Abstract
Increased β-catenin transcriptional activity downstream of the Wnt/Wingless signaling pathway has been observed in many human tumors, most notably colorectal carcinomas. However, β-catenin activation is also observed in many human malignancies with no observable Wnt activity. Wnt-independent pathways that activate β-catenin remain undefined, yet have the potential to play a significant role during tumorigenesis. Here, we report that PIPKIγ, an enzyme that generates the messenger phosphatidylinositol 4,5-bisphosphate, can increase β-catenin activity downstream of growth factor stimulation. In addition to enhancing β-catenin accumulation in the nucleus, PIPKIγ-mediated generation of phosphoinositides increase β-catenin phosphorylation and enhance its transcriptional activity. PIPKIγ expression correlates with a decreased survival in breast cancer patients, an observation that requires β-catenin. Lastly, we show that β-catenin is required for PIPKIγ-dependent increased cell proliferation. These results reveal a novel mechanism where PIPKIγ generation of phosphoinositides enhances β-catenin nuclear translocation and expression of its target genes to promote tumorigenesis.
Keywords: PIPKIγ, β-catenin, phosphoinositide, growth factor, breast cancer
Introduction
β-catenin is a potent oncogene with dual functions in the cell. It was first identified as an essential component in mediating E-cadherin based cell-cell contacts (1). Within the adherens junction, β-catenin links the cytoplasmic domain of E-cadherin to the actin cytoskeleton via its interaction with α-catenin (2). In addition, β-catenin acts as a transcriptional activator in many signaling pathways, including the Wnt/Wingless, epidermal growth factor (EGF), hepatocyte growth factor (HGF) and insulin-like growth factor (IGF) signaling pathways (3-6). In unstimulated cells, cytoplasmic levels of β-catenin are kept in check by a degradation complex consisting of axin, glycogen synthase kinase-3β (GSK-3β) and the adenomatous polyposis coli protein (APC) (1). β-catenin phosphorylation by GSK-3β results in binding β-TRCP/HOS and its ubiquitylation by SCFβ-TRCP leading to proteasomic degradation (7). Wnt stimulation prevents GSK-3β mediated phosphorylation and degradation and instead leads to unique phosphorylation events on β-catenin resulting in its translocation to the nucleus where it interacts with different transcription factors, most notably members of the TCF/LEF family, to control the expression of its target genes including c-jun, c-myc and cyclin D1 (3, 4, 8).
The role of β-catenin in promoting tumorigenesis has been well documented (9). Increased β-catenin activity can lead to uncontrolled cell proliferation, altered cell migration, and changes in cell polarity (9, 10). Activating mutations within the Wnt signaling pathway occur in over 90% of colorectal cancers and have been observed in hepatocellular carcinomas (1). In addition, roles for β-catenin in promoting breast and prostate cancer are emerging (11, 12). However, activating mutations within the Wnt signaling pathway, including CTNNB1 (which encodes β-catenin), are less frequent in many epithelial-derived cancers, including breast cancer (7, 13), although, increased levels of nuclear β-catenin are still frequently observed (14). This indicates that other signaling pathways can induce β-catenin activation during tumorigenesis.
A hallmark of tumorigenesis is the acquisition of invasive capabilities. Emerging evidence supports an epithelial-to-mesenchymal transition (EMT) during tumor progression, which results in the loss of E-cadherin based contacts and increased invasiveness (15, 16). Disruption of E-cadherin mediated adhesions correlates with increased β-catenin-dependent transcription and poor prognosis in cancer patients (17, 18). Forced expression of E-cadherin is able to reduce the tumorigenicity of epithelial-derived cancer cells (19), at least in part due to its ability to sequester β-catenin in the cytoplasm. Stimulation of cells with EGF, HGF or transforming growth factor β (TGF-β) can induce the disruption of E-cadherin based contacts and promote EMT (20, 21). Activation by these growth factors also increases the activity of many kinases including Src, protein kinase B (PKB or AKT) and protein kinase A (PKA), all of which can increase β-catenin nuclear localization and activity (22-25).
Phosphoinositides are a group of signaling molecules that affect a vast array of cellular processes, including polarization, directional migration, gene expression and proliferation (26). Phosphotidylinositol phosphate kinase Iγ (PIPKIγ) phosphorylates phosphatidylinositol 4-phosphate to generate phosphatidylinositol 4,5-bisphosphate (herein referred to as PIP2) in vivo (26). In addition to its use as a precursor to three separate and essential second messengers (inositol 1,4,5-trisphosphate (IP3), 1,2-diacylglycerol (DAG) and PIP3), PIP2 directly binds many protein effectors regulating their activity. PIPKIγ can alternatively splice to give rise to at least 5 different protein isoforms, known as PIPKIγ_i1-5 (27). While the localization of endogenous isoforms are distinct, overexpressed PIPKIγs lose their localization specificity and can be found ubiquitously throughout the cell (27). However, understanding the effects of its overexpression is of critical importance as recently it was shown that PIPKIγ overexpression in breast cancer patients correlated with decreased survival (28), and increased PIPKI activity has been observed in hepatocellular carcinomas (29).
We report a previously undefined role for PIPKIγ in modulating the activation of β-catenin. PIPKIγ expression correlates with poor survival in breast cancer patients only when β-catenin expression is also observed. In defining a cellular mechanism for this clinical observation, we found that PIPKIs associate with β-catenin and that increased PIPKIγ-dependent cellular proliferation requires β-catenin. Phosphoinositide messengers derived from PIPKIγ expression hyperactivate the β-catenin transcriptional machinery. In addition, PIPKIγ_i2 generated phosphoinositide messengers result in the hyperphosphorylation and increased nuclear accumulation of β-catenin. Lastly, we show that PIPKIγ_i2 expression enhances the activation of β-catenin downstream of growth factor stimulation.
Materials and Methods
Please see Supplemental Experimental Procedures for the following sections: “Reagents”; “Cell culture and transfection”; “Lysate preparation and immunoblotting”; “Immunofluorescence and microscopy”; and “GST affinity pull-down assays.”
Luciferase reporter assays
To examine the effects of PIPKIs on β-catenin transcriptional activity, cells were transfected with Super8XTOPFLASH (Millipore), pRL-null Renilla (Promega) and pCMV-HA alone or pCMV-HA containing WT or mutant PIPKI constructs. 18-24 hours after transfection, luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega) and the Pharmingen Monolight 3010 Luminometer. Reporter expression was normalized to co-transfected Renilla luciferase activity. To determine if E-cadherin could inhibit the activation of β-catenin, pCMV-myc or pCMV-myc E-cadherin cytoplasmic domain was included at the time of transfection. For growth factor receptor activation experiments, cells were transfected as stated. Cells were serum starved in DMEM+0.5% FBS for 12-24 h and stimulated with the appropriate growth factor. 24 h later β-catenin activity assayed as stated.
Cell gowth assays
Oligonucleotide sequences used for generation of PIPKIγ short hairpin RNA were: GCCACCTTCTTTCGAAGAA (PIPKIγshRNA) and GCCTTCTTCGCTAAACGAA (ConshRNA). Generation of replication-defective infectious viral particles and the transduction of the cells were carried out following the protocol provided by Addgene and described previously (31, 32). In brief, synthesized oligonucleotides were annealed and cloned into HpaI and XhoI sites of pLL3.7 vector (Addgene). Stabl3 competent cells (Invitrogen) were used for transformation and DNA purification. For generation of viral particles, lentiviral vector with accessory plasmids (pCMV-VSVG, pRSV-Rev and pMD2.G) were cotransfected into HEK 293T cells using calcium phosphate. Conditioned medium was collected 48 hours post-transfection, filtered through 0.45 uM filter and viral particles concentrated by centrifugation at 24,000 rpm in Beckman SW28 centrifuge for 2 h at 4°C. Sub-confluent Hela cells were infected with viral supernatant in the presence of 0.5 μg/ml polybrene (Sigma). Infected cells were either sorted using cell sorter (GFP expression driven by CMV) or individual clones isolated. PIPKIγ knockdown was examined by immunoblotting. To monitor proliferation, infected cells were seeded into 12-well culture plates at a density of 1000 cells/well. Manual cell counting was performed every 48 h for 8 days. Cell numbers were counted from at least three wells for each cell type and expressed as mean ±SD from one representative experiment.
To monitor cell proliferation in Hela-TETOFF cells in conjunction with β-catenin knockdown, Hela-TETOFF cells expressing empty vector or PIPKIγ_i2 were grown in DMEM+10% FBS without doxycycline to induce PIPKIγ_i2 expression. An equal number of cells were transfected with β-catenin validated or control stealth siRNA oligos (Invitrogen) using Oligofectamine™ (Invitrogen) according to the manufacturer's instructions and proliferation monitored for 48 h post-transfection. The extent of β-catenin knockdown was monitored at the 48 h time point. Manual cell counting from at least three wells for each cell type and time point were expressed as mean ±SD.
