Evidence for a novel cell-associated 31-kDa CTGF fragment that is produced by human corneal fibroblasts and regulated by collagen.
Abstract
Purpose.
Connective tissue growth factor (CTGF) is induced by transforming growth factor-beta (TGF-β) after corneal wounding. This study addressed the role of the extracellular matrix in the induction of CTGF by TGF-β.
Methods.
Human corneal fibroblasts (HCFs) were grown on fibronectin (FN), vitronectin (VN), or collagen (CL) in supplemented serum-free media alone or with TGF-β1 or fibroblast growth factor plus heparin. CTGF mRNA was analyzed by qPCR and protein expression by Western blot analysis of Triton X-100 (TX-100)–soluble and TX-100–insoluble cell lysates using antibodies to N-terminal, mid, and C-terminal CTGF regions. Immunocytochemistry was performed on nonconfluent or scrape-wounded confluent HCFs.
Results.
TGF-β–treated HCFs grown on CL produced five times more 38-kDa CTGF than untreated controls (72 hours). TGF-β–treated HCFs on CL secreted twofold more CTGF than those on FN or VN. Furthermore, a 31-kDa CTGF form, lacking the N-terminal domain, was detected in Triton X-100 insoluble fractions in Western blot analysis. Immunodetectable extracellular CTGF formed linear arrays parallel to, but not colocalized with, CL or FN. It also did not colocalize with FAK, vinculin, or integrins αvβ3 and α5β1. Intracellular CTGF was detected in the Golgi apparatus and vesicles, including endosomes.
Conclusions.
Enhanced CTGF secretion induced by TGF-β in CL-grown cells may contribute to positive feedback in which CL is overexpressed in CTGF-induced fibrosis. N-terminal CTGF fragments in the plasma of patients with severe fibrotic disease may be a product of CTGF proteolysis that also produces the newly identified 31-kDa CTGF that remains cell associated and may have its impact by non–integrin signaling pathways.
Healing without scarring is essential to achieve complete repair of corneal function. One factor that is upregulated in both fibroblasts and epithelium after corneal wounding is connective tissue growth factor (CTGF; CCN2).1,2 Although it participates in the regulation of diverse biological processes related to growth and development, the overexpression of CTGF is correlated with severe fibrotic disorders, including fibrosis in skin, kidney, liver, lung, and vasculature (atherosclerosis),3–7 and several forms of cancer in which it may contribute to tumor angiogenesis and anchorage-independent growth.8–10
CTGF was initially identified as a growth factor, then classified as a matricellular protein, and most recently appreciated as a matrix component.3,11 It is a member of the CCN (Cyr61, CTGF, Nov) family of secreted, cell surface, and extracellular matrix (ECM)–associated 35- to 40-kDa proteins.3,12 Early studies of cells in culture media with serum showed that CTGF is produced in response to transforming growth factor-β (TGF-β) as an immediate early gene product.12,13 In the present study, we took advantage of the opportunity to perform in vitro studies with human corneal fibroblasts (HCFs) grown in the absence of serum to clarify the role of growth factors and matrix on CTGF secretion induced by TGF-β. This was of special interest because during wound healing, the matrix composition changes from one that is initially collagen rich to a provisional matrix that is rich in fibronectin and vitronectin.
Experiments using enzymatic dissection of CTGF into its four distinct structural motifs/domains have yielded individual domains of CTGF whose application has helped resolve the source(s) of its great variety of effects in embryogenesis, implantation, angiogenesis, chondrogenesis, tumorigenesis, differentiation, and wound healing.4,14 Thus CTGF domains, individually and in combination, have been reported to be associated with the induction of cell proliferation, chemotaxis, cell adhesion, collagen synthesis, and myofibroblast differentiation.3,4,15–18 Although many of these studies have been performed using CTGF domains expressed in prokaryotic systems, the likelihood that individual domains have a physiological role is supported by the finding of bioactive cleaved forms of CTGF in situ19–22; in particular an N-terminal fragment of CTGF has been detected in the plasma of scleroderma patients.23
We show here that the amount of CTGF secreted by HCFs in response to TGF-β is matrix dependent and that a novel 31-kDa form of CTGF lacking the N terminus is enriched in the detergent-insoluble fraction that includes the matrix and in the conditioned media. Furthermore we present the first evidence that CTGF N-terminal and C-terminal domains are detected in separate vesicles distal from the Golgi apparatus, only some of which have endosomal markers. Our data support the hypothesis that fibroblasts generate different forms of CTGF and that each form may be responsible for a particular profibrotic effect. In addition, since we demonstrated that extracellular CTGF does not overlap with focal adhesions, we suggest that CTGF signaling in HCFs may be initiated through interaction with other surface molecules not primarily localized in focal adhesions.
Materials and Methods
Reagents
TGF-β1 and −β2 were obtained from R&D Systems, Inc. (Minneapolis, MN); fibroblast growth factor-2 (FGF) was from Invitrogen (Carlsbad, CA). Collagen was from Inamed Biomaterials (Fremont, CA); fibronectin and vitronectin were from Sigma (St. Louis, MO). Human CTGF antibodies (see Fig. 5A) anti–mid-CTGF (14939 and control peptide; Santa Cruz Biotechnology, Santa Cruz, CA), anti–N-CTGF rabbit polyclonal antibody (p836), and anti–C-CTGF (p839) were provided by FibroGen, Inc. (San Francisco, CA), and mAb-anti–C-CTGF mouse monoclonal raised to amino acids 247 to 349 in the C terminus (MAB660) was from R&D Systems. Other commercial antibodies were as follows: anti–FAK (05–537; Upstate, Lake Placid, NY), anti–tubulin (T5168; Sigma), anti–vinculin (V-9131; Sigma), anti–collagen I (MCOLL1-abr; Research Diagnostics, Concord, MA), anti–α-smooth muscle actin-Cy3 (C-6198; Sigma), anti–fibronectin (F-3648; Sigma), anti–p230 (611280; BD Biosciences, San Jose, CA), anti–COP (generously provided by Thomas Weber, Mount Sinai School of Medicine, New York, NY), anti–LRP (transmembrane domain; 5A6) (28320; Abcam, Cambridge, MA), anti–EEA1 (ab15846; Abcam), anti–α5β1 (MAB1999; Chemicon, Temecula, CA), and anti–αvβ3 (B36, generously provided by Barry Coller, Rockefeller University, New York, NY).
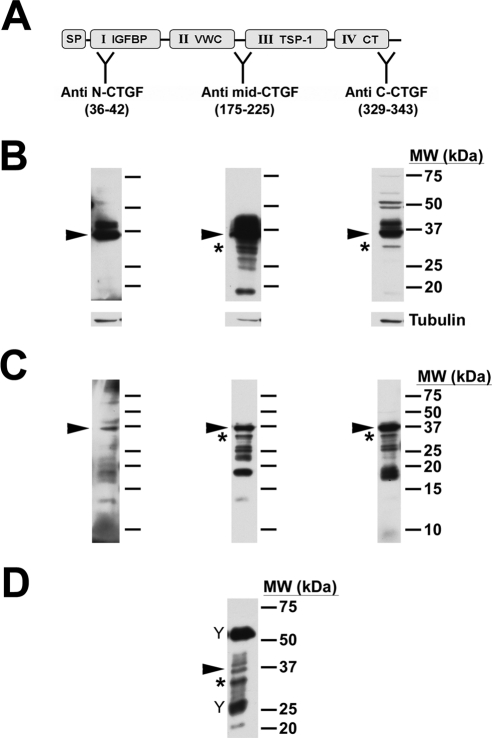
Figure 5.
