Abstract
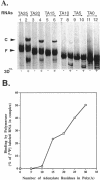
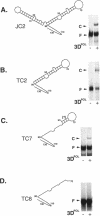
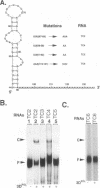
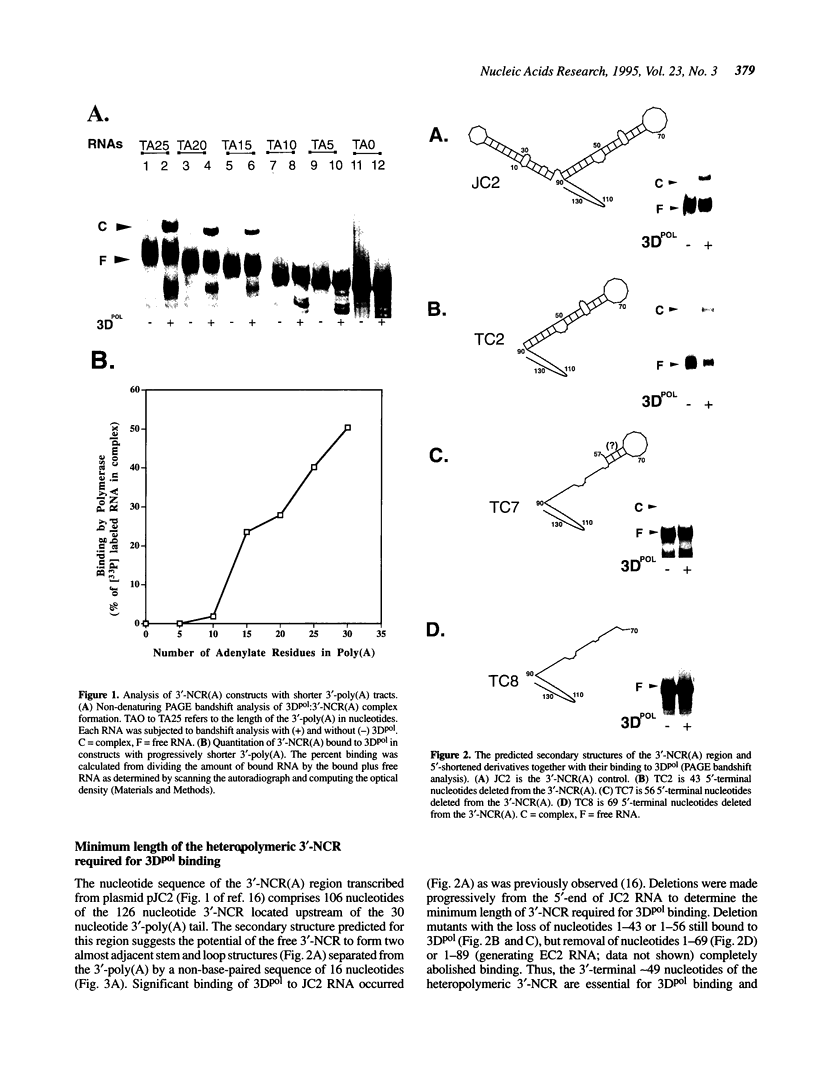
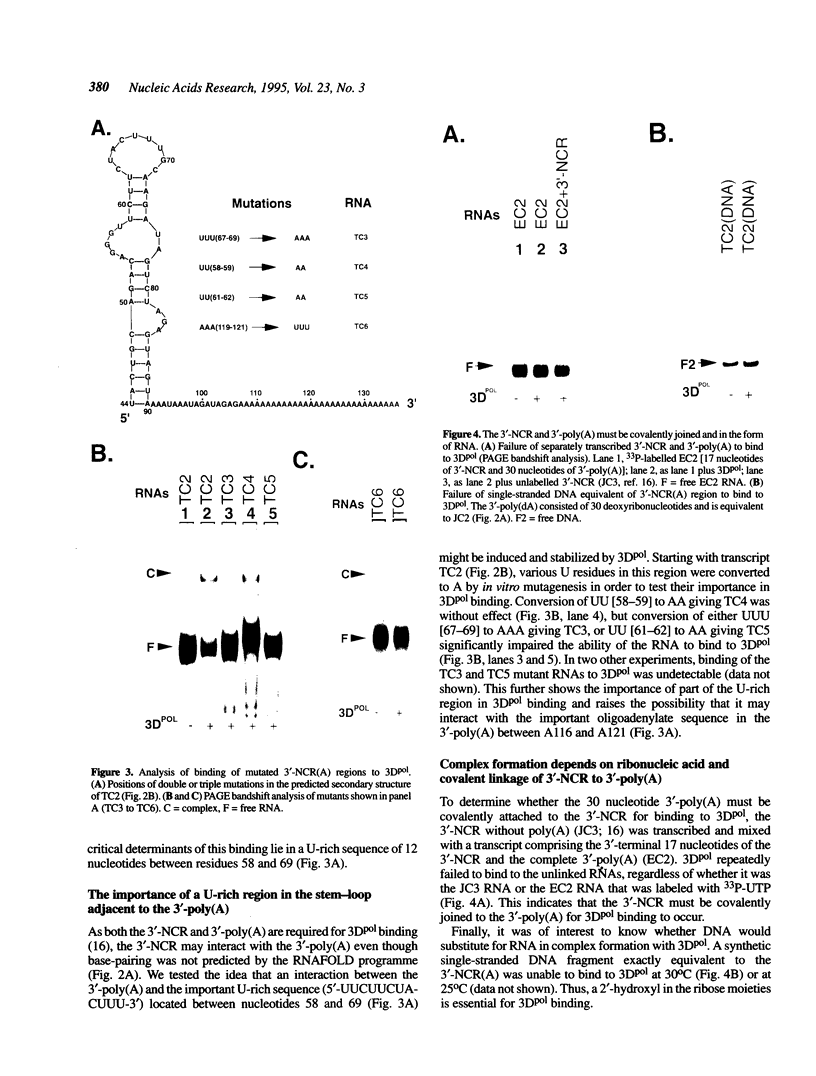
We previously showed that encephalomyocarditis (EMC) virus RNA-dependent RNA polymerase (3Dpol) binds specifically to 3'-terminal segments of EMC virus RNA. This binding, which depends on both the 3'-noncoding region (3'-NCR) and 3'-poly (A) tail [together denoted 3'-NCR(A)], may be an important step in the initiation of virus replication. In this paper, the 3'-NCR and 3'-poly(A) were separately transcribed then mixed, but no complex with 3Dpol was obtained, showing that covalent attachment of the 3'-poly(A) to the 3'-NCR is essential for complex formation. Mutational and deletion analyses localized a critical determinant of 3Dpol binding to a U-rich sequence located 38-49 nucleotides upstream of the 3'-poly(A). Similar analyses led to the identification of a sequence of A residues between positions +10 and +15 of the 3'-poly(A) which are also critical for 3Dpol binding. As U-rich and A-rich regions are important for 3Dpol binding, a speculative model is proposed in which 3Dpol induces and stabilizes the base-pairing of the 3'-poly(A) with the adjacent U-rich sequence to form an unusual pseudoknot structure to which 3Dpol binds with high affinity.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrera I., Schuppli D., Sogo J. M., Weber H. Different mechanisms of recognition of bacteriophage Q beta plus and minus strand RNAs by Q beta replicase. J Mol Biol. 1993 Jul 20;232(2):512–521. doi: 10.1006/jmbi.1993.1407. [DOI] [PubMed] [Google Scholar]
- Cui T., Sankar S., Porter A. G. Binding of encephalomyocarditis virus RNA polymerase to the 3'-noncoding region of the viral RNA is specific and requires the 3'-poly(A) tail. J Biol Chem. 1993 Dec 15;268(35):26093–26098. [PubMed] [Google Scholar]
- Eggen R., Verver J., Wellink J., Pleij K., van Kammen A., Goldbach R. Analysis of sequences involved in cowpea mosaic virus RNA replication using site-specific mutants. Virology. 1989 Dec;173(2):456–464. doi: 10.1016/0042-6822(89)90558-8. [DOI] [PubMed] [Google Scholar]
