Abstract
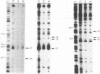
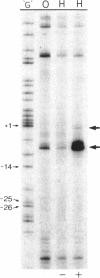
The complexes forming between the alternative sigma factor protein sigma N (sigma 54), its holoenzyme and promoter DNA were analysed using the hydroxyl radical probe and by photochemical footprinting of bromouridine-substituted DNA. Close contacts between the promoter, sigma N and its holoenzyme appear to be restricted predominantly to one face of the DNA helix, extending from -31 to -5. They all appear attributable to sigma N and no extra close contacts from the core RNA polymerase subunits in the holoenzyme-promoter DNA complex were detected. We suggest that the apparent absence of close core RNA polymerase contacts in the region of the promoter DNA to be melted during open complex formation is important for maintaining the closed complex. Results of the hydroxyl radical footprinting imply that sigma N makes multiple DNA backbone contacts across and beyond the -12, -24 consensus promoter elements, and the photochemical footprints indicate that consensus thymidine residues contribute important major groove contacts to sigma N. Formation of the open complex is shown to involve a major structural transition in the DNA contacted by sigma N, establishing a direct role for sigma N in formation of the activated promoter complex.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Buck M., Cannon W. A simple procedure for visualising protein-nucleic acid complexes by photochemical crosslinking. Nucleic Acids Res. 1994 Mar 25;22(6):1119–1120. doi: 10.1093/nar/22.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M., Cannon W. Activator-independent formation of a closed complex between sigma 54-holoenzyme and nifH and nifU promoters of Klebsiella pneumoniae. Mol Microbiol. 1992 Jun;6(12):1625–1630. doi: 10.1111/j.1365-2958.1992.tb00887.x. [DOI] [PubMed] [Google Scholar]
- Buck M., Cannon W. Specific binding of the transcription factor sigma-54 to promoter DNA. Nature. 1992 Jul 30;358(6385):422–424. doi: 10.1038/358422a0. [DOI] [PubMed] [Google Scholar]
- Buck M., Cannon W., Woodcock J. Transcriptional activation of the Klebsiella pneumoniae nitrogenase promoter may involve DNA loop formation. Mol Microbiol. 1987 Sep;1(2):243–249. doi: 10.1111/j.1365-2958.1987.tb00518.x. [DOI] [PubMed] [Google Scholar]
- Cannon W., Buck M. Central domain of the positive control protein NifA and its role in transcriptional activation. J Mol Biol. 1992 May 20;225(2):271–286. doi: 10.1016/0022-2836(92)90921-6. [DOI] [PubMed] [Google Scholar]
- Cannon W., Claverie-Martin F., Austin S., Buck M. Core RNA polymerase assists binding of the transcription factor sigma 54 to promoter DNA. Mol Microbiol. 1993 Apr;8(2):287–298. doi: 10.1111/j.1365-2958.1993.tb01573.x. [DOI] [PubMed] [Google Scholar]
- Cannon W., Claverie-Martin F., Austin S., Buck M. Identification of a DNA-contacting surface in the transcription factor sigma-54. Mol Microbiol. 1994 Jan;11(2):227–236. doi: 10.1111/j.1365-2958.1994.tb00303.x. [DOI] [PubMed] [Google Scholar]
- Charlton W., Cannon W., Buck M. The Klebsiella pneumoniae nifJ promoter: analysis of promoter elements regulating activation by the NifA promoter. Mol Microbiol. 1993 Mar;7(6):1007–1021. doi: 10.1111/j.1365-2958.1993.tb01192.x. [DOI] [PubMed] [Google Scholar]
- Choy H. A., Romeo J. M., Geiduschek E. P. Activity of a phage-modified RNA polymerase at hybrid promoters. Effects of substituting thymine for hydroxymethyluracil in a phage SP01 middle promoter. J Mol Biol. 1986 Sep 5;191(1):59–73. doi: 10.1016/0022-2836(86)90422-5. [DOI] [PubMed] [Google Scholar]
- Claverie-Martin F., Magasanik B. Role of integration host factor in the regulation of the glnHp2 promoter of Escherichia coli. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1631–1635. doi: 10.1073/pnas.88.5.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowing D. W., Mecsas J., Record M. T., Jr, Gross C. A. Intermediates in the formation of the open complex by RNA polymerase holoenzyme containing the sigma factor sigma 32 at the groE promoter. J Mol Biol. 1989 Dec 5;210(3):521–530. doi: 10.1016/0022-2836(89)90128-9. [DOI] [PubMed] [Google Scholar]
- Dixon W. J., Hayes J. J., Levin J. R., Weidner M. F., Dombroski B. A., Tullius T. D. Hydroxyl radical footprinting. Methods Enzymol. 1991;208:380–413. doi: 10.1016/0076-6879(91)08021-9. [DOI] [PubMed] [Google Scholar]
- Dombroski A. J., Walter W. A., Record M. T., Jr, Siegele D. A., Gross C. A. Polypeptides containing highly conserved regions of transcription initiation factor sigma 70 exhibit specificity of binding to promoter DNA. Cell. 1992 Aug 7;70(3):501–512. doi: 10.1016/0092-8674(92)90174-b. [DOI] [PubMed] [Google Scholar]
- Harrison S. C., Aggarwal A. K. DNA recognition by proteins with the helix-turn-helix motif. Annu Rev Biochem. 1990;59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- Hoover T. R., Santero E., Porter S., Kustu S. The integration host factor stimulates interaction of RNA polymerase with NIFA, the transcriptional activator for nitrogen fixation operons. Cell. 1990 Oct 5;63(1):11–22. doi: 10.1016/0092-8674(90)90284-l. [DOI] [PubMed] [Google Scholar]
- Hsieh M., Tintut Y., Gralla J. D. Functional roles for the glutamines within the glutamine-rich region of the transcription factor sigma 54. J Biol Chem. 1994 Jan 7;269(1):373–378. [PubMed] [Google Scholar]
