complex 1 (mTORC1) by raptor Ser863 and multisite phosphorylation. J Biol Chem. 2010;285:80–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Smith JR, Pereira-Smith OM. Replicative senescence: implications for in vivo aging and tumor suppression. Science. 1996;273:63–67 [DOI] [PubMed] [Google Scholar]
- 32. Matsunaga H, Handa JT, Aotaki-Keen A, Sherwood SW, West MD, Hjelmeland LM. Beta-galactosidase histochemistry and telomere loss in senescent retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 1999;40:197–202 [PubMed] [Google Scholar]
- 33. Dimri GP, Lee X, Basile G, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Janzen V, Forkert R, Fleming HE, et al. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443:421–426 [DOI] [PubMed] [Google Scholar]
- 35. Alcorta DA, Xiong Y, Phelps D, Hannon G, Beach D, Barrett JC. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc Natl Acad Sci U S A. 1996;93:13742–13747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Crozier SJ, Anthony JC, Schworer CM, et al. Tissue-specific regulation of protein synthesis by insulin and free fatty acids. Am J Physiol Endocrinol Metab. 2003;285:E754–E762 [DOI] [PubMed] [Google Scholar]
- 37. Corradetti MN, Guan KL. Upstream of the mammalian target of rapamycin: do all roads pass through mTOR? Oncogene. 2006;25:6347–6360 [DOI] [PubMed] [Google Scholar]
- 38. Sancak Y, Peterson TR, Shaul YD, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657 [DOI] [PubMed] [Google Scholar]
- 40. Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell. 2002;10:151–162 [DOI] [PubMed] [Google Scholar]
- 41. Harrison DE, Strong R, Sharp ZD, et al. Rapamycin fed late in life extends life span in genetically heterogeneous mice. Nature. 2009;460:392–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hamel CP, Tsilou E, Harris E, et al. A developmentally regulated microsomal protein specific for the pigment epithelium of the vertebrate retina. J Neurosci Res. 1993;34:414–425 [DOI] [PubMed] [Google Scholar]
- 43. Punzo C, Kornacker K, Cepko CL. Stimulation of the insulin/mTOR pathway delays cone death in a mouse model of retinitis pigmentosa. Nat Neurosci. 2009;12:44–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pattingre S, Espert L, Biard-Piechaczyk M, Codogno P. Regulation of macroautophagy by mTOR and Beclin 1 complexes. Biochimie (Paris). 2008;90:313–323 [DOI] [PubMed] [Google Scholar]
- 46. Cuervo AM. Autophagy and aging: keeping that old broom working. Trends Genet. 2008;24:604–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391 [DOI] [PubMed] [Google Scholar]
- 48. Toth ML, Sigmond T, Borsos E, et al. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4:330–338 [DOI] [PubMed] [Google Scholar]
- 49. Jia K, Levine B. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy. 2007;3:597–599 [DOI] [PubMed] [Google Scholar]
- 50. Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–84 [DOI] [PubMed] [Google Scholar]
- 51. Honda S, Hjelmeland LM, Handa JT. Senescence associated beta galactosidase activity in human retinal pigment epithelial cells exposed to mild hyperoxia in vitro. Br J Ophthalmol. 2002;86:159–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Glotin AL, Debacq-Chainiaux F, Brossas JY, et al. Prematurely senescent ARPE-19 cells display features of age-related macular degeneration. Free Radic Biol Med. 2008;44:1348–1361 [DOI] [PubMed] [Google Scholar]
- 53. Yang P, Peairs JJ, Tano R, Jaffe GJ. Oxidant-mediated Akt activation in human RPE cells. Invest Ophthalmol Vis Sci. 2006;47:4598–4606 [DOI] [PubMed] [Google Scholar]
- 54. Alexander A, Cai SL, Kim J, et al. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc Natl Acad Sci U S A. 2010;107:4153–4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dunn KC, Aotaki-Keen AE, Putkey FR, Hjelmeland LM. ARPE-19, a human retinal pigment epithelial cell line with differentiated properties. Exp Eye Res. 1996;62:155–169 [DOI] [PubMed] [Google Scholar]