31-kDa CTGF was not detected by the anti–N-CTGF antibody. (A) Diagram of CTGF showing its modular architecture and the domains to which the specific antibodies were raised, indicated at “Y”: signal peptide (SP), insulin-like growth factor binding protein (IGFBP), von Willebrand factor type C repeat (VWC), thrombospondin type 1 (TSP-1), and C-terminal (CT). (B) HCFs were treated with TGF-β for 24 hours before lysis and detection with CTGF antibodies on Western blot. Full-length CTGF (arrowheads) was detected by all three antibodies. 31-kDa CTGF (asterisk) was detected in the blots probed with anti–mid-CTGF or anti–C-CTGF antibodies. In contrast, anti–N-CTGF did not detect the 31-kDa form. Tubulin was used as a loading control (lower panels). (C) TX-100–insoluble fractions were probed with the same CTGF antibodies and yielded similar results. (D) Immunoprecipitation by anti–mid-CTGF antibody of conditioned media of HCFs cultured on CL with TGF-β for 72 hours yielded multiple forms of CTGF. Full-length CTGF (arrowhead) and the 31-kDa form (asterisk), as well as other molecular weight forms, including 20 and 26 to 28 kDa, were immunodetected. (“Y” symbols indicate bands from heavy and light IgG chains of the immunoprecipitation antibody.)
Preparation of Human Corneal Fibroblast Cultures
Human corneas from donors between the ages of 22 and 65 and not suitable for transplantation were obtained from National Disease Research Interchange (Philadelphia, PA) or from the Lion's Eye Bank (Manhasset, NY). All human tissue was handled in accordance to the tenets of the Declaration of Helsinki. Corneal stroma was separated from the epithelium and endothelium and corneal fibroblasts (keratocytes) released as described in Bernstein et al.24 The resultant corneal fibroblasts were amplified by culturing in DMEM/F-12 plus 10% FBS (Atlas Biologicals, Fort Collins, CO), ABAM (Sigma), and gentamicin (Gibco). Media were renewed every 2 to 3 days.
Cell Harvesting, Lysis, and Western Blot Analysis
Cultured HCFs were trypsinized and replated at approximately 50% to 80% confluence in culture dishes coated with either VN (1–10 μg/mL), FN (10 μg/mL), or CL (10 μg/mL) in supplemented serum-free media (SSFM): DMEM/F-12 with ABAM, gentamicin, 1× RPMI-1640 vitamin mix, 1 μg/mL glutathione (Sigma), 1 mM sodium pyruvate, 0.1 mM MEM nonessential amino acids (Gibco), and ITS supplement (Sigma).25 TGF-β (0.5 ng/mL) was used and consistently upregulated CTGF and α-SMA production; this concentration is within the range that induces the myofibroblast phenotype (0.25–1 ng/mL).12,24,26–28 To augment the fibroblast phenotype, FGF (10 ng/mL) and heparin (5 μg/mL) were added.27 Media with growth factors were replaced at 48 to 72 hours.
To harvest and lyse cells, cells were detached with detachment buffer (250 mM sucrose, 10 mM Tris pH 7.6, 1 mM EDTA, 1 mM EGTA, 1× protease inhibitor cocktail [Roche], 1 mM phenylmethylsulfonyl fluoride [PMSF]), lysed in lysis buffer (140 mM NaCl, 10 mM Tris pH 7.6, 1 mM EDTA, 1% Triton X-100 [TX-100]), 0.05% sodium dodecyl sulfate, 1× protease inhibitor cocktail, 1 mM PMSF), and incubated on ice for 20 minutes. Centrifugation at 13,000g for 12 minutes separated TX-100–soluble fraction and TX-100–insoluble pellet. The pellet was solubilized in 2% SDS sample buffer. Both Triton X-soluble and Triton X-insoluble fractions were subjected to SDS-PAGE (10%–15%) and Western blot transfer to PVDF membranes (Millipore, Bedford, MA). Membranes were probed with primary antibodies to CTGF, followed by peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA) and detection by enhanced chemiluminescence (Pierce, Rockford, IL). For protein normalization, the blots were stripped and reprobed with primary antibody to tubulin. Radiographs were quantified (Image Station 440CF; Eastman Kodak, Rochester, NY). Experiments were performed at least three times with similar results, and images of representative blots are shown. All subsequent experiments were conducted with cells plated on CL after we found that CTGF production was greater in that condition.
Reverse Transcription–Polymerase Chain Reaction
HCFs were plated onto CL-coated dishes in SSFM + TGF-β1 (0.5 ng/mL). The next day, extracted RNA (RNeasy Mini Kit; Qiagen, Valencia, CA) was evaluated by the Microarray Shared Research Facility at Mount Sinai School of Medicine (New York, NY), which follows a SYBR Green protocol for RT-PCR reactions using a thermocycler (Prism 7900HT; Applied Biosystems). Primers used were human CTGF forward (5′-TTGGCAGGCTGATTTCTAGG-3′) and reverse (5′-GGTGCAAACATGTAACTTTTGG-3′).
Immunoprecipitation of CTGF from Conditioned Media
HCFs were plated onto CL in SSFM plus TGF-β (0.5 ng/mL). After 24, 48, or 72 hours, supernatants were harvested and frozen at −20°C. Conditioned media were precleared with Protein G beads. Anti–mid-CTGF or nonspecific IgG antibody (4 μg) was prebound to Protein G beads, which were then added to the precleared media and incubated overnight at 4°C, followed by washing the beads once with IP buffer (150 mM NaCl, 25 mM Tris pH 7.4, 0.5% TX-100, and protease inhibitor cocktail) and three times with PBS. Sample protein was eluted from the beads by resuspension in 1× SDS sample buffer and heating for 5 minutes at approximately 90°C, before separation by SDS-PAGE and Western blot detection.
Scrape Wounding of Cell Monolayers
HCFs were plated onto CL-coated coverslips in SSFM plus 0.2% FBS. After 6 to 8 hours, the monolayers were scraped and incubated in SSFM plus TGF-β (0.5 ng/mL) to which we added 0.5% FBS which supported a defined leading edge of the migrating fibroblasts without affecting TGF-β response. After 8 to 10 hours, coverslips were then processed for immunocytochemistry as described.
Immunocytochemistry
HCFs were fixed in 3% paraformaldehyde, permeabilized in 0.1% TX-100, and blocked in 0.1% BSA in PBS (PBSA) plus 3% normal serum. To provide a visual representation of the Triton X-100–insoluble fraction (see description of Western blot protocol), HCFs on coverslips were incubated in prechilled 1% TX-100 on ice for 20 minutes, then washed, fixed, and blocked with 3% serum in PBSA, followed by incubation with primary and secondary antibodies (Alexa 488 and Alexa 568; Invitrogen) diluted in PBSA. Coverslips were stained with Hoechst 33342 (Sigma) and mounted. Microscopy was performed on a laser scanning confocal microscope (AxioPhot2 [Zeiss, Oberkochen, Germany]or TCS-SP [Leica, Wetzlar, Germany]) equipped with CCD cameras. Images were processed using image editing software (PhotoShop; Adobe, Mountain View, CA). Experiments were performed at least three times with similar results, and representative images are shown.