Results
PIPKIγ associates with β–catenin independent of E-cadherin expression
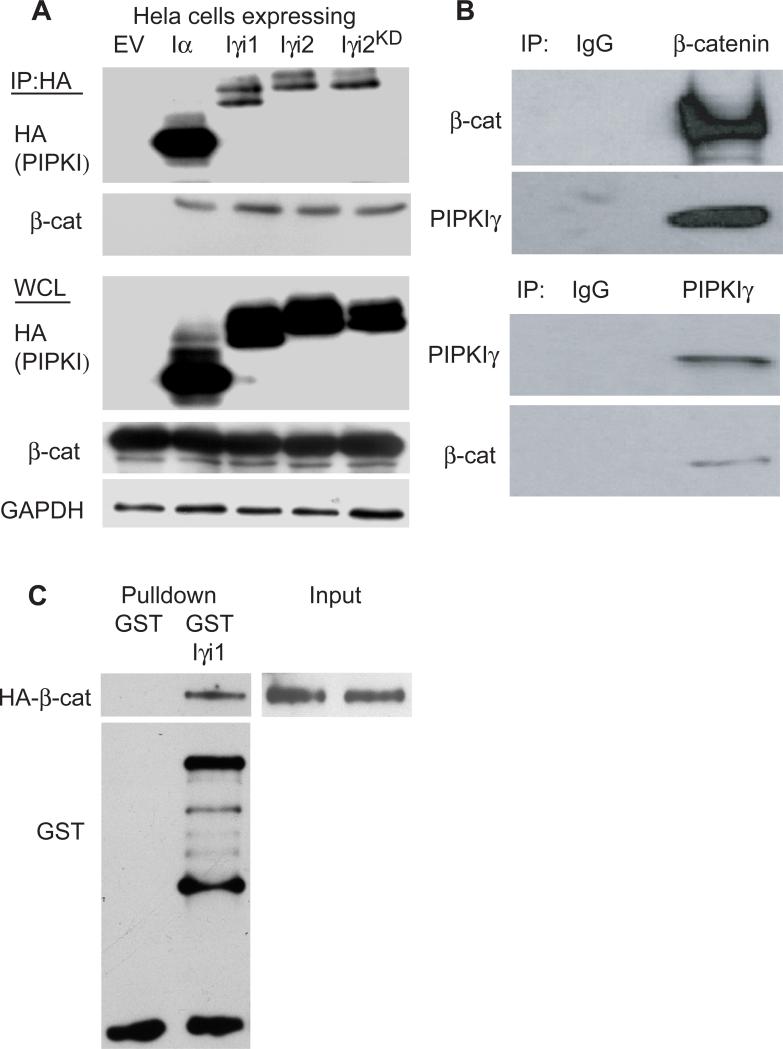
PIPKIγ associates with a region on E-cadherin that lies within the previously defined β-catenin binding region (33). Because of this, an interaction between PIPKIγ and β-catenin was investigated. β-catenin co-immunoprecipitated with transiently transfected HA-tagged PIPKIα, PIPKIγ_i1, PIPKIγ_i2 and PIPKIγ_i2KD in Hela cells but not in lysates taken from cells transfected with empty vector (EV) (Fig. 1A). Because both β-catenin and PIPKIγ splice variants associate with E-cadherin, and E-cadherin is expressed at low levels in these cells (data not shown), we determined if E-cadherin was required to generate an association between β-catenin and PIPKIs. Endogenous PIPKIγ and β-catenin coimmunoprecipitated from A431D and CHO-K1 cells, which lack cadherins (34, 35), demonstrating that this interaction is independent of the classical cadherins in vivo (Fig. 1B and data not shown). Finally, to test whether or not β-catenin and PIPKIγ were able to directly associate, GST pulldown assays using recombinant proteins were performed and showed that β-catenin and PIPKIγ_i1 are able to directly interact (Fig. 1C). The PIPKIγ_i1 protein does not contain a C-terminal extension and its sequence is conserved in all PIPKIγ splice variants (27). The combined data demonstrate that PIPKIγ interacts directly and separately with both β-catenin and E-cadherin, and that the β-catenin interacting region on PIPKIγ lies within the conserved region.
Figure 1. PIPKIγ directly associates with β–catenin independent of E-cadherin.
(A) Hela cells were transiently transfected with empty vector (EV) or the HA-tagged PIPKI shown and an interaction with β-catenin assayed by immunoprecipitation (IP) using anti-HA antibody. Whole cell lysates (WCL) were probed with anti-HA, anti-β-catenin and anti-GAPDH to monitor transfection and protein levels. (B) Endogenous β-catenin and PIPKIγ_i1 were co-IP’ed from A431D cells. (C) Recombinant GST or GST-β-catenin was assayed for interaction with purified PIPKIγ_i1 by in vitro pulldown assays.
PIPKIγ stimulates β-catenin transcriptional activity
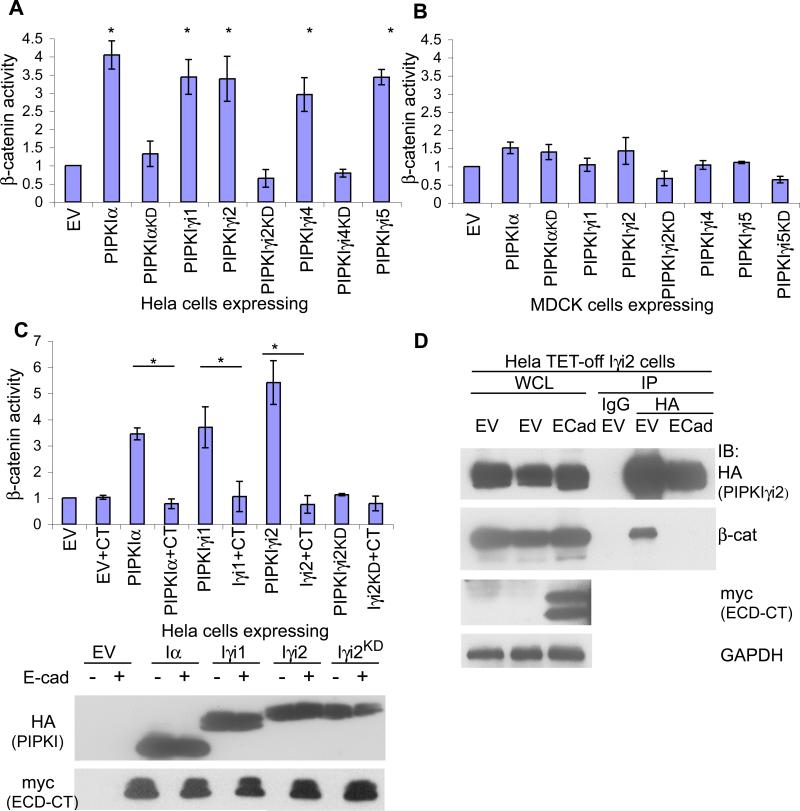
To examine the effect of PIPKIγ on the transcriptional activity of β-catenin, TOPflash/FOPflash luciferase reporter constructs(36) were transfected into HeLa cells expressing PIPKIs and transcriptional activity quantified (23). When grown in the presence of serum, PIPKIα and each PIPKIγ isoform (PIPKIγ_i1-5) stimulated β-catenin transcriptional activity in Hela cells (Fig. 2A). This increased activity required the generation of phosphoinositides. Kinase-inactive (‘kinase-dead’,KD) mutants of PIPKIs, PIPKIαKD, PIPKIγ_i2KD or PIPKIγ_i4KD , failed to stimulate β-catenin transcriptional activity. Furthermore, PIPKIγKD isoforms diminished β-catenin activity indicating that they act in a dominant-negative fashion (Fig. 2A). Expression of PIPKIγ isoforms with tetracycline-inducible cell lines (Hela TET-OFF) demonstrated that PIPKIγ stimulated β-catenin activity to the same or greater extent as transient transfection experiments and PIPKIγKD mutants diminished β-catenin activity to that of uninduced cells (Fig. S1A). Transient transfection of PIPKIs in HEK293T cells also stimulated β-catenin transcriptional activity, demonstrating that this signaling event was not exclusive to Hela cells (Fig. S1B).
Figure 2. PIPKIγ expression stimulates β-catenin transcriptional activity in mesenchymal-like cells lacking E-cadherin.
β-catenin transcriptional activity was measured in (A) Hela or (B) MDCK cells transiently transfected with either empty vector (EV) or the HA-tagged PIPKI shown (* represents p-value ≤ 0.001). (C) Hela cells were treated as in (A) however either empty vector (EV) or E-cadherin cytoplasmic domain (CT) included in the transfection where indicated (* represents pvalue ≤ 0.01). Representive western blots show the expression of PIPKIs with myc-tagged E-cadherin C-terminus. (D) Hela TET-off cells expressing PIPKIγ_i2 were transfected with either empty vector or myc-E-cadherinCT where indicated. Lysates of equal protein concentration were IP’d using anti-HA to determine how the interaction between PIPKIγ_i2 and β-catenin was affected upon expression of E-cadherin. For all graphs n≥3, error bars=std dev.
PIPKIγ can modulate the adherens junction and E-cadherin trafficking in epithelial cells (33). In addition, E-cadherin expression can inhibit β-catenin activity in mesenchymal-like cells (37). We determined whether PIPKI expression could stimulate β-catenin transcriptional activity in polarized epithelial cells. Surprisingly, expression of PIPKIα or PIPKIγ isoforms in MDCK cells did not significantly increase β-catenin transcriptional activity (Fig. 2B). We then tested whether re-expression of E-cadherin could block β-catenin activity in Hela cells grown in the presence of serum. Expression of the E-cadherin cytoplasmic domain (ECD-CT) (33) in Hela cells blocked PIPKI activation of β-catenin (Fig. 2C). Furthermore, ECD-CT expression in cells overexpressing PIPKIγ_i2 significantly reduced the association between PIPKIγ_i2 and β-catenin (Fig. 2D). These data support a model where the PIPKIγ association with β-catenin leads to stimulation of its activity, which is dependent upon the generation of phosphoinositides and is blocked by E-cadherin.
