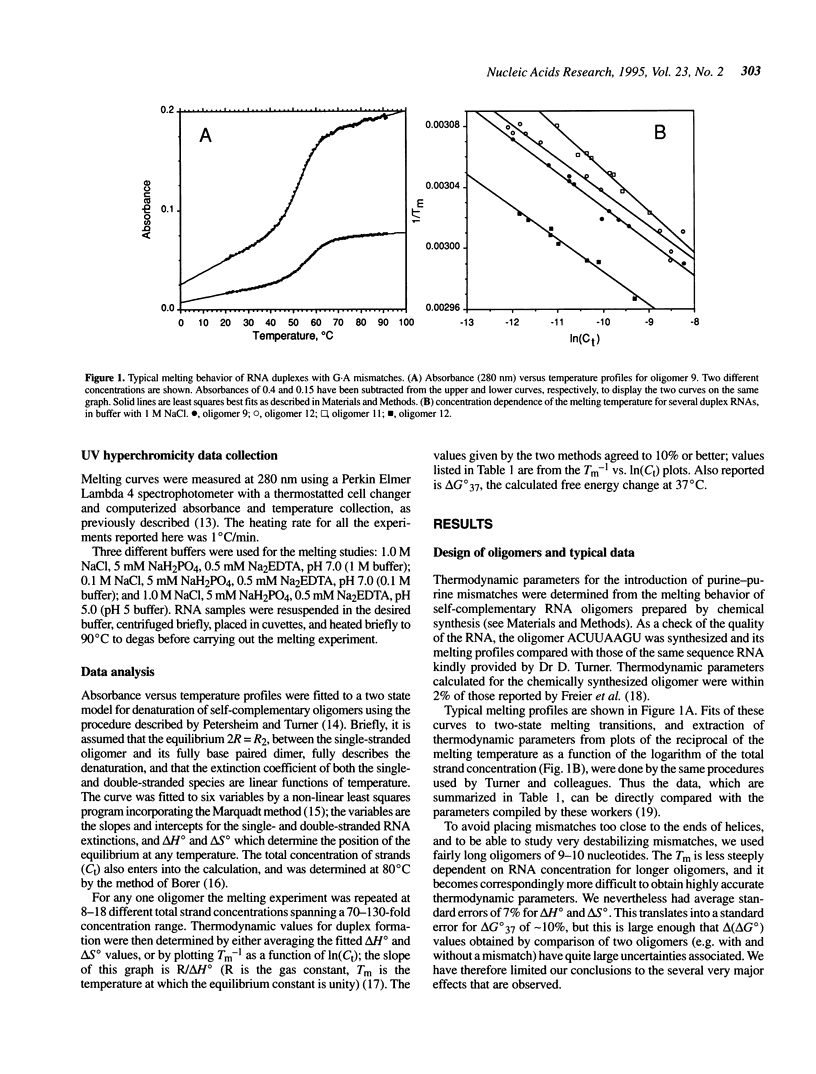
Abstract
Thermodynamic parameters are presented for 12 different RNA duplexes containing A.A, A.G, G.A and G.G mismatches flanked by C-G base pairs. UV melting studies were conducted under three different buffer conditions in order to evaluate the effects of salt concentration and pH on the stability of each mismatch-containing duplex. The main findings are: (i) the mismatches have a wide range of effects on duplex stability, decreasing delta G degrees 37 of denaturation by approximately 0-7 kcal/mol; (ii) the nearest-neighbor assumption commonly used to calculate helix stability breaks down for G.A mismatches; and (iii) G.A mismatches separated by 2 bp form a protonated structure.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Battiste J. L., Tan R., Frankel A. D., Williamson J. R. Binding of an HIV Rev peptide to Rev responsive element RNA induces formation of purine-purine base pairs. Biochemistry. 1994 Mar 15;33(10):2741–2747. doi: 10.1021/bi00176a001. [DOI] [PubMed] [Google Scholar]
- Borer P. N., Dengler B., Tinoco I., Jr, Uhlenbeck O. C. Stability of ribonucleic acid double-stranded helices. J Mol Biol. 1974 Jul 15;86(4):843–853. doi: 10.1016/0022-2836(74)90357-x. [DOI] [PubMed] [Google Scholar]
- Brown T., Leonard G. A., Booth E. D., Chambers J. Crystal structure and stability of a DNA duplex containing A(anti).G(syn) base-pairs. J Mol Biol. 1989 May 20;207(2):455–457. doi: 10.1016/0022-2836(89)90268-4. [DOI] [PubMed] [Google Scholar]
- Brown T., Leonard G. A., Booth E. D., Kneale G. Influence of pH on the conformation and stability of mismatch base-pairs in DNA. J Mol Biol. 1990 Apr 5;212(3):437–440. doi: 10.1016/0022-2836(90)90320-L. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freier S. M., Petersheim M., Hickey D. R., Turner D. H. Thermodynamic studies of RNA stability. J Biomol Struct Dyn. 1984 Mar;1(5):1229–1242. doi: 10.1080/07391102.1984.10507514. [DOI] [PubMed] [Google Scholar]
- Gautheret D., Konings D., Gutell R. R. A major family of motifs involving G.A mismatches in ribosomal RNA. J Mol Biol. 1994 Sep 9;242(1):1–8. doi: 10.1006/jmbi.1994.1552. [DOI] [PubMed] [Google Scholar]
- Gutell R. R., Larsen N., Woese C. R. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol Rev. 1994 Mar;58(1):10–26. doi: 10.1128/mr.58.1.10-26.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Woese C. R. Higher order structural elements in ribosomal RNAs: pseudo-knots and the use of noncanonical pairs. Proc Natl Acad Sci U S A. 1990 Jan;87(2):663–667. doi: 10.1073/pnas.87.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heus H. A., Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991 Jul 12;253(5016):191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- Laing L. G., Draper D. E. Thermodynamics of RNA folding in a conserved ribosomal RNA domain. J Mol Biol. 1994 Apr 15;237(5):560–576. doi: 10.1006/jmbi.1994.1255. [DOI] [PubMed] [Google Scholar]