Results
Modulation of CTGF Expression in Human Corneal Fibroblasts by TGF-β and FGF
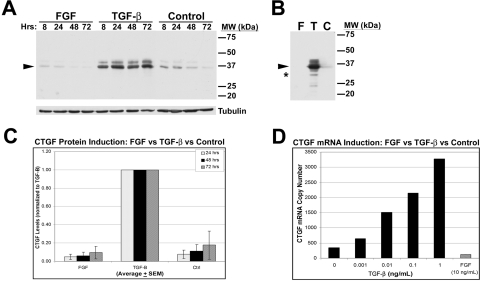
Previous studies have shown that TGF-β is a major regulator of CTGF.4,12 The earlier studies were performed on cells cultured in the presence of serum, which has many cytokines, growth factors, and matrix molecules that confound the impact of TGF-β and specific matrix signals. To clarify the influence of TGF-β and of matrix, we performed the current studies on cells cultured in supplemented serum-free media and grown on defined matrix. HCFs grown on CL and treated with TGF-β, FGF, or neither (control) were harvested after 8, 24, 48, and 72 hours. Cells treated with TGF-β for 24 hours produced more than fivefold full-length (Mr ∼ 38 kDa) CTGF (containing domains I–IV) compared to non-treated controls (Figs. 1A, 1B respectively, arrowhead). TGF-β–induced CTGF expression remained elevated over 72 hours. In contrast, FGF-treated cells had little detectable CTGF at 8 hours that was decreased further at 48 and 72 hours (Figs. 1A, 1B, and quantified in 1C).
Figure 1.
CTGF production by HCFs is stimulated by TGF-β. HCFs were plated on collagen in SSFM only or with FGF or TGF-β for 8, 24, 48, and 72 hours. (A) Anti–mid-CTGF antibody detected a doublet of 36- to 38-kDa CTGF (38 kDa) in TX-100–soluble lysates in Western blot analysis (arrowhead). Tubulin controls confirm equal loading. (B) Triton X-100–insoluble fractions from HCFs at 24 hours were solubilized in 2% SDS buffer followed by sonication and SDS-PAGE and Western blot analysis. As with the soluble lysates, increased levels of CTGF were detected in the TGF-β–treated cells and not from those treated with FGF or with no growth factor. In addition to the 38-kDa CTGF (arrowhead), a previously undescribed band at 31 kDa (asterisk) was strongly immunodetected in this fraction with anti–mid-CTGF antibody, as were other lower MWt bands (e.g., 24 kDa). (C) Quantification by densitometry of TGF-β–induced production of 38-kDa CTGF at 24, 48, and 72 hours in the TX-100–soluble fraction (n = 20). TGF-β–treated HCF cultures had a fivefold increase in immunodetected CTGF production compared with FGF-treated or nontreated control cultures. (D) qPCR analysis showed a dose-response increase of CTGF mRNA in HCFs treated for 24 hours with TGF-β (black bars). In contrast, 24-hour FGF (10 ng/mL) treatment (gray bar) decreased CTGF mRNA levels to below the level of control cultures grown without addition of growth factors (“0” TGF-β).
RT-PCR analysis also demonstrated CTGF mRNA expression was regulated oppositely by these two growth factors. There was a linear relationship between TGF-β dose (0.001–1 ng/mL) and production of CTGF mRNA at 24 hours (Fig. 1D, dark bars), whereas FGF treatment (10 ng/mL) decreased CTGF mRNA compared with nontreated controls (Fig. 1D, light bar). These data confirm and extend the results from our studies on primary rabbit corneal fibroblasts.12 We confirm that in HCFs, TGF-β stimulates, but FGF inhibits, the production of both CTGF mRNA and protein. Thus, unlike other growth factors that have been shown to transiently increase CTGF mRNA without increasing CTGF protein,29,30 we found that TGF-β induced an increase in CTGF protein levels over 72 hours.
Effect of Matrix on TGF-β–Stimulated CTGF Production
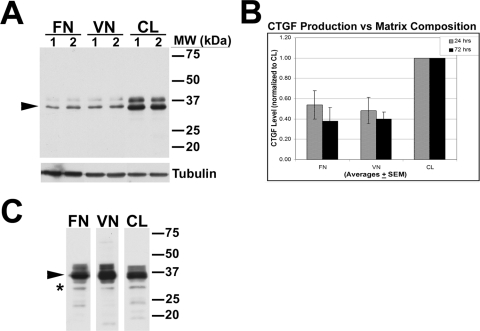
After corneal wounding, fibroblasts in situ are exposed to a changing matrix that includes fibronectin (FN), vitronectin (VN), and collagen (CL). We plated HCFs onto FN, VN, or CL to determine whether signaling from these ECM components affects TGF-β–induced CTGF production. Cells on CL synthesized about twice as much of the 38-kDa CTGF as cells on FN or VN (Figs. 2A, arrowhead; 2B), as detected by anti–mid-CTGF in Western blot analysis of equivalent amounts of TX-100–soluble lysates. Thus, CL signals enhance TGF-β–stimulated CTGF production by HCFs. In the TX-100–insoluble fractions of these cells grown on the three different matrices, we immunodetected CTGF in comparable patterns suggesting that CTGF can accumulate on these matrices and is in a position to affect adhesion, migration, and signaling (Fig. 2C, arrowhead).
Figure 2.
TGF-β–induced CTGF was greater in HCFs growing on CL than on FN or VN. HCFs were plated on FN, VN, or CL in duplicate experiments (lanes 1 and 2) in the presence of TGF-β for 24 hours. (A) Anti–mid-CTGF Ab was used to detect 38-kDa CTGF in TX-100 soluble lysates by Western blot. An increasing amount of CTGF (arrowhead) was detected in cells plated on CL compared with cells plated on FN and VN. Tubulin controls confirm equal loading. (B) Quantification of a 38-kDa CTGF doublet in HCFs treated with TGF-β for 24 or 72 hours on the three different matrices. Bars represent averages + SEM normalized to CTGF levels of samples plated on CL. (C) In TX-100–insoluble fractions of lysates of HCFs grown on FN, VN, or CL, anti–mid-CTGF immunodetected 38-kDa CTGF (arrowhead). Novel 31-kDa form of immunodetectable CTGF was enriched in the TX-100–insoluble fraction in cells grown on all three matrices (asterisk). Because we loaded these lanes based on equal sample volume rather than protein concentration, quantitative comparisons cannot be made. Previously described 24-kDa and 18- to 20-kDa CTGF bands were also detected.