PIPKIγ generation of phosphoinositides enhances the nuclear accumulation of β-catenin
Wnt and growth factor activation of β-catenin results in its nuclear accumulation where it regulates gene expression (10). β-catenin localization changes were observed upon expression of PIPKIγ. The activation of β-catenin by PIPKIγ_i1 and PIPKIγ_i2 was the focus, but we do not discount the possibility that PIPKIα, PIPKIγ_i4 or PIPKIγ_i5 can also result in similar cellular phenotypes. Wildtype or PIPKIγ mutants were expressed in Hela cells and immunofluorescence used to visualize changes in the localization of endogenous levels of β-catenin. Transient expression of PIPKIγ_i2 increased nuclear accumulation of β-catenin (Fig. 3). This accumulation required catalytic activity as expression of PIPKIγ_i2KD failed to increase nuclear β-catenin levels (Fig. 3). Similar results were observed in Hela TET-off cells stably expressing PIPKIγ_i2 or PIPKIγ_i2KD (Fig. S2). Additionally, both transient and stable expression of PIPKIγ_i1 or PIPKIγ_i1KD in Hela cells revealed a similar phenotype (data not shown) demonstrating that PIPKIγ overexpression stimulated the nuclear accumulation of β-catenin in cells grown in the presence of serum.
Figure 3. PIPKIγ_i2 generation of phosphoinositides increases the nuclear accumulation of β-catenin.
Hela cells grown on glass coverslips for 24 h were transiently transfected with empty vector (EV) or HA-tagged PIPKIγ_i2 or PIPKIγ_i2KD where indicated. 24 h after transfection, cells were fixed and stained with anti-HA to monitor PIPKIγ expressing cells, anti-β-catenin and DAPI (pseudocolored red). All images were taken with a 60X objective.
PIPKIγ stimulates the phosphorylation of β-catenin at sites known to initiate nuclear translocation and transcriptional activation
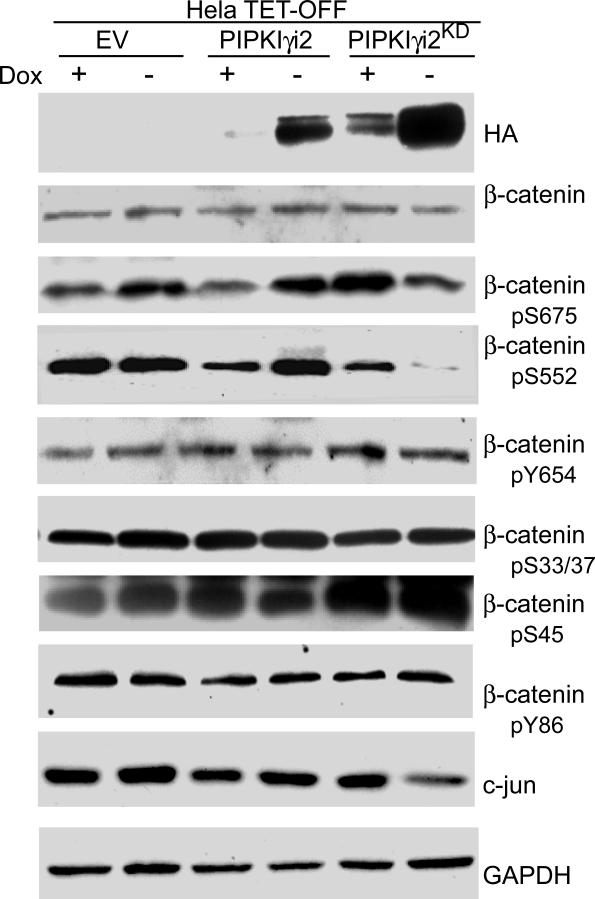
Accumulation of cytoplasmic levels of β-catenin, its nuclear importation and transcriptional activation are all enhanced by phosphorylation (22). Using Hela TET-OFF cells, we found that PIPKIγ_i2 expression increased β-catenin phosphorylation on Ser552 and Ser 675 (Fig. 4). Furthermore, phosphorylation levels at these sites decreased upon expression of PIPKIγ_i2KD (Fig. 4). Surprisingly, neither PIPKIγ_i2 nor PIPKIγ_i2KD expression significantly altered β-catenin protein levels, nor did their expression alter β-catenin phosphorylation on Ser33, 37 and 45, sites known to be involved in the β-catenin proteasomal degradation pathway (Fig. 4). β-catenin activation results in transcriptional changes to its target genes. Consistent with this, we observed an increase in the levels of c-jun protein, a known target of β-catenin transcriptional acitvity, upon expression of PIPKIγ_i2 and saw a decrease in c-jun levels in cells expressing PIPKIγ_i2KD (Fig. 4). This confirms that PIPKIγ generation of phosphoinositides enhances β-catenin activity to induce transcriptional changes in its target genes.
Figure 4. PIPKIγ expression stimulates the phosphorylation of β-catenin at sites known to initiate nuclear translocation and transcriptional activation.
Hela TETOFF cells stably expressing empty vector (EV) or HA-tagged PIPKIγ_i2 or PIPKIγ_i2KD were maintained in DMEM+10%FBS+doxycycline. To initiate PIPKIγ_i2 expression, media was replaced with fresh media +/- dox. where indicated. 24 h later, whole cells lysates were prepared and probed with the antibodies mentioned.
PIPKIγ enhances β-catenin activity upon growth factor stimulation
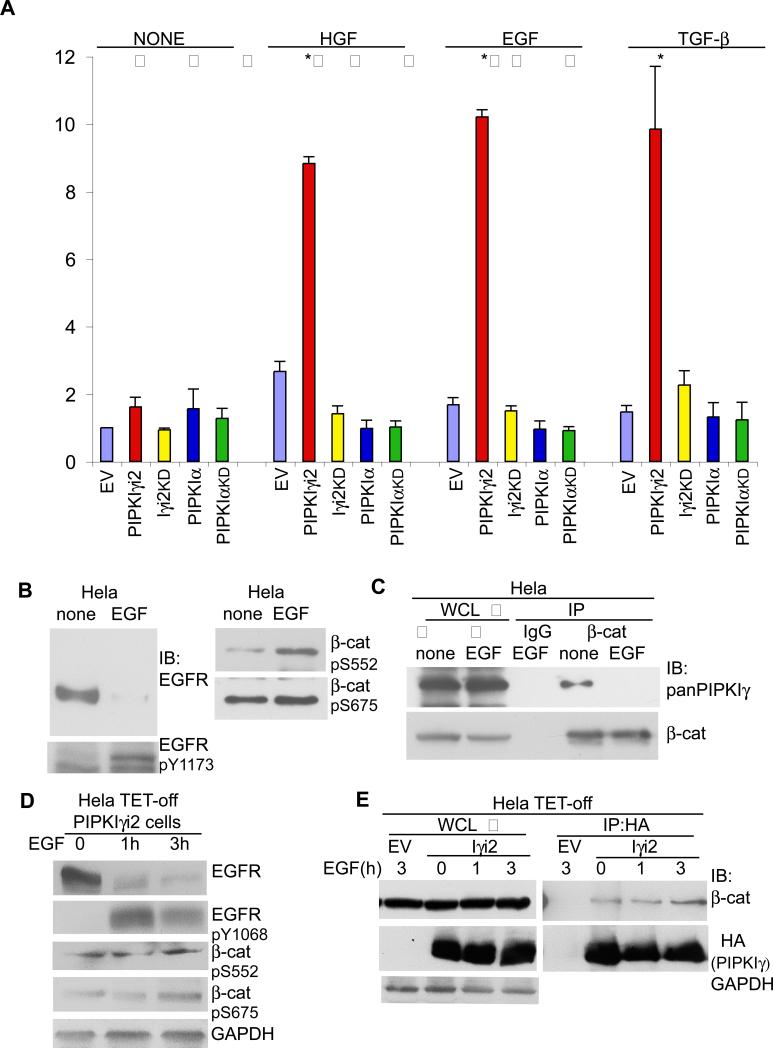
Overexpression of PIPKIγ correlates with increased metastasis and poor prognosis in breast cancer patients and this correlated with the expression of the EGF receptor HER1 and HER2 (28). In addition, many growth factors such as EGF and TGF-β can stimulate the nuclear translocation and transcriptional activation of β-catenin (24). We set out to determine if PIPKIγ played a role in the activation of β-catenin downstream of growth factor receptor activation. For this, β-catenin transcriptional activity was quantified in Hela cells transiently expressing wildtype or kinase inactive mutants of PIPKIα or PIPKIγ_i2. After transfection of the PIPKI, TOPFLASH and renilla reporter constructs, cells were serum starved and then stimulated with EGF, HGF, or TGF-β1/2. Increased PIPKIγ_i2 expression greatly enhanced β-catenin dependent transcriptional activity following treatment with growth factors (Fig. 5A). This increase in β-catenin activity was dependent on its catalytic activity, as PIPKIγ_i2KD failed to increase transcriptional activity (Fig. 5A). Surprisingly, the activation of β-catenin following stimulation with EGF, HGF or TGF-β was PIPKIγ specific, as PIPKIα expression did not enhance β-catenin activity (Fig. 5A). EGF-mediated activation of β-catenin was also enhanced in Hela TET-OFF cells stably expressing PIPKIγ_i2 and severely diminished in cells expressing PIPKIγ_i2KD (Fig. S3C). Stimulation of Hela cells with EGF, HGF or TGF-β resulted in expected activation of signaling molecules within these pathways as evidenced by western blotting using activation specific antibodies (Fig. S3B). These results indicate that PIPKIγ expression and phosphoinositide generation enhance β-catenin transcriptional activation downstream of growth factor receptor stimulation.