- Larsen N., Zwieb C. SRP-RNA sequence alignment and secondary structure. Nucleic Acids Res. 1991 Jan 25;19(2):209–215. doi: 10.1093/nar/19.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990 Dec 5;216(3):585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Pace N. R., Smith D. K., Olsen G. J., James B. D. Phylogenetic comparative analysis and the secondary structure of ribonuclease P RNA--a review. Gene. 1989 Oct 15;82(1):65–75. doi: 10.1016/0378-1119(89)90031-0. [DOI] [PubMed] [Google Scholar]
- Petersheim M., Turner D. H. Base-stacking and base-pairing contributions to helix stability: thermodynamics of double-helix formation with CCGG, CCGGp, CCGGAp, ACCGGp, CCGGUp, and ACCGGUp. Biochemistry. 1983 Jan 18;22(2):256–263. doi: 10.1021/bi00271a004. [DOI] [PubMed] [Google Scholar]
- Peterson R. D., Bartel D. P., Szostak J. W., Horvath S. J., Feigon J. 1H NMR studies of the high-affinity Rev binding site of the Rev responsive element of HIV-1 mRNA: base pairing in the core binding element. Biochemistry. 1994 May 10;33(18):5357–5366. doi: 10.1021/bi00184a001. [DOI] [PubMed] [Google Scholar]
- Rich A., RajBhandary U. L. Transfer RNA: molecular structure, sequence, and properties. Annu Rev Biochem. 1976;45:805–860. doi: 10.1146/annurev.bi.45.070176.004105. [DOI] [PubMed] [Google Scholar]
- SantaLucia J., Jr, Kierzek R., Turner D. H. Effects of GA mismatches on the structure and thermodynamics of RNA internal loops. Biochemistry. 1990 Sep 18;29(37):8813–8819. doi: 10.1021/bi00489a044. [DOI] [PubMed] [Google Scholar]
- SantaLucia J., Jr, Turner D. H. Structure of (rGGCGAGCC)2 in solution from NMR and restrained molecular dynamics. Biochemistry. 1993 Nov 30;32(47):12612–12623. doi: 10.1021/bi00210a009. [DOI] [PubMed] [Google Scholar]
- Stern S., Wilson R. C., Noller H. F. Localization of the binding site for protein S4 on 16 S ribosomal RNA by chemical and enzymatic probing and primer extension. J Mol Biol. 1986 Nov 5;192(1):101–110. doi: 10.1016/0022-2836(86)90467-5. [DOI] [PubMed] [Google Scholar]
- Szewczak A. A., Moore P. B., Chang Y. L., Wool I. G. The conformation of the sarcin/ricin loop from 28S ribosomal RNA. Proc Natl Acad Sci U S A. 1993 Oct 15;90(20):9581–9585. doi: 10.1073/pnas.90.20.9581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub W., Sussman J. L. Adenine-guanine base pairing ribosomal RNA. Nucleic Acids Res. 1982 Apr 24;10(8):2701–2708. doi: 10.1093/nar/10.8.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D. H., Sugimoto N., Freier S. M. RNA structure prediction. Annu Rev Biophys Biophys Chem. 1988;17:167–192. doi: 10.1146/annurev.bb.17.060188.001123. [DOI] [PubMed] [Google Scholar]
- Walter A. E., Wu M., Turner D. H. The stability and structure of tandem GA mismatches in RNA depend on closing base pairs. Biochemistry. 1994 Sep 20;33(37):11349–11354. doi: 10.1021/bi00203a033. [DOI] [PubMed] [Google Scholar]
- Wimberly B., Varani G., Tinoco I., Jr The conformation of loop E of eukaryotic 5S ribosomal RNA. Biochemistry. 1993 Feb 2;32(4):1078–1087. doi: 10.1021/bi00055a013. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Gutell R., Gupta R., Noller H. F. Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol Rev. 1983 Dec;47(4):621–669. doi: 10.1128/mr.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]