- Gargouri-Bouzid R., David C., Haenni A. L. The 3' promoter region involved in RNA synthesis directed by the turnip yellow mosaic virus genome in vitro. FEBS Lett. 1991 Dec 2;294(1-2):56–58. doi: 10.1016/0014-5793(91)81342-6. [DOI] [PubMed] [Google Scholar]
- Jacobson S. J., Konings D. A., Sarnow P. Biochemical and genetic evidence for a pseudoknot structure at the 3' terminus of the poliovirus RNA genome and its role in viral RNA amplification. J Virol. 1993 Jun;67(6):2961–2971. doi: 10.1128/jvi.67.6.2961-2971.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore J., De Geus B., Jackson R. J., Pouwels P. H., Enger-Valk B. E. Poliovirus protein 3CD is the active protease for processing of the precursor protein P1 in vitro. J Gen Virol. 1988 Jul;69(Pt 7):1627–1636. doi: 10.1099/0022-1317-69-7-1627. [DOI] [PubMed] [Google Scholar]
- Jupin I., Bouzoubaa S., Richards K., Jonard G., Guilley H. Multiplication of beet necrotic yellow vein virus RNA 3 lacking a 3' poly(A) tail is accompanied by reappearance of the poly(A) tail and a novel short U-rich tract preceding it. Virology. 1990 Sep;178(1):281–284. doi: 10.1016/0042-6822(90)90404-f. [DOI] [PubMed] [Google Scholar]
- Kuhn R. J., Hong Z., Strauss J. H. Mutagenesis of the 3' nontranslated region of Sindbis virus RNA. J Virol. 1990 Apr;64(4):1465–1476. doi: 10.1128/jvi.64.4.1465-1476.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W. A., Bujarski J. J., Dreher T. W., Hall T. C. Minus-strand initiation by brome mosaic virus replicase within the 3' tRNA-like structure of native and modified RNA templates. J Mol Biol. 1986 Feb 20;187(4):537–546. doi: 10.1016/0022-2836(86)90332-3. [DOI] [PubMed] [Google Scholar]