Figure 5. PIPKIγ activates β-catenin upon growth factor stimulation.
(A) β-catenin transcriptional activity was measured in Hela cells transiently transfected with empty vector (EV) or the HA-tagged PIPKI shown. 24 h after transfection, cells were serum starved and then left untreated (none) or stimulated with 1nM EGF, 50 ng/ml HGF or 2ng/ml TGF-β for 24 h where indicated followed by cell lysis to measure β-catenin activity (n≥3, error bars=std dev, * represents p ≤ 0.01). (B) Hela cells were serum starved and then left untreated (none) or stimulated with 1nM EGF for 2 h where indicated. Whole cells lysates (WCL) were prepared and probed with the antibodies shown. (C) Cells were treated as in (B). Following cell lysis, equal protein concentrations from none and EGF treated whole cell lysates were incubated with anti-β-catenin antibody. Immunoprecipitated β-catenin and co-IPed PIPKIγ were detected with specific antibodies. (D) Hela TET-off PIPKIγ_i2 cells were maintained in medium without doxycycline to induce PIPKIγ_i2 expression. Cells were serum starved and stimulated with 1nM EGF for 0, 1 or 3 h where indicated. Whole cells lysates (WCL) were prepared and probed with the antibodies shown. (E) Cells were treated as in (D). Following cell lysis, whole cell lysates of equal protein concentrations were incubated with anti-HA antibody. Immunoprecipitated HA-PIPKIγ_i2 and co-IP'd β-catenin were detected with specific antibodies. Hela TET-off cells transfected with the empty vector were used as a control during the IP.
The classical Wnt pathway leads to the activation of β-catenin. Increased expression of PIPKIγ_i2 or PIPKIγ_i2KD did not significantly affect β-catenin activity following Wnt stimulation (Fig. S3A). However, Wnt-dependent activation of β-catenin was moderately but consistently enhanced upon PIPKIα expression and decreased upon PIPKIαKD expression, though the regulation was not statistically significant (Fig. S3A). Recently, PI4KIIα and PIP5KIβ were shown to be required for LRP6 phosphorylation downstream of Wnt stimulation (38).
Next, we determined whether growth factor stimulation induced β-catenin phosphorylation patterns similar to PIPKIγ_i2 expression. EGF stimulation increased EGFR phosphorylation on Tyr1173 and Tyr1086 (Fig. 5B and Supp. Fig. S3B). In addition, EGF stimulation increased β-catenin phosphorylation on Ser552 and Ser675 (Fig. 5B). Surprisingly, the stimulation of Hela cells with EGF resulted in a decreased association between endogenous PIPKIγ and β-catenin as evidenced by coimmunoprecipitation experiments using β-catenin specific and pan-PIPKIγ antibodies (Fig. 5C). In cells overexpressing PIPKIγ_i2, EGF treatment resulted in enhanced EGFR phosphorylation and β-catenin phosphorylation on Ser552 and Ser675 (Fig. 5D). Furthermore, EGF treatment enhanced the association between overexpressed PIPKIγ_i2 and β-catenin (Fig. 5E).
β-catenin is required for tumorigenic phenotypes associated with PIPKIγ expression
The cellular mechanisms by which PIPKIγ activates β-catenin suggest that these molecules could cooperate to promote tumorigenesis. To investigate whether PIPKIγ and β-catenin cooperate to alter tumorigenesis, a tissue microarray database was probed and Kaplan-Meier survival curves generated (28). In patients with no detectable β-catenin expression, PIPKIγ expression did not have a significant outcome on the survival of patients (data not shown). However, in patients where β-catenin expression was observed, higher PIPKIγ expression correlated with a reduced survival rate in breast cancer patients (data not shown). This suggests that β-catenin and PIPKIγ can act synergistically to promote tumorigenesis.
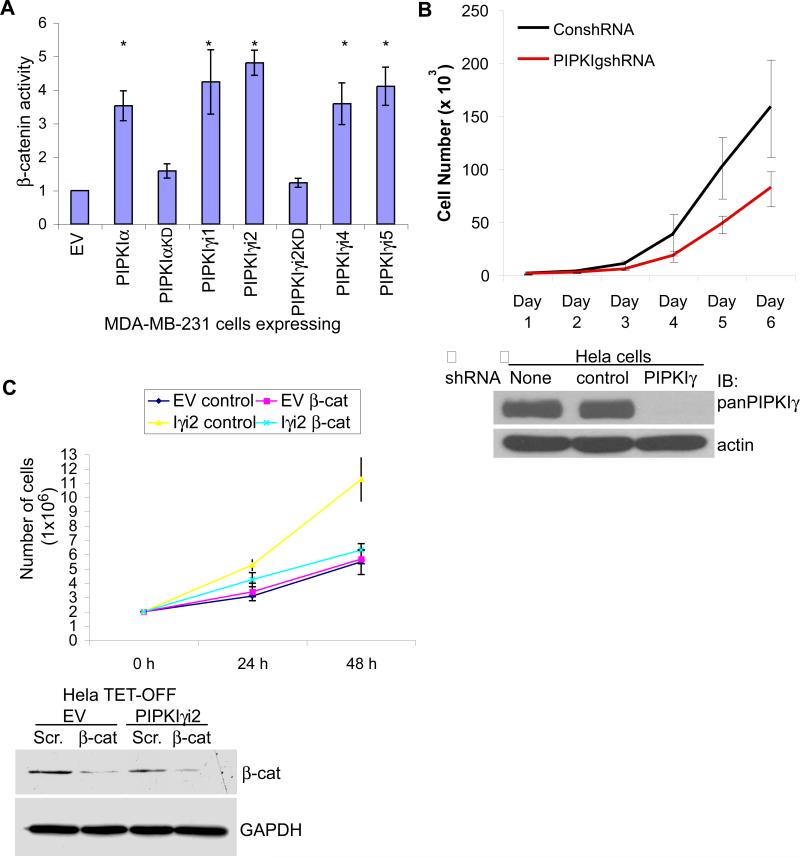
To further explore this possibility, the growth rate of cells expressing PIPKIγ was monitored. Knockdown of PIPKIγ using siRNA in MDA-MB-231 breast cancer cells results in a significant decrease in proliferation (28). Furthermore, PIPKIγ expression in these cells enhanced β-catenin activity, a result dependent on its catalytic activity (Fig. 6A). In addition, lentiviral-mediated knockdown of PIPKIγ in Hela cells also decreased proliferation (Fig. 6B). siRNA was used to knockdown β-catenin in Hela TET-OFF cells expressing empty vector (EV) or PIPKIγ_i2. When grown in the presence of serum, PIPKIγ_i2 expression significantly increased cell proliferation (Fig. 6C, Iγi2 control vs. EV control). This increase diminished upon treatment with β-catenin specific siRNA (Fig. 6C). Surprisingly, knockdown of β-catenin in Hela TET-OFF cells expressing the empty vector had little effect on the growth rate when grown in the presence of serum (Fig. 6C). This shows that PIPKIγ expression enhances cell proliferation in epithelial derived tumorigenic cells through its ability to activate β-catenin.
Figure 6. β-catenin is required for tumorigenic phenotypes associated with PIPKIγ expression.
(A) β-catenin transcriptional activity was measured in MDA-MB-231 cells transiently transfected with empty vector (EV) or the HA-tagged PIPKI shown (* represents p-value ≤ 0.01; n≥3; error bars=std dev). (B) Hela cells were infected with lentivirus containing scrambled or PIPKIγ specific shRNA and cell proliferation monitored for 6 days post-infection. Western blot analysis revealed the efficiency of PIPKIγ knockdown 72 h post-infection. (C) Hela TET-OFF parental and PIPKIγ_i2 expressing cells were treated with control or β-catenin specific siRNA and cell proliferation monitored for 48 h post-transfection. Western blot analysis revealed knockdown efficiency. Shown are graphical representations of average (n=3) cell growth at each time point. Error bars=st dev.
Discussion
The findings reported here represent a previously unrecognized role for PIPKIγ in regulating β-catenin transcriptional activity, providing a potential mechanism for understanding how PIPKIγ promotes tumorigenesis. Emerging evidence is defining a significant role for signaling pathways that promote β-catenin activation independent of Wnt and GSK-3β. Our findings mimic those of other groups in that the increased trascriptional activity of β-catenin is not accompanied by significant changes to its overall protein levels (4, 6, 23, 47, 48). While this differs from the classical view of β-catenin activation via inhibition of its degradation, the overall effect is the same. Understanding how these pathways modulate β-catenin transcriptional acitivity is extremely important, as β-catenin can multiple aspects of tumorigenesis, including proliferation, migration, loss of cell polarity and the establishment of a less differentiated state. In particular, β-catenin transcriptional activity enhances the epithelial-to-mesenchymal transition, a process resulting in increased cellular migration and invasion associated with metastatic formation (39). β-catenin activation via unique signaling pathways provides novel therapeutic targets for the treatment of malignancies. A recent report showed that β-catenin translocation to the nucleus occurs primarily in the peripheral cells of a tumor mass and correlates with an increased propensity for migration in these cells (40).