A Novel 31-kDa Form of CTGF
We consistently detected a previously unreported Mr ∼31-kDa form of CTGF that was prominent in Western blot analysis of the TX-100–insoluble fraction and was solubilized by sonicating in the presence of SDS (Figs. 1B, 2C, asterisk). We confirmed that all bands detected on Western blot analysis were derived from CTGF by preincubating the anti–CTGF antibody with a competing peptide (data not shown). In addition to the novel 31-kDa band, we confirmed the presence of lower molecular weight CTGF forms previously reported.4,19,20,23,31,32
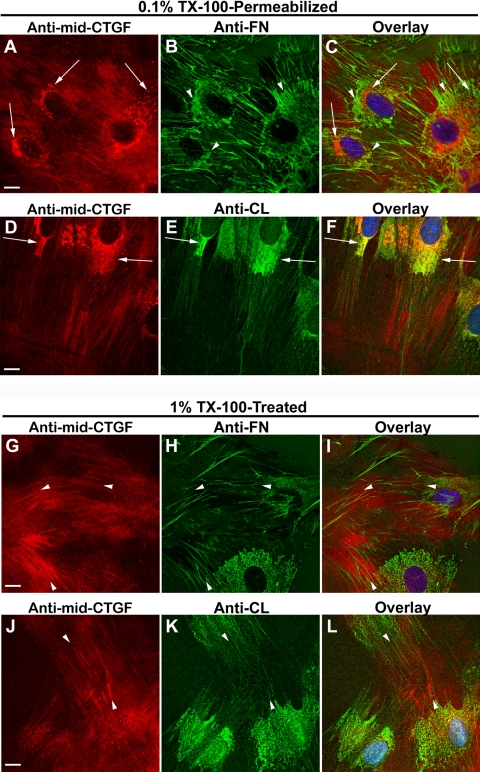
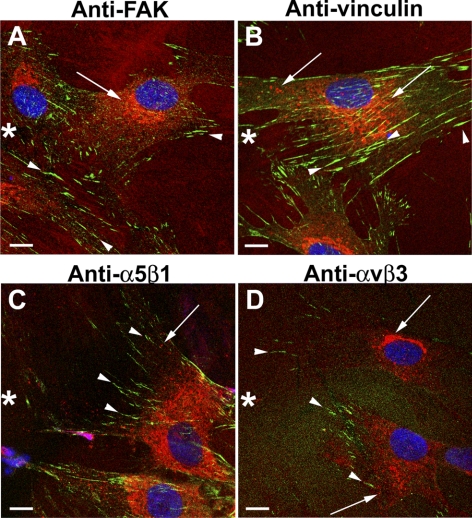
To visualize by immunocytochemistry CTGF in the TX-100–insoluble fraction, HCFs were subjected to prolonged exposure of 1% TX-100 to release the TX-100–soluble components before fixation (Figs. 3G–L), leaving behind the TX-100–insoluble material, which was then immunodetected for CTGF and matrix proteins. TX-100–insoluble CTGF was found in the ECM in a linear arrangement (Figs. 3G, 3J, arrowheads) that aligned with but did not overlap with FN (Figs. 3H, 3K, arrowheads) or with CL (Figs. 3I, 3L, arrowheads). Furthermore, CTGF did not overlap with integrin α5β1 (Fig. 4C) or αvβ3 (Fig. 4D), nor did it overlap with FAK (Fig. 4A) or vinculin (Fig. 4B), which mark integrin-rich focal adhesions by which cells adhere to CL, FN, and VN. Figures 3 and 4 together suggest that TX-100–insoluble extracellular CTGF did not bind to the integrins that bind to FN and CL. In fixed cells briefly permeabilized with 0.1% TX-100 after fixation, CTGF was immunodetected in the Golgi apparatus, as shown previously21 (Figs. 3A, 3D).
Figure 3.
Organization of CTGF with fibronectin or collagen. HCFs grown on FN-coated or CL-coated coverslips in SSFM plus TGF-β for 24 hours were fixed and then permeabilized with 0.1% TX-100 for 1.5 minutes in a standard immunocytochemical protocol (A–F). To “visualize” the TX-100–insoluble fraction, the coverslips were permeabilized with 1% TX-100 on ice for 20 minutes before fixation (G–L). (A–C, arrows) CTGF (red) detected in the ER/Golgi that does not colocalize with intracellular regions of FN (green) enrichment (arrowheads). (D–F) Juxtanuclear ER/Golgi (arrows) in which detectable CTGF (red) and CL (green) overlap (yellow). (G–L) CTGF (red) was detected in linear arrangement outside of cells (G, J, arrowheads) and aligned with fibrillar FN (H, I, arrowheads) or fibrillar CL but did not colocalize with either (K, L, arrowheads). Vesicular and Golgi-localized CTGF staining (seen in A–F) is absent after the prolonged and stronger TX-100 permeabilization (G–J). Scale bars, 10 μm.
Figure 4.
CTGF did not colocalize at focal adhesions with FAK, vinculin, or integrin α5β1 or αvβ3. HCFs growing on CL-coated coverslips were scrape-wounded and incubated with TGF-β for 10 hours and then fixed, permeabilized, and immunodetected for CTGF (red) and FAK, vinculin, and integrin α5β1 or αvβ3 (green). In the cells migrating into the wound (asterisk), CTGF was detected in intracellular vesicles and the Golgi (arrows). Focal adhesions enriched in FAK, vinculin, α5β1, or αvβ3 (arrowheads) did not colocalize with intracellular or extracellular CTGF (A–D). Scale bars, 10 μm.
Given that CTGF is known to have a single transcript and to lack alternative spliced forms,33 it is likely that 31-kDa CTGF arises from the proteolytic cleavage and release of either N- or C- terminal portions from the full-length CTGF. To distinguish between N- and C-terminal cleavage, we used antibodies to epitopes in the two terminal domains, anti–N-CTGF against amino acid residues 36 to 42 in domain I and anti–C-CTGF against amino acid residues 329 to 343 in domain IV. In addition, we used another CTGF antibody, anti–mid-CTGF against amino acid residues 175 to 225, composing the hinge between domains II and III (Fig. 5A). TX-100–soluble lysates from cultures of HCFs grown on CL in SSFM plus TGF-β were subjected to Western blot analysis (Fig. 5B). All three antibodies detected the full-length forms of CTGF (Fig. 5B, arrowheads). The N-terminal antibody did not detect 31-kDa CTGF, suggesting that the 31-kDa fragment lacks the N-terminal epitope (both light and dark exposures were analyzed). Anti–mid-CTGF and anti–C-CTGF detected a prominent band at 31 kDa (Fig. 5B, asterisks). Furthermore, reprobing with the anti–mid-CTGF antibody revealed the presence of a 31-kDa band on the anti–N-CTGF blot (data not shown).
Similarly, in TX-100–insoluble fractions, anti–N-CTGF did not detect a 31-kDa form, whereas anti–mid-CTGF and anti–C-CTGF did (Fig. 5C, asterisks). Each of the antibodies detected lower molecular weight CTGF bands that have been previously described, though not in identical patterns, suggesting that these smaller forms lack various CTGF domains.
To identify which CTGF forms were secreted into the media, we immunoprecipitated conditioned media after 72 hours of TGF-β treatment using the anti–mid-CTGF antibody. The 38-kDa CTGF (Fig. 5D, arrowhead) and 31-kDa CTGF (Fig. 5D, asterisk) forms were detected along with a 20-kDa form, consistent with reports of CTGF cleavage products in vivo that are composed of one or more of its modules/domains.19–22 The relative proportion of 38-kDa and 31-kDa CTGF is reversed compared with that of cell lysates (compare Fig. 5D with Figs. 5B and 5C), consistent with the hypothesis that little full-length CTGF exists in the extracellular space because it has been processed to smaller forms.