- Kleiner D., Paul W., Merrick M. J. Construction of multicopy expression vectors for regulated over-production of proteins in Klebsiella pneumoniae and other enteric bacteria. J Gen Microbiol. 1988 Jul;134(7):1779–1784. doi: 10.1099/00221287-134-7-1779. [DOI] [PubMed] [Google Scholar]
- Klement J. F., Moorefield M. B., Jorgensen E., Brown J. E., Risman S., McAllister W. T. Discrimination between bacteriophage T3 and T7 promoters by the T3 and T7 RNA polymerases depends primarily upon a three base-pair region located 10 to 12 base-pairs upstream from the start site. J Mol Biol. 1990 Sep 5;215(1):21–29. doi: 10.1016/s0022-2836(05)80091-9. [DOI] [PubMed] [Google Scholar]
- Kovacic R. T. The 0 degree C closed complexes between Escherichia coli RNA polymerase and two promoters, T7-A3 and lacUV5. J Biol Chem. 1987 Oct 5;262(28):13654–13661. [PubMed] [Google Scholar]
- Kustu S., Santero E., Keener J., Popham D., Weiss D. Expression of sigma 54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol Rev. 1989 Sep;53(3):367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. S., Ishihama A., Kustu S. The C terminus of the alpha subunit of RNA polymerase is not essential for transcriptional activation of sigma 54 holoenzyme. J Bacteriol. 1993 Apr;175(8):2479–2482. doi: 10.1128/jb.175.8.2479-2482.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonetto M., Gribskov M., Gross C. A. The sigma 70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992 Jun;174(12):3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mecsas J., Cowing D. W., Gross C. A. Development of RNA polymerase-promoter contacts during open complex formation. J Mol Biol. 1991 Aug 5;220(3):585–597. doi: 10.1016/0022-2836(91)90102-c. [DOI] [PubMed] [Google Scholar]
- Merrick M. J. In a class of its own--the RNA polymerase sigma factor sigma 54 (sigma N). Mol Microbiol. 1993 Dec;10(5):903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- Merrick M., Chambers S. The helix-turn-helix motif of sigma 54 is involved in recognition of the -13 promoter region. J Bacteriol. 1992 Nov;174(22):7221–7226. doi: 10.1128/jb.174.22.7221-7226.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minchin S. D., Austin S., Dixon R. A. Transcriptional activation of the Klebsiella pneumoniae nifLA promoter by NTRC is face-of-the-helix dependent and the activator stabilizes the interaction of sigma 54-RNA polymerase with the promoter. EMBO J. 1989 Nov;8(11):3491–3499. doi: 10.1002/j.1460-2075.1989.tb08514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morett E., Buck M. In vivo studies on the interaction of RNA polymerase-sigma 54 with the Klebsiella pneumoniae and Rhizobium meliloti nifH promoters. The role of NifA in the formation of an open promoter complex. J Mol Biol. 1989 Nov 5;210(1):65–77. doi: 10.1016/0022-2836(89)90291-x. [DOI] [PubMed] [Google Scholar]
- Morris L., Cannon W., Claverie-Martin F., Austin S., Buck M. DNA distortion and nucleation of local DNA unwinding within sigma-54 (sigma N) holoenzyme closed promoter complexes. J Biol Chem. 1994 Apr 15;269(15):11563–11571. [PubMed] [Google Scholar]
- Ogata R., Gilbert W. Contacts between the lac repressor and the thymines in the lac operator. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4973–4976. doi: 10.1073/pnas.74.11.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popham D. L., Szeto D., Keener J., Kustu S. Function of a bacterial activator protein that binds to transcriptional enhancers. Science. 1989 Feb 3;243(4891):629–635. doi: 10.1126/science.2563595. [DOI] [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J Biol Chem. 1989 May 15;264(14):8074–8081. [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. Probing the Escherichia coli glnALG upstream activation mechanism in vivo. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8934–8938. doi: 10.1073/pnas.85.23.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. Role of eukaryotic-type functional domains found in the prokaryotic enhancer receptor factor sigma 54. Cell. 1990 Sep 7;62(5):945–954. doi: 10.1016/0092-8674(90)90269-k. [DOI] [PubMed] [Google Scholar]
- Schickor P., Metzger W., Werel W., Lederer H., Heumann H. Topography of intermediates in transcription initiation of E.coli. EMBO J. 1990 Jul;9(7):2215–2220. doi: 10.1002/j.1460-2075.1990.tb07391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa R., Chung Y. J., Rose J. P., Wang B. C. Crystal structure of bacteriophage T7 RNA polymerase at 3.3 A resolution. Nature. 1993 Aug 12;364(6438):593–599. doi: 10.1038/364593a0. [DOI] [PubMed] [Google Scholar]
- Su W., Porter S., Kustu S., Echols H. DNA-looping and enhancer activity: association between DNA-bound NtrC activator and RNA polymerase at the bacterial glnA promoter. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5504–5508. doi: 10.1073/pnas.87.14.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintut Y., Wong C., Jiang Y., Hsieh M., Gralla J. D. RNA polymerase binding using a strongly acidic hydrophobic-repeat region of sigma 54. Proc Natl Acad Sci U S A. 1994 Mar 15;91(6):2120–2124. doi: 10.1073/pnas.91.6.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C., Tintut Y., Gralla J. D. The domain structure of sigma 54 as determined by analysis of a set of deletion mutants. J Mol Biol. 1994 Feb 11;236(1):81–90. doi: 10.1006/jmbi.1994.1120. [DOI] [PubMed] [Google Scholar]