Previously, PIPKIγ splice variants have been shown to interact directly with E-cadherin, modulating its trafficking to and from the plasma membrane (33). In normal epithelial cells, β-catenin is incorporated into cell-cell adhesions where it provides a physical link between the cytoplasmic domain of E-cadherin and the actin cytoskeleton. Indeed, β-catenin is required for the establishment of cell polarity in normal epithelia (41). Upon depletion of Ca2+ or stimulation with growth factors such as EGF, HGF or TGF-β, E-cadherin is rapidly internalized, increasing cytoplasmic levels of β-catenin (22, 42). PIPKIγ could activate β-catenin by enhancing the endocytosis and degradation of E-cadherin thereby preventing E-cadherin from sequestering β-catenin at the cell membrane. We observe that Hela cells express low levels of E-cadherin that localizes to the plasma membrane at sites of cell-cell contact when cells are confluent. Expression of PIPKIγ in these cells could lead to higher levels of cytoplasmic β-catenin thus allowing for a greater activation. Consistent with this model, overexpression of the E-cadherin C-terminal domain blocked PIPKIγ-mediated increase in β-catenin transcriptional activity. However, expression of PIPKIγ in normal epithelial cells, such as MDCKs, does not prevent E-cadherin translocation to the membrane or the formation of cell-cell contacts (33), nor did it significantly increase β-catenin activity, suggesting that other mechanisms are required to initiate E-cadherin endocytosis and/or β-catenin activation. In support of this, E-cadherin endocytosis is not sufficient to activate β-catenin when the β-catenin degradation complex is present (43).
Activation of some growth factor-stimulated signaling pathways has been shown to inhibit β-catenin degradation and/or enhance β-catenin stabilization and nuclear translocation (22). Because these pathways are often hyperactivated in cancers, this could explain why we see increased β-catenin activity upon PIPKIγ expression only in transformed cells. Therefore, under circumstances such as increased growth factor signaling, higher levels of the PIPKIγ protein results in a more enhanced activation of β-catenin. It is likely that PIPKIγ can increase the activation of β-catenin on more than one level, as it could lead to its disassociation from E-cadherin and act within signaling pathways to increase its nuclear localization and transcriptional activity.
PIPKIγ enhances β-catenin activity through its ability to generate phosphoinositide messengers, most likely PI4,5P2, as the other messenger known to be generated by PIPKIs in vivo, PI3,4,5P3, utilizes a substrate that is present at miniscule cellular concentrations (26, 44). A spatial and temporal increase in PI4,5P2 could enhance signaling pathways which utilize this messenger and also activate β-catenin. For example, increased pools of PIP2 could be utilized by PI3K to generate higher amounts of PIP3, thereby increasing or prolonging the activation of this signal. We observe an increase in the phosphorylation of β-catenin at Ser552 and Ser675 upon PIPKIγ expression. Ser552 can be phosphorylated by AKT (23) resulting in β-catenin associating with 14-3-3zeta, its accumulation in the nucleus and increased transcriptional activity. Additionally, AKT is activated downstream of PIP3 generation by PI3K, whose activation within cells following stimulation by growth factors such as EGF, HGF and TGF-β has been well documented (45).
Phosphorylation of β-catenin at Ser552 and Ser675 is also mediated by cyclic AMP-dependent protein kinase (PKA), enhancing β-catenin activity (46), and Ser675 has been shown to be phosphorylated downstream of p-21-activated protein kinase-1 (PAK-1) (47). Hydrolysis of PIP2 generated by PIPKIγ can induce many physiological responses depending on the cell type, most notably increasing cellular Ca2+ levels (26). As PKA and Ca2+ synergize to induce cellular responses, this may explain how PIPKIγ enhances β-catenin activity. In the future, it will be important to further define the kinase(s) responsible for β-catenin phosphorylation downstream of PIPKIγ activity. In addition to playing a potential role in the activation of kinases that activate β-catenin, PIP2 generation might be required for the trafficking of E-cadherin containing vesicles to the lysosome (49). A dominant-negative PIPKIγ could block E-cadherin degradation leading to its recycling back to the membrane where it could associate with β-catenin, decreasing its nuclear activity.
It has yet to be determined if PIPKIγ activation of β-catenin requires their association. Surprisingly, the association between endogenous PIPKIγ and β-catenin decreased upon stimulation of Hela cells with EGF. This suggests that β-catenin activation by PIPKIγ splice variants was primarily an indirect effect. However, in cells overexpressing PIPKIγ, treatment with EGF for 3 h resulted in an enhanced association between the two proteins. The physiological relevance to our finding that PIPKIγ and β-catenin can directly associate remains to be defined. As endogenous levels of PIPKIγ splice-variants localize to different cellular sites, it will be important to further define this interaction to gain insight into its functional role. In addition, further studies are needed to characterize the expression of PIPKIγ splice-variants during tumorigenesis.
In summary, we have uncovered a novel component within signaling pathways that can be used to activate β-catenin. PIPKIγ generation of phosphoinositide messengers leads to the nuclear accumulation of β-catenin, thereby increasing its ability to induce transcriptional changes. Furthermore, we have shown that in response to growth factor stimulation, PIPKIγ expression markedly increases β-catenin activity and that the increase in cell proliferation observed in mesenchymal-like cells overexpressing PIPKIγ requires β-catenin. Lastly, we have observed that the decreased survival rate amongst breast cancer patients that express PIPKIγ correlates with β-catenin expression suggesting a role for this pathway in tumor progression.
Supplementary Material
Supplemental Information
PIPKIγ regulates β-catenin transcriptional activity in mesenchymal-like cells downstream of growth factor receptor activation
Mark Schramp, Narendra Thapa, Jessica Heck and Richard Anderson
Supplemental Experimental Procedures
Reagents
Polyclonal antibodies to PIPKIγ were created as previously described (1). Anti–HA was obtained from Covance; anti-EGFR(LA22), anti-GAPDH, anti-β-catenin, anti-actin and anti-phospho-EGFR(Y1173) were obtained from Millipore; anti-β-catenin was obtained from BD Transduction Laboratories; anti-phospho-β-catenin(Y86), anti–mouse and anit-rabbit Alexa Fluor 488 and 555 conjugates, and SlowFade reagent were obtained from Invitrogen; anti-phospho-β-catenin(S675), anti-phospho-β-catenin(S552), anti-phospho-β-catenin(S33/37), anti-LRP6, anti-phospho-LRP6, anti-c-met, anti-c-jun and anti-phospho-c-met(Y1234/1235) were obtained from Cell Signaling Technology; anti-phospho-β-catenin(Y654), anti-phospho-EGFR(Y1086), anti-HA, anti-Smad2/3 and anti-phospho-Smad2/3 were obtained from Abcam; anti-phospho-β-catenin(S45) was obtained from Biosource; anti-myc (9E10) was obtained from Santa Cruz Biotechnology; Doxycycline was obtained from Clontech; Wnt-3a and TGF-β were obtained from R&D Systems; EGF and HGF were obtained from Sigma; Horseradish peroxidase-conjugated anti-GST antibody was purchased from Amersham Biosciences and horseradish peroxidase-conjugated anti-T7 antibody was obtained from Novagen. Secondary horseradish-peroxidase-conjugated antibodies for Western blotting were obtained from Jackson Immunoresearch Laboratories. PIPKIγ and PIPKIα mammalian and bacterial expression vectors were described previously (2). pCMV-HA PIPKIγi4_KD, PIPKIγ_i5KD and PIPKIαKD were generated using the QuikChange II Site-Directed Mutagenesis kit (Stratagene). pCMV-myc containing the E-cadherin C-terminus was previously described (2).
Cell culture and transfection
All cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% FBS. Hela TET-OFF cells stably expressing various PIPKIγ constructs have been described elsewhere (2). Hela and MDCK cells were obtained from ATCC. For growth factor stimulation experiments, cells were serum starved in DMEM+0.5% FBS for 12-24 h and then stimulated with the appropriate growth factor.
Adherent cells were transfected with Lipofectamine™ 2000 (Invitrogen) or FUGENE® 6 transfection reagents according to the manufacturer's instructions; transient transfection conditions were optimized for maximum expression and minimal toxicity.