Immunocytochemical Localization of CTGF N-terminal and C-terminal Forms
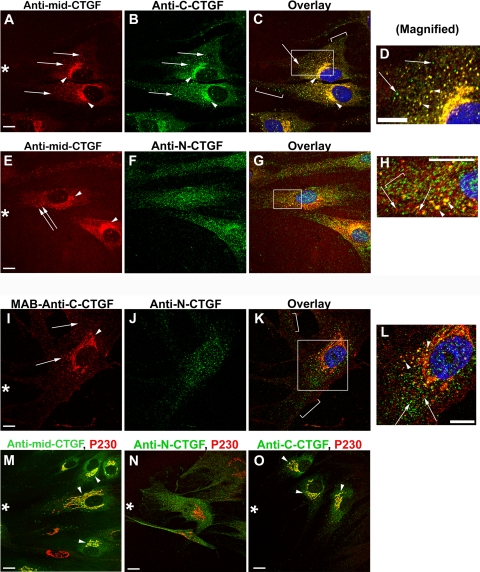
To complement the Western blot data and to provide clues to the localization of sites of CTGF processing, we used a scrape-wounding model to maximize new CTGF secretion and immunodetected with the same three CTGF antibodies. All three antibodies localized in cytoplasmic vesicles proximal to the Golgi apparatus (Figs. 6A, 6B, 6E, 6F, arrows). Anti–mid-CTGF and anti–C-CTGF colocalized in the Golgi apparatus (Figs. 6A–C, arrowheads) and with p230, which localizes in the trans-Golgi network (Figs. 6M, 6O, arrowheads). Anti–N-CTGF did not colocalize with the Golgi apparatus or the trans-Golgi network (Fig. 6N). The complete colocalization in the Golgi for anti–mid-CTGF and anti–C-CTGF suggested that both of their epitopes were readily accessible at that point (Figs. 6C, 6D, yellow). Although anti–N-CTGF did not label Golgi lamellae, it did label Golgi-proximal vesicles (Fig. 6H, arrowheads), suggesting that the N-terminal domain of CTGF was not accessible to antibody detection in the lamellae, possibly because of a secondary structure. This possibility was supported by the fact that the anti–N-CTGF antibody detected 38-kDa CTGF in Western blot analysis (Fig. 5B) run under denaturing, reducing conditions. Vesicles distal from the Golgi typically stained with either anti–C-CTGF or with anti–N-CTGF, but not both (Figs. 6I–L, arrows and brackets). Furthermore, the distal vesicles did not stain with anti–mid-CTGF (Figs. 6C, 6H, arrows and brackets). We conclude that the epitope recognized by anti–mid-CTGF (in the CTGF linker region and domain III) was intact in the initial compartments of the secretory pathway but not in the distal vesicles.
Figure 6.
Immunocytochemical localizations using anti–mid-CTGF, anti–N-CTGF, anti–C-CTGF, and anti–p230 Golgi antibodies in HCFs. HCFs growing on CL-coated coverslips were scrape-wounded and incubated with TGF-β before fixation and immunodetection with antibodies used for the Western blot analysis in Figure 5. Although the images are from cells adjacent to the wound (asterisk), they are representative of CTGF domain staining seen in cells not adjacent to a scrape wound. (A–D) Antibody detection of CTGF central domain (A, red) and C-terminal domain (B, green) colocalized to the Golgi apparatus (A, B, arrowheads; C, yellow) and some adjacent vesicles (D, arrowheads). In addition, the C-terminal CTGF domain was detected in vesicles that were distal from the Golgi apparatus (arrow and brackets in C [an overlay of A and B]) and in (D [magnification of boxed region in C]). (E–H) Antibody to the N terminus of CTGF was seen in vesicles adjacent to the Golgi and throughout the cell rather than in the Golgi lamellae (F, J, arrows). Anti–mid-CTGF and anti–N-CTGF colocalized in vesicles in the Golgi region (G; magnified in H, yellow vesicles, arrowheads). However, many vesicles stained only for either N-CTGF (green, bracket in H) or mid-CTGF (red, arrows in H). (I–L) Most vesicles had either immunodetectable C-CTGF or immunodetectable N-CTGF, but not both. Using an mAb, anti–C-CTGF was immunodetected in the Golgi lamellae and vesicles (I, red) but was usually not colocalized with anti–N-CTGF (K, L), suggesting that each terminal fragment may be taken up into separate vesicles. Arrows: vesicles that stain only with C-terminal domain (red) or with N-terminal domain (green). As in (G), antibody to N-CTGF did not colocalize with Golgi lamellae (K). Colocalization with antibodies to trans-Golgi protein p230 (M–O, red) confirms that anti–mid-CTGF (M, green) and anti–C-CTGF (O, green) detect CTGF in the Golgi apparatus. However, anti–N-CTGF (N, green) does not colocalize with antibodies to Golgi (N, red). Scale bars, 10 μm.
Immunocytochemical Detection of CTGF with Endocytic Markers
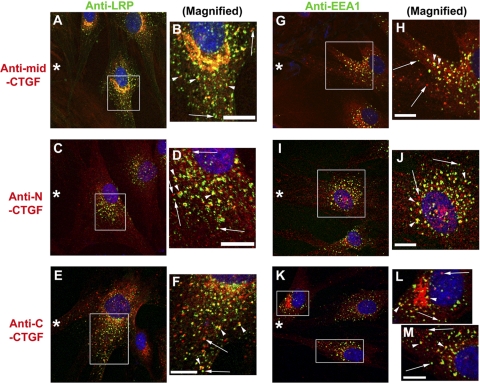
Chen et al.21 provided biochemical evidence that CTGF is taken up and degraded in endosomes,21 and they immunolocalized CTGF only in the Golgi, not in vesicles. In our study, we used the more sensitive confocal microscopy and CTGF domain-specific antibodies in conjunction with antibodies to endosomal markers and were successful in detecting CTGF in endosomes (Fig. 7). We colocalized CTGF in endosomal vesicles identified by antibodies to low-density lipoprotein receptor–related protein (LRP), a scavenger receptor reported to bind CTGF34,35 (Figs. 7A–F) and to the early endosome antigen (EEA1) (Figs. 7G-M). These endosomal markers were immunodetected in vesicles throughout the cell body and adjacent to the Golgi apparatus. Overlay images showed three patterns of vesicular costaining of CTGF and LRP or EEA1 antibodies, better seen after magnification of the boxed regions within these images (Figs. 7B, 7D, 7F, 7H, 7J, 7L, 7M). In the first pattern, colocalization of CTGF and LRP or EEA1 is indicated by yellow vesicles. In the second pattern, two adjacent vesicles (arrowheads), red from CTGF antibody and green from LRP or EEA1 antibody (sometimes with a yellow region of partial colocalization) produced a “traffic light” pattern. The traffic light pattern was not the result of nonalignment of images because the orientation of red to green vesicles was random. In the third pattern, vesicles (arrows) were stained only with CTGF antibody (red) or with LRP or EEA1 antibody (green), indicating no overlap of contents. In summary, this is the first immunodetection of CTGF in endocytic vesicles. Because many of the CTGF-containing vesicles do not have endosomal markers, they may represent vesicles in the secretory pathway.