- Newman J. F., Piatti P. G., Gorman B. M., Burrage T. G., Ryan M. D., Flint M., Brown F. Foot-and-mouth disease virus particles contain replicase protein 3D. Proc Natl Acad Sci U S A. 1994 Jan 18;91(2):733–737. doi: 10.1073/pnas.91.2.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste M. S., Flanegan J. B. Measurement of poliovirus RNA polymerase binding to poliovirion and nonviral RNAs using a filter-binding assay. Nucleic Acids Res. 1988 Nov 11;16(21):10339–10352. doi: 10.1093/nar/16.21.10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter A. G. Picornavirus nonstructural proteins: emerging roles in virus replication and inhibition of host cell functions. J Virol. 1993 Dec;67(12):6917–6921. doi: 10.1128/jvi.67.12.6917-6921.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankar S., Porter A. G. Expression, purification, and properties of recombinant encephalomyocarditis virus RNA-dependent RNA polymerase. J Virol. 1991 Jun;65(6):2993–3000. doi: 10.1128/jvi.65.6.2993-3000.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankar S., Porter A. G. Point mutations which drastically affect the polymerization activity of encephalomyocarditis virus RNA-dependent RNA polymerase correspond to the active site of Escherichia coli DNA polymerase I. J Biol Chem. 1992 May 15;267(14):10168–10176. [PubMed] [Google Scholar]
- Sarnow P., Bernstein H. D., Baltimore D. A poliovirus temperature-sensitive RNA synthesis mutant located in a noncoding region of the genome. Proc Natl Acad Sci U S A. 1986 Feb;83(3):571–575. doi: 10.1073/pnas.83.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D. H., Baltimore D. Requirement of 3'-terminal poly(adenylic acid) for the infectivity of poliovirus RNA. Proc Natl Acad Sci U S A. 1974 Aug;71(8):2983–2987. doi: 10.1073/pnas.71.8.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamatsu N., Watanabe Y., Meshi T., Okada Y. Mutational analysis of the pseudoknot region in the 3' noncoding region of tobacco mosaic virus RNA. J Virol. 1990 Aug;64(8):3686–3693. doi: 10.1128/jvi.64.8.3686-3693.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ypma-Wong M. F., Dewalt P. G., Johnson V. H., Lamb J. G., Semler B. L. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology. 1988 Sep;166(1):265–270. doi: 10.1016/0042-6822(88)90172-9. [DOI] [PubMed] [Google Scholar]