Lysate preparation, and immunoblotting
Whole-cell extracts were prepared in RIPA lysis buffer (20mM Tris-HCl, pH 7.5, 1% sodium deoxycholate, 2 mM EDTA, 1% [vol/vol] Nonidet P-40, 150 mM NaCl, 0.1% SDS, 50 mM NaF, supplemented with protease inhibitors). For immunoprecipitation experiments, lysates were prepared in a modified RIPA buffer (50mM Tris-HCl, pH 7.2, 0.5% sodium deoxycholate, 2 mM EDTA, 1% [vol/vol] Triton X-100, 50 mM NaCl, 0.1% SDS, supplemented with Complete Protease Inhibitor Mixture (Roche)). Lysates were sonicated, cleared of debris by centrifuation at 15,0000xg for 10 min at 4°C and protein concentrations calculated using the BCA (bicinchoninic acid) protein quantification assay (Bio-Rad Laboratories) according to the manufacturer's instructions. For all IPs, samples of equal protein concentration and volume were pre-cleared with normal mouse or rabbit IgG (Millipore) and Protein A/G sepharose beads (GE Healthcare). IPs were carried out overnight at 4°C followed by washing in modified RIPA buffer 5X. For immunoblot analysis, proteins were resolved by SDS-PAGE and transferred to Immobilon polyvinylidene fluoride filters (Millipore). Blots were incubated 30 min in PBS-T (PBS + 0.1% Tween-20) containing 5% nonfat dry milk or 3% BSA. The blots were incubated with primary antibody overnight at 4°C followed by incubation with HRP-conjugated secondary antibodies, and immunolabeled proteins were visualized with Western Lightning Chemiluminescence Reagent Plus (PerkinElmer).
Immunofluorescence and microscopy
HeLa or Hela TET-Off in DME + 10% FBS cells were grown on glass coverslips placed inside six-well plates 24 h prior to transfection. Cells growing on glass coverslips were washed in PBS and fixed in PBS plus 4% PFA for 30 min at room temperature or overnight at 4°C, permeabilized in PBS containing 0.2% Triton X-100, and incubated for 30 min in PBS containing 3% BSA (Jackson Immunoresearch Laboratories) and 2% normal goat serum (Calbiochem). Samples were incubated with primary antibodies for 1 h at room temperature. Incubation with fluorophore-conjugated secondary antibodies was performed at 37°C for 30 min. Signals were developed using either Alexa Fluor 488 goat anti–rabbit or anti–mouse IgG for 1 h at room temperature. Samples were mounted in SlowFade antifade reagent containing DAPI. Indirect immunofluorescence microscopy was performed on a Nikon Eclipse TE2000U instrument equipped with a Photometrics CoolSNAP CCD (charged coupled device) camera. Images were captured and further processed using MetaMorph (Molecular Devices) cellular imaging software. Images were exported to Photoshop CS2 (Adobe) for final processing and assembly.
GST affinity pull-down assays
Biochemical affinity precipitation assays to measure an association between β-catenin and PIPKIγ were performed essentially as described previously (3). In brief, constructs in pET28 and pET42 were transformed into BL21 (DE3) competent cells (Novagen). Proteins were expressed and purified using His-Bind Resin following manufacturer's instructions (Novagen) or using glutathione-Sepharose 4B Fast Flow as per the manufacturer's instructions (Amersham Biosciences). Equal amounts of purified recombinant GST-tagged PIPKIγ and T7-tagged β-catenin splice variants were incubated with 50 ul 1:1 diluted glutathione-Sepharose 4B Fast Flow (Amersham Biosciences) in 500 ul of pulldown buffer (PBS, 0.5% NP-40, and 2mM DTT) overnight at 4°C while rotating. Purified GST alone incubated with T7-tagged β-catenin splice variants was used as a negative control for non-specific protein binding. The beads were washed with 1 ml ice-cold pulldown buffer four times, resolved by SDS-PAGE, and analyzed via Western blot. All tubes, reagents, and rotors were prechilled on ice before use and all steps were performed in a cold room.
References
1. Ling K, Doughman RL, Firestone AJ, Bunce MW, Anderson RA. Type I gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature 2002;420(6911):89-93.
2. Ling K, Bairstow SF, Carbonara C, Turbin DA, Huntsman DG, Anderson RA. Type Igamma phosphatidylinositol phosphate kinase modulates adherens junction and E-cadherin trafficking via a direct interaction with mu 1B adaptin. J Cell Biol 2007;176(3):343-53.
3. Ling K, Doughman RL, Iyer VV, et al. Tyrosine phosphorylation of type Igamma phosphatidylinositol phosphate kinase by Src regulates an integrin-talin switch. J Cell Biol 2003;163(6):1339-49.
Figure S1. PIPKIγ expression stimulates β-catenin transcriptional activity in Hela TET-OFF and HEK293T cells. (A) Hela TET-OFF cells stably transfected with empty vector, PIPKIγ_i1, PIPKIγ_i2 or PIPKIγ_i2KD were grown for 24 h in DMEM+10%FBS +/- doxycycline where indicated. Cells were transfected with the TOP-FLASH reporter construct and β-catenin activity quantified 24 h later. Western blotting using anti-HA confirmed the expression of the PIPKIγ isoform and anti-β-catenin monitored the effects of PIPKIγ expression on overall protein levels. (B) HEK293T cells were transiently transfected with the TOP-FLASH luciferase β-catenin reporter construct and with either empty vector (EV) or the HA-tagged type I PIPK shown and β-catenin activity assayed (n=3, error bars=std dev.).
Figure S2. PIPKIγ generation of PIP2 increases the nuclear accumulation of β-catenin. Hela TET-OFF cells stably expressing empty vector (EV) or HA-tagged PIPKIγ_i2 or PIPKIγ_i2KD were grown on glass coverslips in DMEM+ 10%FBS +doxycycline. To initiate PIPKIγ_i2 expression, media was replaced with fresh media +/-dox. where indicated. 24 h later, cells were fixed and stained with dapi, anti-HA and anti-β-catenin. All images were taken with a 60X objective.
Figure S3. Effect of PIPKIα or PIPKIγ expression on Wnt-dependent β-catenin activity. β-catenin activity was measured in Hela cells transiently transfected with empty vector (EV) or the HA-tagged PIPKI shown. 24 h after transfection, cells were serum starved and then left untreated (none) or stimulated with 100ng/ml wnt-3a where indicated followed by cell lysis to measure β-catenin activity. (B) The efficacy of growth factor stimulation in transiently transfected Hela cells was monitored by western blotting using activation specific antibodies. Shown are representive blots of at least 3 experiments. (D) β-catenin activity was measured in Hela TET-OFF cells stably transfected with empty vector, PIPKIγ_i2 or PIPKIγ_i2KD. Cells were serum starved in DMEM+0.5%FBS without doxycycline. Cells were treated with 1nM EGF and β-catenin activity quantified 24 h later (n=3, error bars=std dev.).
Financial Support
This work was supported by NIH grants GM057549, GM051968 and CA104708.
References
- 1.Brembeck FH, Rosario M, Birchmeier W. Balancing cell adhesion and Wnt signaling, the key role of beta-catenin. Curr Opin Genet Dev. 2006;16(1):51–9. doi: 10.1016/j.gde.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Kemler R. From cadherins to catenins: cytoplasmic protein interactions and regulation of cell adhesion. Trends Genet. 1993;9(9):317–21. doi: 10.1016/0168-9525(93)90250-l. [DOI] [PubMed] [Google Scholar]
- 3.Desbois-Mouthon C, Cadoret A, Blivet-Van Eggelpoel MJ, et al. Insulin and IGF-1 stimulate the beta-catenin pathway through two signalling cascades involving GSK-3beta inhibition and Ras activation. Oncogene. 2001;20(2):252–9. doi: 10.1038/sj.onc.1204064. [DOI] [PubMed] [Google Scholar]
- 4.Lu Z, Ghosh S, Wang Z, Hunter T. Downregulation of caveolin-1 function by EGF leads to the loss of E-cadherin, increased transcriptional activity of beta-catenin, and enhanced tumor cell invasion. Cancer Cell. 2003;4(6):499–515. doi: 10.1016/s1535-6108(03)00304-0. [DOI] [PubMed] [Google Scholar]
- 5.Moon RT, Bowerman B, Boutros M, Perrimon N. The promise and perils of Wnt signaling through beta-catenin. Science. 2002;296(5573):1644–6. doi: 10.1126/science.1071549. [DOI] [PubMed] [Google Scholar]
- 6.Monga SP, Mars WM, Pediaditakis P, et al. Hepatocyte growth factor induces Wnt-independent nuclear translocation of beta-catenin after Met-beta-catenin dissociation in hepatocytes. Cancer Res. 2002;62(7):2064–71. [PubMed] [Google Scholar]
- 7.Lu Z, Hunter T. Wnt-independent beta-catenin transactivation in tumor development. Cell Cycle. 2004;3(5):571–3. [PubMed] [Google Scholar]
- 8.Mann B, Gelos M, Siedow A, et al. Target genes of beta-catenin-T cell-factor/lymphoid-enhancer-factor signaling in human colorectal carcinomas. Proc Natl Acad Sci USA. 1999;96(4):1603–8. doi: 10.1073/pnas.96.4.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peifer M, Polakis P. Wnt signaling in oncogenesis and embryogenesis--a look outside the nucleus. Science. 2000;287(5458):1606–9. doi: 10.1126/science.287.5458.1606. [DOI] [PubMed] [Google Scholar]
- 10.Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11(24):3286–305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 11.Schlange T, Matsuda Y, Lienhard S, Huber A, Hynes NE. Autocrine WNT signaling contributes to breast cancer cell proliferation via the canonical WNT pathway and EGFR transactivation. Breast Cancer Res. 2007;9(5):R63. doi: 10.1186/bcr1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du C, Jaggi M, Zhang C, Balaji KC. Protein kinase D1-mediated phosphorylation and subcellular localization of beta-catenin. Cancer Res. 2009;69(3):1117–24. doi: 10.1158/0008-5472.CAN-07-6270. [DOI] [PubMed] [Google Scholar]
- 13.van de Wetering M, Barker N, Harkes IC, et al. Mutant E-cadherin breast cancer cells do not display constitutive Wnt signaling. Cancer Res. 2001;61(1):278–84. [PubMed] [Google Scholar]