Figure 7.
Immunodetection of the scavenger receptor LRP and the endosome marker EEA1 in vesicles: colocalization with CTGF and its N- or C-terminal domain. HCFs on CL-coated coverslips were scrape-wounded (asterisk) and incubated with TGF-β for 8 to 10 hours and then fixed, permeabilized, and immunostained with antibodies, as indicated. (A–F) CTGF central (A) or N-terminal (C) or C-terminal (E) domains (all red) were detected in vesicles, including those proximal to the Golgi apparatus. LRP (green) was similarly localized to vesicles (arrows) distributed throughout the cell. In these overlay images, there were three patterns of vesicular localization of CTGF antibody and LRP antibody (better seen in B, D, F, which are magnifications of the boxed regions in A, C, E). Many CTGF-stained vesicles were also stained with LRP antibody. The two other vesicular localization patterns were vesicles that were either green or red (arrows) and that had a “traffic light pattern” of adjacent vesicles (arrowheads), one stained with CTGF antibody (red) and the other stained with LRP antibody (green), sometimes with regions of apparent colocalization between them (yellow). The traffic light vesicles were not the result of the nonalignment of images because the orientation of the red to the green varies within a small region as seen in (B, D, F). Similar patterns of CTGF and LRP staining were seen in cells not adjacent to a scrape wound (data not shown). Scale bars, 10 μm. (G–M) Anti–EEA1-identified endosomal vesicles (green). Some endosomes were colocalized by antibodies to CTGF central (G) or N-terminal (I) or C-terminal (K) domains (all red), better seen in magnified regions (H, J, L, M). The same three patterns of overlap/nonoverlap of EEA1 and CTGF domains are seen, as was described for LRP and CTGF. Scale bars, 10 μm.
Discussion
In this study, we confirmed in HCFs that CTGF production is regulated by two opposing growth factors: TGF-β stimulates and FGF inhibits CTGF production. Additionally, we report that TGF-β–induced CTGF production was enhanced by plating on CL more than on FN or VN. Through detergent-based fractionation and Western blot analyses, we detected a novel 31-kDa form of CTGF that was highly enriched in the TX-100–insoluble fraction of HCF lysate and was also immunoprecipitated from conditioned media. This 31-kDa form lacks the extreme N-terminal portion of CTGF and is likely a product of posttranslational modification.
We have identified the impact of matrix on the induction of CTGF secretion by growing the cells on a specific matrix in the absence of serum. Previous studies were performed in the presence of serum, containing growth factors, cytokines, and matrix components that had masked the impact of matrix.4,12,14 The influence of matrix tractional forces on the cells through an impact on cytoskeletal organization is consistent with the report that RhoA signaling modulates CTGF expression.36,37 Our present finding that CL enhances TGF-β–stimulated CTGF production by HCFs plated on CL, more than on FN or VN, is significant because the matrix early after wounding is largely the collagen of the quiescent corneal stroma (types I and V).38 The “stiffness” of collagen promotes traction, suggesting that a positive feedback system could contribute to fibrosis in which CTGF stimulates collagen production by fibroblasts39 and collagen enhances CTGF production. In normal healing, CTGF stimulation of fibronectin production39 by activated fibroblasts contributes to a “provisional” or “repair” ECM (which includes fibronectin, fibrin, and tenascin), a matrix that also supports TGF-β induction of CTGF. Experiments are under way to evaluate CTGF production and processing by HCFs in a three-dimensional environment.40
The 31-kDa form of CTGF we detected in the Triton X-100–insoluble fraction of TGF-β–treated corneal fibroblasts has not been previously reported. It is likely that it results from proteolysis of the full-length CTGF because no alternatively spliced forms of CTGF (CCN2) have been found, although alternative spice forms of CCN1 and CCN5 have been discovered.33 Proteolytic generation of individual CTGF domains from full-length could occur through ECM metalloproteases upregulated by TGF-β treatment,41 endosomal hydrolases,21 or proteases in the secretory pathway.42 Arguing against extracellular protease activity after CTGF secretion are our findings that the generation of 31-kDa CTGF was not prevented by a battery of broad-spectrum protease inhibitors in the culture media that we had previously found effective in preventing extracellular proteolysis of uPAR in HCFs (data not shown).24 This is not conclusive because there may be other extracellular proteases involved for which we lack inhibitors. Arguing against endosomal hydrolysis as the mechanism of CTGF proteolysis was our finding that 31-kDa CTGF was generated in spite of chloroquine treatment, which inhibits lysosomal hydrolases (data not shown).21 However, we did find by immunocytochemistry that CTGF colocalizes in vesicles with endocytic markers, thus visualizing on a cellular level confirmation of the biochemical data for CTGF endocytosis.21 Because vesicles stain with antibodies to N-terminal or C-terminal CTGF epitopes, separate from each other and from those stained with antibody to the central epitope, it is likely that each of the regions of CTGF can be endocytosed separately and, therefore, may be proteolyzed before endocytosis. It is notable that there are many vesicles that stain for only C-terminal or N-terminal CTGF and do not have detectable endosomal markers. This is consistent with the possibility that smaller versions of CTGF, such as 31-kDa CTGF, could arise from proteolytic processing in the exocytotic pathway by a subtilisin-like pro-protein convertase, demonstrated with secreted FGF23 processing.43 More experiments will be necessary to firmly establish the enzymes and the site of generation of 31-kDa CTGF.
Our Western blot data suggest that after the N terminus is released, much of the cell-associated 31-kDa CTGF is matricellular (Triton X-100–insoluble). The association observed with FN may involve the reported molecular interaction with CTGF domain 4.44,45 Furthermore, the release of the N terminus is consistent with a newly developed ELISA based on the detection of amino terminal fragments of CTGF in plasma or vitreous as predictive of the severity of fibrotic disease.46 We do not know the impact on signaling of a CTGF molecule that is composed of domains II, III, and IV. In previous investigations of cell adhesion and signaling, in which CTGF fragments were tested,17,18 domain 4 promoted cell adhesion,16 and the C-terminal half promoted fibroblast proliferation.17 Given that 31-kDa CTGF includes the C-terminal half, it could promote the increase of the local fibroblast population, thereby contributing to the expansion of fibrotic lesions.
The mechanism of signaling from CTGF has been under active investigation. CTGF is known to be a “sticky” protein and reportedly binds with cell surface receptors, including integrins α4β1, α5β1,47 α6β1,48 and αvβ3.5 (Recent studies have shown that CTGF also binds to aggrecan.49) However, we could not colocalize CTGF with the previously suggested CTGF integrin partners α5β1 and αvβ3, and we did not find colocalization between CTGF and vinculin or FAK, which are components of integrin-based focal adhesions. Of particular interest, we found a previously unreported ECM organization: immunodetectable CTGF was arranged parallel to linear arrays of FN or CL. This organization of matrix-associated CTGF is consistent with the recent report that cultured human corneal fibroblasts assemble their ECM into parallel arrays40 and suggests that CTGF may interact with FN or CL (through additional binding partners) as a component of the provisional ECM. We were able to clearly visualize this matrix-associated CTGF organization by prolonged TX-100 exposure before fixation, though it was also detectable after brief permeabilization of fixed cells with a low concentration of TX-100. Thus, although it is likely that integrin signaling from collagen in the matrix enhances the TGF-β induction of CTGF, the effect of CTGF may not be directly integrin dependent.