- 14.Lin SY, Xia W, Wang JC, et al. Beta-catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression. Proc Natl Acad Sci USA. 2000;97(8):4262–6. doi: 10.1073/pnas.060025397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7(2):131–42. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 16.Stockinger A, Eger A, Wolf J, Beug H, Foisner R. E-cadherin regulates cell growth by modulating proliferation-dependent beta-catenin transcriptional activity. J Cell Biol. 2001;154(6):1185–96. doi: 10.1083/jcb.200104036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottardi CJ, Wong E, Gumbiner BM. E-cadherin suppresses cellular transformation by inhibiting beta-catenin signaling in an adhesion-independent manner. J Cell Biol. 2001;153(5):1049–60. doi: 10.1083/jcb.153.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gottardi CJ, Gumbiner BM. Adhesion signaling: how beta-catenin interacts with its partners. Curr Biol. 2001;11(19):R792–4. doi: 10.1016/s0960-9822(01)00473-0. [DOI] [PubMed] [Google Scholar]
- 19.Frixen UH, Behrens J, Sachs M, et al. E-cadherin-mediated cell-cell adhesion prevents invasiveness of human carcinoma cells. J Cell Biol. 1991;113(1):173–85. doi: 10.1083/jcb.113.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol. 2005;6(8):622–34. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- 21.Graham NA, Asthagiri AR. Epidermal growth factor-mediated T-cell factor/lymphoid enhancer factor transcriptional activity is essential but not sufficient for cell cycle progression in nontransformed mammary epithelial cells. J Biol Chem. 2004;279(22):23517–24. doi: 10.1074/jbc.M314055200. [DOI] [PubMed] [Google Scholar]
- 22.Daugherty RL, Gottardi CJ. Phospho-regulation of Beta-catenin adhesion and signaling functions. Physiology (Bethesda) 2007;22:303–9. doi: 10.1152/physiol.00020.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang D, Hawke D, Zheng Y, et al. Phosphorylation of beta-catenin by AKT promotes beta-catenin transcriptional activity. J Biol Chem. 2007;282(15):11221–9. doi: 10.1074/jbc.M611871200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin T, George Fantus I, Sun J. Wnt and beyond Wnt: multiple mechanisms control the transcriptional property of beta-catenin. Cell Signal. 2008;20(10):1697–704. doi: 10.1016/j.cellsig.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 25.Qi J, Wang J, Romanyuk O, Siu CH. Involvement of Src family kinases in N-cadherin phosphorylation and beta-catenin dissociation during transendothelial migration of melanoma cells. Mol Biol Cell. 2006;17(3):1261–72. doi: 10.1091/mbc.E05-10-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oude Weernink PA, Schmidt M, Jakobs KH. Regulation and cellular roles of phosphoinositide 5-kinases. Eur J Pharmacol. 2004;500(1-3):87–99. doi: 10.1016/j.ejphar.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 27.Schill NJ, Anderson RA. Two novel phosphatidylinositol 4-phosphate, 5-kinase type I gamma splice variants expressed in human cells display distinctive cellular targeting. Biochem J. 2009 doi: 10.1042/BJ20090638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Y, Turbin DA, Ling K, et al. Type I gamma phosphatidylinositol phosphate kinase modulates invasion and proliferation and its expression correlates with poor prognosis in breast cancer. Breast Cancer Res. 12(1):R6. doi: 10.1186/bcr2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singhal RL, Yeh YA, Look KY, Sledge GW, Jr., Weber G. Coordinated increase in activities of the signal transduction enzymes PI kinase and PIP kinase in human cancer cells. Life Sci. 1994;55(19):1487–92. doi: 10.1016/0024-3205(94)00690-3. [DOI] [PubMed] [Google Scholar]
- 30.Makretsov NA, Huntsman DG, Nielsen TO, et al. Hierarchical clustering analysis of tissue microarray immunostaining data identifies prognostically significant groups of breast carcinoma. Clin Cancer Res. 2004;10(18 Pt 1):6143–51. doi: 10.1158/1078-0432.CCR-04-0429. [DOI] [PubMed] [Google Scholar]
- 31.Rubinson DA, Dillon CP, Kwiatkowski AV, et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33(3):401–6. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 32.Lee JS, Hmama Z, Mui A, Reiner NE. Stable gene silencing in human monocytic cell lines using lentiviral-delivered small interference RNA. Silencing of the p110alpha isoform of phosphoinositide 3-kinase reveals differential regulation of adherence induced by 1alpha,25-dihydroxycholecalciferol and bacterial lipopolysaccharide. J Biol Chem. 2004;279(10):9379–88. doi: 10.1074/jbc.M310638200. [DOI] [PubMed] [Google Scholar]
- 33.Ling K, Bairstow SF, Carbonara C, Turbin DA, Huntsman DG, Anderson RA. Type Igamma phosphatidylinositol phosphate kinase modulates adherens junction and E-cadherin trafficking via a direct interaction with mu 1B adaptin. J Cell Biol. 2007;176(3):343–53. doi: 10.1083/jcb.200606023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roczniak-Ferguson A, Reynolds AB. Regulation of p120-catenin nucleocytoplasmic shuttling activity. J Cell Sci. 2003;116(Pt 20):4201–12. doi: 10.1242/jcs.00724. [DOI] [PubMed] [Google Scholar]
- 35.Suriano G, Mulholland D, de Wever O, et al. The intracellular E-cadherin germline mutation V832 M lacks the ability to mediate cell-cell adhesion and to suppress invasion. Oncogene. 2003;22(36):5716–9. doi: 10.1038/sj.onc.1206672. [DOI] [PubMed] [Google Scholar]
- 36.Korinek V, Barker N, Morin PJ, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275(5307):1784–7. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 37.Birchmeier W, Behrens J. Cadherin expression in carcinomas: role in the formation of cell junctions and the prevention of invasiveness. Biochim Biophys Acta. 1994;1198(1):11–26. doi: 10.1016/0304-419x(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 38.Pan W, Choi SC, Wang H, et al. Wnt3a-mediated formation of phosphatidylinositol 4,5-bisphosphate regulates LRP6 phosphorylation. Science. 2008;321(5894):1350–3. doi: 10.1126/science.1160741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmalhofer O, Brabletz S, Brabletz T. E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev. 2009;28(1-2):151–66. doi: 10.1007/s10555-008-9179-y. [DOI] [PubMed] [Google Scholar]
- 40.Brabletz T, Jung A, Reu S, et al. Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc Natl Acad Sci USA. 2001;98(18):10356–61. doi: 10.1073/pnas.171610498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cain S, Martinez G, Kokkinos MI, et al. Differential requirement for beta-catenin in epithelial and fiber cells during lens development. Dev Biol. 2008;321(2):420–33. doi: 10.1016/j.ydbio.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 42.Conacci-Sorrell M, Zhurinsky J, Ben-Ze'ev A. The cadherin-catenin adhesion system in signaling and cancer. J Clin Invest. 2002;109(8):987–91. doi: 10.1172/JCI15429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kam Y, Quaranta V. Cadherin-bound beta-catenin feeds into the Wnt pathway upon adherens junctions dissociation: evidence for an intersection between beta-catenin pools. PLoS One. 2009;4(2):e4580. doi: 10.1371/journal.pone.0004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X, Loijens JC, Boronenkov IV, et al. Phosphatidylinositol-4-phosphate 5-kinase isozymes catalyze the synthesis of 3-phosphate-containing phosphatidylinositol signaling molecules. J Biol Chem. 1997;272(28):17756–61. doi: 10.1074/jbc.272.28.17756. [DOI] [PubMed] [Google Scholar]
- 45.Bianco R, Melisi D, Ciardiello F, Tortora G. Key cancer cell signal transduction pathways as therapeutic targets. Eur J Cancer. 2006;42(3):290–4. doi: 10.1016/j.ejca.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 46.Hino S, Tanji C, Nakayama KI, Kikuchi A. Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase stabilizes beta-catenin through inhibition of its ubiquitination. Mol Cell Biol. 2005;25(20):9063–72. doi: 10.1128/MCB.25.20.9063-9072.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun J, Khalid S, Rozakis-Adcock M, Fantus IG, Jin T. P-21-activated protein kinase-1 functions as a linker between insulin and Wnt signaling pathways in the intestine. Oncogene. 2009;28(35):3132–44. doi: 10.1038/onc.2009.167. [DOI] [PubMed] [Google Scholar]
- 48.Ji H, Wang J, Nika H, et al. EGF-induced ERK activation promotes CK2-mediated disassociation of alpha-Catenin from beta-Catenin and transactivation of beta-Catenin. Mol Cell. 2009;36(4):547–59. doi: 10.1016/j.molcel.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schill NJ, Anderson RA. Out, in and back again: PtdIns(4,5)P(2) regulates cadherin trafficking in epithelial morphogenesis. Biochem J. 2009;418(2):247–60. doi: 10.1042/BJ20081844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Information
PIPKIγ regulates β-catenin transcriptional activity in mesenchymal-like cells downstream of growth factor receptor activation
Mark Schramp, Narendra Thapa, Jessica Heck and Richard Anderson
Supplemental Experimental Procedures
Reagents
Polyclonal antibodies to PIPKIγ were created as previously described (1). Anti–HA was obtained from Covance; anti-EGFR(LA22), anti-GAPDH, anti-β-catenin, anti-actin and anti-phospho-EGFR(Y1173) were obtained from Millipore; anti-β-catenin was obtained from BD Transduction Laboratories; anti-phospho-β-catenin(Y86), anti–mouse and anit-rabbit Alexa Fluor 488 and 555 conjugates, and SlowFade reagent were obtained from Invitrogen; anti-phospho-β-catenin(S675), anti-phospho-β-catenin(S552), anti-phospho-β-catenin(S33/37), anti-LRP6, anti-phospho-LRP6, anti-c-met, anti-c-jun and anti-phospho-c-met(Y1234/1235) were obtained from Cell Signaling Technology; anti-phospho-β-catenin(Y654), anti-phospho-EGFR(Y1086), anti-HA, anti-Smad2/3 and anti-phospho-Smad2/3 were obtained from Abcam; anti-phospho-β-catenin(S45) was obtained from Biosource; anti-myc (9E10) was obtained from Santa Cruz Biotechnology; Doxycycline was obtained from Clontech; Wnt-3a and TGF-β were obtained from R&D Systems; EGF and HGF were obtained from Sigma; Horseradish peroxidase-conjugated anti-GST antibody was purchased from Amersham Biosciences and horseradish peroxidase-conjugated anti-T7 antibody was obtained from Novagen. Secondary horseradish-peroxidase-conjugated antibodies for Western blotting were obtained from Jackson Immunoresearch Laboratories. PIPKIγ and PIPKIα mammalian and bacterial expression vectors were described previously (2). pCMV-HA PIPKIγi4_KD, PIPKIγ_i5KD and PIPKIαKD were generated using the QuikChange II Site-Directed Mutagenesis kit (Stratagene). pCMV-myc containing the E-cadherin C-terminus was previously described (2).