In conclusion, our studies have facilitated identification of the impact of ECM on the generation of CTGF, discovery of a new form of CTGF that may be associated with fibrosis rather than healing, and realization that CTGF may have its impact through nonintegrin signaling pathways. Future studies exploring the impact 31-kDa CTGF and how it is generated will help us determine whether this is a target for antifibrotic intervention.
Acknowledgments
The authors thank Liliana Ossowski for insightful discussions.
Footnotes
Supported by National Institutes of Health Grants R01 EY009414 (SKM) and EYO17030 (AMB), Core Grant P30-EY01867, and a Research to Prevent Blindness grant to the Department of Ophthalmology. Microscopy was performed at the Mount Sinai School of Medicine-Microscopy Shared Research Facility, supported, in part, with funding from National Institutes of Health, National Cancer Institute, Shared Resources Grant 5R24 CA095823–04, National Science Foundation Major Research Instrumentation Grant DBI-9724504, and National Institutes of Health Shared Instrumentation Grant 1 S10 RR0, 9145–01.
Disclosure: E.G. Tall, FibroGen, Inc. (F); A.M. Bernstein, FibroGen, Inc. (F); N. Oliver, FibroGen, Inc. (F, E, R); J.L. Gray, FibroGen, Inc. (F, R); S.K. Masur, FibroGen, Inc. (F)
References
- 1. Blalock TD, Duncan MR, Varela JC, et al. Connective tissue growth factor expression and action in human corneal fibroblast cultures and rat corneas after photorefractive keratectomy. Invest Ophthalmol Vis Sci. 2003;44:1879–1887 [DOI] [PubMed] [Google Scholar]
- 2. Secker GA, Shortt AJ, Sampson E, Schwarz QP, Schultz GS, Daniels JT. TGFβ stimulated re-epithelialisation is regulated by CTGF and Ras/MEK/ERK signalling. Exp Cell Res. 2008;314:131–142 [DOI] [PubMed] [Google Scholar]
- 3. Grotendorst GR. Connective tissue growth factor: a mediator of TGF-β action on fibroblasts. Cytokine Growth Factor Rev. 1997;8:171–179 [DOI] [PubMed] [Google Scholar]
- 4. Brigstock DR. The connective tissue growth factor/cysteine-rich 61/nephroblastoma overexpressed (CCN) family. Endocr Rev. 1999;20:189–206 [DOI] [PubMed] [Google Scholar]
- 5. Gao R, Brigstock DR. Connective tissue growth factor (CCN2) induces adhesion of rat activated hepatic stellate cells by binding of its C-terminal domain to integrin alpha (v) beta(3) and heparan sulfate proteoglycan. J Biol Chem 2004;279:8848–8855 [DOI] [PubMed] [Google Scholar]
- 6. Leask A. Transcriptional profiling of the scleroderma fibroblast reveals a potential role for connective tissue growth factor (CTGF) in pathological fibrosis. Keio J Med. 2004;53:74–77 [DOI] [PubMed] [Google Scholar]
- 7. Wahab NA, Yevdokimova N, Weston BS, et al. Role of connective tissue growth factor in the pathogenesis of diabetic nephropathy. Biochem J. 2001;359:77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dornhofer N, Spong S, Bennewith K, et al. Connective tissue growth factor-specific monoclonal antibody therapy inhibits pancreatic tumor growth and metastasis. Cancer Res. 2006;66:5816–5827 [DOI] [PubMed] [Google Scholar]
- 9. Pan LH, Beppu T, Kurose A, et al. Neoplastic cells and proliferating endothelial cells express connective tissue growth factor (CTGF) in glioblastoma. Neurol Res. 2002;24:677–683 [DOI] [PubMed] [Google Scholar]
- 10. De Winter P, Leoni P, Abraham D. Connective tissue growth factor: structure-function relationships of a mosaic, multifunctional protein. Growth Factors. 2008;26:80–91 [DOI] [PubMed] [Google Scholar]
- 11. Bornstein P. Matricellular proteins: an overview. Matrix Biol. 2000;19:555–556 [DOI] [PubMed] [Google Scholar]
- 12. Folger PA, Zekaria D, Grotendorst G, Masur SK. Transforming growth factor-beta-stimulated connective tissue growth factor expression during corneal myofibroblast differentiation. Invest Ophthalmol Vis Sci. 2001;42:2534–2541 [PubMed] [Google Scholar]
- 13. Grotendorst GR, Okochi H, Hayashi N. A novel transforming growth factor beta response element controls the expression of the connective tissue growth factor gene. Cell Growth Differ. 1996;7:469–480 [PubMed] [Google Scholar]
- 14. Lau LF, Lam SC. The CCN family of angiogenic regulators: the integrin connection. Exp Cell Res. 1999;248:44–57 [DOI] [PubMed] [Google Scholar]
- 15. Rachfal AW, Brigstock DR. Structural and functional properties of CCN proteins. Vitam Horm. 2005;70:69–103 [DOI] [PubMed] [Google Scholar]
- 16. Ball DK, Rachfal AW, Kemper SA, Brigstock DR. The heparin-binding 10 kDa fragment of connective tissue growth factor (CTGF) containing module 4 alone stimulates cell adhesion. J Endocrinol. 2003;176:R1–R7 [DOI] [PubMed] [Google Scholar]
- 17. Grotendorst GR, Duncan MR. Individual domains of connective tissue growth factor regulate fibroblast proliferation and myofibroblast differentiation. FASEB J. 2005;19:729–738 [DOI] [PubMed] [Google Scholar]
- 18. Kubota S, Kawaki H, Kondo S, et al. Multiple activation of mitogen-activated protein kinases by purified independent CCN2 modules in vascular endothelial cells and chondrocytes in culture. Biochimie (Paris). 2006;88:1973–1981 [DOI] [PubMed] [Google Scholar]
- 19. Ball DK, Surveyor GA, Diehl JR, et al. Characterization of 16- to 20-kilodalton (kDa) connective tissue growth factors (CTGFs) and demonstration of proteolytic activity for 38-kDa CTGF in pig uterine luminal flushings. Biol Reprod. 1998;59:828–835 [DOI] [PubMed] [Google Scholar]
- 20. Brigstock DR, Steffen CL, Kim GY, Vegunta RK, Diehl JR, Harding PA. Purification and characterization of novel heparin-binding growth factors in uterine secretory fluids: identification as heparin-regulated Mr 10,000 forms of connective tissue growth factor. J Biol Chem. 1997;272:20275–20282 [DOI] [PubMed] [Google Scholar]
- 21. Chen Y, Segarini P, Raoufi F, Bradham D, Leask A. Connective tissue growth factor is secreted through the Golgi and is degraded in the endosome. Exp Cell Res. 2001;271:109–117 [DOI] [PubMed] [Google Scholar]
- 22. Kubota S, Eguchi T, Shimo T, et al. Novel mode of processing and secretion of connective tissue growth factor/ecogenin (CTGF/Hcs24) in chondrocytic HCS-2/8 cells. Bone. 2001;29:155–161 [DOI] [PubMed] [Google Scholar]