Cell culture and transfection
All cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% FBS. Hela TET-OFF cells stably expressing various PIPKIγ constructs have been described elsewhere (2). Hela and MDCK cells were obtained from ATCC. For growth factor stimulation experiments, cells were serum starved in DMEM+0.5% FBS for 12-24 h and then stimulated with the appropriate growth factor.
Adherent cells were transfected with Lipofectamine™ 2000 (Invitrogen) or FUGENE® 6 transfection reagents according to the manufacturer's instructions; transient transfection conditions were optimized for maximum expression and minimal toxicity.
Lysate preparation, and immunoblotting
Whole-cell extracts were prepared in RIPA lysis buffer (20mM Tris-HCl, pH 7.5, 1% sodium deoxycholate, 2 mM EDTA, 1% [vol/vol] Nonidet P-40, 150 mM NaCl, 0.1% SDS, 50 mM NaF, supplemented with protease inhibitors). For immunoprecipitation experiments, lysates were prepared in a modified RIPA buffer (50mM Tris-HCl, pH 7.2, 0.5% sodium deoxycholate, 2 mM EDTA, 1% [vol/vol] Triton X-100, 50 mM NaCl, 0.1% SDS, supplemented with Complete Protease Inhibitor Mixture (Roche)). Lysates were sonicated, cleared of debris by centrifuation at 15,0000xg for 10 min at 4°C and protein concentrations calculated using the BCA (bicinchoninic acid) protein quantification assay (Bio-Rad Laboratories) according to the manufacturer's instructions. For all IPs, samples of equal protein concentration and volume were pre-cleared with normal mouse or rabbit IgG (Millipore) and Protein A/G sepharose beads (GE Healthcare). IPs were carried out overnight at 4°C followed by washing in modified RIPA buffer 5X. For immunoblot analysis, proteins were resolved by SDS-PAGE and transferred to Immobilon polyvinylidene fluoride filters (Millipore). Blots were incubated 30 min in PBS-T (PBS + 0.1% Tween-20) containing 5% nonfat dry milk or 3% BSA. The blots were incubated with primary antibody overnight at 4°C followed by incubation with HRP-conjugated secondary antibodies, and immunolabeled proteins were visualized with Western Lightning Chemiluminescence Reagent Plus (PerkinElmer).
Immunofluorescence and microscopy
HeLa or Hela TET-Off in DME + 10% FBS cells were grown on glass coverslips placed inside six-well plates 24 h prior to transfection. Cells growing on glass coverslips were washed in PBS and fixed in PBS plus 4% PFA for 30 min at room temperature or overnight at 4°C, permeabilized in PBS containing 0.2% Triton X-100, and incubated for 30 min in PBS containing 3% BSA (Jackson Immunoresearch Laboratories) and 2% normal goat serum (Calbiochem). Samples were incubated with primary antibodies for 1 h at room temperature. Incubation with fluorophore-conjugated secondary antibodies was performed at 37°C for 30 min. Signals were developed using either Alexa Fluor 488 goat anti–rabbit or anti–mouse IgG for 1 h at room temperature. Samples were mounted in SlowFade antifade reagent containing DAPI. Indirect immunofluorescence microscopy was performed on a Nikon Eclipse TE2000U instrument equipped with a Photometrics CoolSNAP CCD (charged coupled device) camera. Images were captured and further processed using MetaMorph (Molecular Devices) cellular imaging software. Images were exported to Photoshop CS2 (Adobe) for final processing and assembly.
GST affinity pull-down assays
Biochemical affinity precipitation assays to measure an association between β-catenin and PIPKIγ were performed essentially as described previously (3). In brief, constructs in pET28 and pET42 were transformed into BL21 (DE3) competent cells (Novagen). Proteins were expressed and purified using His-Bind Resin following manufacturer's instructions (Novagen) or using glutathione-Sepharose 4B Fast Flow as per the manufacturer's instructions (Amersham Biosciences). Equal amounts of purified recombinant GST-tagged PIPKIγ and T7-tagged β-catenin splice variants were incubated with 50 ul 1:1 diluted glutathione-Sepharose 4B Fast Flow (Amersham Biosciences) in 500 ul of pulldown buffer (PBS, 0.5% NP-40, and 2mM DTT) overnight at 4°C while rotating. Purified GST alone incubated with T7-tagged β-catenin splice variants was used as a negative control for non-specific protein binding. The beads were washed with 1 ml ice-cold pulldown buffer four times, resolved by SDS-PAGE, and analyzed via Western blot. All tubes, reagents, and rotors were prechilled on ice before use and all steps were performed in a cold room.
References
1. Ling K, Doughman RL, Firestone AJ, Bunce MW, Anderson RA. Type I gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature 2002;420(6911):89-93.
2. Ling K, Bairstow SF, Carbonara C, Turbin DA, Huntsman DG, Anderson RA. Type Igamma phosphatidylinositol phosphate kinase modulates adherens junction and E-cadherin trafficking via a direct interaction with mu 1B adaptin. J Cell Biol 2007;176(3):343-53.
3. Ling K, Doughman RL, Iyer VV, et al. Tyrosine phosphorylation of type Igamma phosphatidylinositol phosphate kinase by Src regulates an integrin-talin switch. J Cell Biol 2003;163(6):1339-49.
Figure S1. PIPKIγ expression stimulates β-catenin transcriptional activity in Hela TET-OFF and HEK293T cells. (A) Hela TET-OFF cells stably transfected with empty vector, PIPKIγ_i1, PIPKIγ_i2 or PIPKIγ_i2KD were grown for 24 h in DMEM+10%FBS +/- doxycycline where indicated. Cells were transfected with the TOP-FLASH reporter construct and β-catenin activity quantified 24 h later. Western blotting using anti-HA confirmed the expression of the PIPKIγ isoform and anti-β-catenin monitored the effects of PIPKIγ expression on overall protein levels. (B) HEK293T cells were transiently transfected with the TOP-FLASH luciferase β-catenin reporter construct and with either empty vector (EV) or the HA-tagged type I PIPK shown and β-catenin activity assayed (n=3, error bars=std dev.).
Figure S2. PIPKIγ generation of PIP2 increases the nuclear accumulation of β-catenin. Hela TET-OFF cells stably expressing empty vector (EV) or HA-tagged PIPKIγ_i2 or PIPKIγ_i2KD were grown on glass coverslips in DMEM+ 10%FBS +doxycycline. To initiate PIPKIγ_i2 expression, media was replaced with fresh media +/-dox. where indicated. 24 h later, cells were fixed and stained with dapi, anti-HA and anti-β-catenin. All images were taken with a 60X objective.
Figure S3. Effect of PIPKIα or PIPKIγ expression on Wnt-dependent β-catenin activity. β-catenin activity was measured in Hela cells transiently transfected with empty vector (EV) or the HA-tagged PIPKI shown. 24 h after transfection, cells were serum starved and then left untreated (none) or stimulated with 100ng/ml wnt-3a where indicated followed by cell lysis to measure β-catenin activity. (B) The efficacy of growth factor stimulation in transiently transfected Hela cells was monitored by western blotting using activation specific antibodies. Shown are representive blots of at least 3 experiments. (D) β-catenin activity was measured in Hela TET-OFF cells stably transfected with empty vector, PIPKIγ_i2 or PIPKIγ_i2KD. Cells were serum starved in DMEM+0.5%FBS without doxycycline. Cells were treated with 1nM EGF and β-catenin activity quantified 24 h later (n=3, error bars=std dev.).