- 23. Dziadzio M, Usinger W, Leask A, et al. N-terminal connective tissue growth factor is a marker of the fibrotic phenotype in scleroderma. QJM. 2005;98:485–492 [DOI] [PubMed] [Google Scholar]
- 24. Bernstein AM, Twining SS, Warejcka DJ, Tall E, Masur SK. Urokinase receptor cleavage: a crucial step in fibroblast-to-myofibroblast differentiation. Mol Biol Cell. 2007;18:2716–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jester JV, Barry-Lane PA, Cavanagh HD, Petroll WM. Induction of alpha-smooth muscle actin expression and myofibroblast transformation in cultured corneal keratocytes. Cornea. 1996;15:505–516 [PubMed] [Google Scholar]
- 26. Greenberg RS, Bernstein AM, Benezra M, Gelman IH, Taliana L, Masur SK. FAK-dependent regulation of myofibroblast differentiation. FASEB J. 2006;20:1006–1008 [DOI] [PubMed] [Google Scholar]
- 27. Maltseva O, Folger P, Zekaria D, Petridou S, Masur SK. Fibroblast growth factor reversal of the corneal myofibroblast phenotype. Invest Ophthalmol Vis Sci. 2001;42:2490–2495 [PubMed] [Google Scholar]
- 28. Taliana L, Benezra M, Greenberg RS, Masur SK, Bernstein AM. ZO-1: lamellipodial localization in a corneal fibroblast wound model. Invest Ophthalmol Vis Sci. 2005;46:96–103 [DOI] [PubMed] [Google Scholar]
- 29. Igarashi A, Okochi H, Bradham DM, Grotendorst GR. Regulation of connective tissue growth factor gene expression in human skin fibroblasts and during wound repair. Mol Biol Cell. 1993;4:637–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tamatani T, Kobayashi H, Tezuka K, et al. Establishment of the enzyme-linked immunosorbent assay for connective tissue growth factor (CTGF) and its detection in the sera of biliary atresia. Biochem Biophys Res Commun. 1998;251:748–752 [DOI] [PubMed] [Google Scholar]
- 31. Perbal B. CCN proteins: multifunctional signalling regulators. Lancet. 2004;363:62–64 [DOI] [PubMed] [Google Scholar]
- 32. Yang DH, Kim HS, Wilson EM, Rosenfeld RG, Oh Y. Identification of glycosylated 38-kDa connective tissue growth factor (IGFBP-related protein 2) and proteolytic fragments in human biological fluids, and up-regulation of IGFBP-rP2 expression by TGF-beta in Hs578T human breast cancer cells. J Clin Endocrinol Metab. 1998;83:2593–2596 [DOI] [PubMed] [Google Scholar]
- 33. Perbal B. Alternative splicing of CCN mRNAs: it has been upon us. J Cell Commun Signal. 2009;3:153–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Leask A, Abraham DJ. All in the CCN family: essential matricellular signaling modulators emerge from the bunker. J Cell Sci. 2006;119:4803–4810 [DOI] [PubMed] [Google Scholar]
- 35. Segarini PR, Nesbitt JE, Li D, Hays LG, Yates JR, 3rd, Carmichael DF. The low density lipoprotein receptor-related protein/alpha2-macroglobulin receptor is a receptor for connective tissue growth factor. J Biol Chem. 2001;276:40659–40667 [DOI] [PubMed] [Google Scholar]
- 36. Ott C, Iwanciw D, Graness A, Giehl K, Goppelt-Struebe M. Modulation of the expression of connective tissue growth factor by alterations of the cytoskeleton. J Biol Chem. 2003;278:44305–44311 [DOI] [PubMed] [Google Scholar]
- 37. Janmey PA, Winer JP, Murray ME, Wen Q. The hard life of soft cells. Cell Motil Cytoskeleton. 2009;66:597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fini ME. Keratocyte and fibroblast phenotypes in the repairing cornea. Prog Retin Eye Res. 1999;18:529–551 [DOI] [PubMed] [Google Scholar]
- 39. Frazier K, Williams S, Kothapalli D, Klapper H, Grotendorst GR. Stimulation of fibroblast cell growth, matrix production, and granulation tissue formation by connective tissue growth factor. J Invest Dermatol. 1996;107:404–411 [DOI] [PubMed] [Google Scholar]
- 40. Guo X, Hutcheon AE, Melotti SA, Zieske JD, Trinkaus-Randall V, Ruberti JW. Morphologic characterization of organized extracellular matrix deposition by ascorbic acid-stimulated human corneal fibroblasts. Invest Ophthalmol Vis Sci. 2007;48:4050–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Daniels JT, Schultz GS, Blalock TD, et al. Mediation of transforming growth factor-beta(1)-stimulated matrix contraction by fibroblasts: a role for connective tissue growth factor in contractile scarring. Am J Pathol. 2003;163:2043–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang WM, Lee S, Steiglitz BM, et al. Transforming growth factor-beta induces secretion of activated ADAMTS-2: a procollagen III N-proteinase. J Biol Chem. 2003;278:19549–19557 [DOI] [PubMed] [Google Scholar]
- 43. Fukumoto S. Post-translational modification of fibroblast growth factor 23. Ther Apher Dial. 2005;9:319–322 [DOI] [PubMed] [Google Scholar]
- 44. Hoshijima M, Hattori T, Inoue M, et al. CT domain of CCN2/CTGF directly interacts with fibronectin and enhances cell adhesion of chondrocytes through integrin alpha5beta1. FEBS Lett. 2006;580:1376–1382 [DOI] [PubMed] [Google Scholar]
- 45. Yoshida K, Munakata H. Connective tissue growth factor binds to fibronectin through the type I repeat modules and enhances the affinity of fibronectin to fibrin. Biochim Biophys Acta. 2007;1770:672–680 [DOI] [PubMed] [Google Scholar]
- 46. Leask A, Parapuram SK, Shi-Wen X, Abraham DJ. Connective tissue growth factor (CTGF, CCN2) gene regulation: a potent clinical bio-marker of fibroproliferative disease? J Cell Commun Signal. 2009;3:89–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen Y, Abraham DJ, Shi-Wen X, Black CM, Lyons KM, Leask A. CCN2 (connective tissue growth factor) promotes fibroblast adhesion to fibronectin. Mol Biol Cell. 2004;15:5635–5646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Heng EC, Huang Y, Black SA, Jr, Trackman PC. CCN2, connective tissue growth factor, stimulates collagen deposition by gingival fibroblasts via module 3 and alpha6- and beta1 integrins. J Cell Biochem. 2006;98:409–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Aoyama E, Hattori T, Hoshijima M, et al. N-terminal domains of CCN family 2/connective tissue growth factor bind to aggrecan. Biochem J. 2009;420:413–420 [DOI] [PubMed] [Google Scholar]