Abstract
In studies using subtraction cloning to screen for alterations in mRNA expression in skeletal muscle from humans with Type 2 diabetes mellitus and control subjects, one of the most prominent differences was in the mRNA for elongation factor (EF)-1α. With Northern blot analysis, EF-1α expression was enhanced by 2- to 6-fold in both Types 1 and 2 human diabetics. In contrast, no changes in expression of EF-1β or -γ were noted. We observed similar results in animal models of Type 1 diabetes. EF-1α expression, but not EF-1β or -γ expression, was also enhanced in streptozotocin-induced diabetic rats, and this effect was reversed by insulin treatment. An increased level of EF-1α mRNA was also observed in nonobese diabetic mice. This unbalanced regulation of the expression of the different subunits of EF-1 may contribute to alterations not only in protein synthesis but also in other cellular events observed in the diabetic state.
Keywords: gene expression, insulin, diabetes mellitus
Uncontrolled diabetes mellitus results in complex metabolic and structural alterations, leading to abnormal carbohydrate, lipid, and protein metabolism, as well as long-term complications involving vascular tissue, kidney, and nerve. The molecular mechanisms for many of these changes are not completely understood but involve changes in both the level and posttranslational modification of a number of proteins. The decreased levels of insulin and the insulin resistance present in Types 1 and 2 diabetes lead to a reduction in total protein synthesis, as well as changes in expression of a number of insulin and/or metabolically regulated genes (1–3).
Although there is considerable information about the effects of diabetes and insulin on regulation of genes in liver and adipose tissue, much less is known about their effects on gene expression in skeletal muscle. In an attempt to identify changes in gene expression at the level of skeletal muscle in humans with diabetes mellitus, we have used the technique of subtraction cloning (4). Using this approach, we demonstrated increased expression of several mitochondrially encoded genes (5) and glycogen phosphorylase (6) in muscle from diabetic patients. We also identified Rad, a member of the Ras/GTPase proteins family, whose expression was selectively increased in muscle of some patients with Type 2 diabetes as compared with muscle from normal individuals or patients with Type 1 diabetes (7).
This report details another GTP-binding protein whose expression is increased in diabetes that was uncovered by using this technique, namely eukaryotic elongation factor (EF)-1α (also named EF-1A) (8). EF-1, the primary factor involved in the elongation reaction during protein synthesis, is composed of two distinct parts: a nucleotide-binding subunit, EF-1α, and a nucleotide exchange complex composed of the EF-1β and -γ subunits (8, 9). EF-1α binds GTP and aminoacyl-tRNA and leads to the codon-dependent placement of the aminoacyl-tRNA at the acceptor site on the ribosome. After release of EF-1α-GDP from the ribosome, the complex EF-1βγ facilitates the exchange of GTP for bound GDP. In the present study, we have characterized the effects of diabetes on the mRNA level of EF-1α, as well as EF-1β and -γ, the two subunits involved in a complex modulating the activity of EF-1α.
Research Design and Methods
Strategy of Subtraction Cloning and Identification of Diabetes-Regulated Clones.
Two subtraction libraries were prepared from human skeletal muscle cDNAs as previously described (7); one was enriched in mRNA species preferentially expressed in muscle of normal individuals and the other in mRNA species preferentially expressed in muscle of a Type 2 diabetic patient. Portions of each subtraction library were used to transform competent XL-1 blue cells (Stratagene), which were plated on a medium containing ampicillin. Individual colonies were then picked and grown in 96-well dishes. A replicate of each 96-well archive plate was made with a replicator beaded lid (FAST System, Falcon Labware, Becton Dickinson), and the archive plate was stored in 20% glycerol at −70°C. Duplicate dot-blots were prepared on Biotrans nylon membrane (ICN) from the copy plate of the normal- and diabetic-enriched colonies and hybridized with subtracted probes prepared by the PCR with the two subtraction libraries as templates and with SK and KS primers. About 5,000 colonies from these two libraries were individually screened, and 29 clones of these (about 0.7% of the two subtraction libraries) appeared to be differentially expressed and represented potential diabetes-related changes in gene expression. These were then used as probes on comparative Northern blots containing RNA samples from several normal and diabetic individuals. Fourteen clones (≈0.3% of those initially screened) were confirmed to be consistently differentially expressed, whereas the remainder showed variable patterns of expression among individuals. The cDNA inserts showing differential hybridization were sequenced by using the dideoxynucleotide chain termination method and a Sequenase kit (United States Biochemical). Both strands of the clones were sequenced by using T3 and T7 primers, as well as synthetic oligonucleotide primers deduced from the partially determined sequence and selected at convenient intervals. Sequences were aligned and analyzed by using the eugene and sam programs (Molecular Biology Computing Research Resource, Dana Farber Cancer Institute and Harvard School of Public Health).
Human and Rodent Muscle Samples.
Gastrocnemius and quadriceps human muscle samples were obtained at surgery from nondiabetic and diabetic patients undergoing above or below the knee amputation, as previously described (7). All muscle samples were dissected from the viable margin of the muscle and immediately frozen in liquid nitrogen.
Male Sprague–Dawley rats (130–180 g) were from Charles River Breeding Laboratories. Male obese hyperglycemic mice (C57BL/6J ob/ob, 6–8 wk) and their lean matched controls (ob/+) were purchased from The Jackson Laboratory. Rats and mice were fed standard rodent chow and water ad libitum. Diabetes was induced in the rats with streptozotocin (STZ) in citrate buffer, pH 4.5, administered i.p. as a single dose of 100 mg/kg body weight after an overnight fast. Diabetic rats were studied 7 d after STZ injection. The STZ-treated rats became glycosuric within 48 h, and both control and diabetic rats were maintained with free access to food and water. On the fourth day after the injection, a subgroup of diabetic animals was given twice daily s.c. injections of insulin (2.5 units of soluble + 2.5 units of NPH insulin) for the next 3–1/2 days. All three groups of animals were then killed at 1 week (4 h after the last injection of insulin).
Rats and mice were anesthetized with sodium amobarbital (15 and 20 mg/kg body weight i.p., respectively) and were used in experiments 10–15 min later, i.e., as soon as anesthesia was assured by loss of pedal and corneal reflexes. Hindlimb muscles were quickly excised and frozen in liquid nitrogen.
Plasma glucose levels were determined with a Beckman Glucose Analyzer on blood samples obtained when the animals were killed. Insulin was determined by a standard RIA, as previously described (10).
Northern Blot Analysis.
Frozen human or rodent muscles were ground in liquid nitrogen into a fine powder, and total RNA was extracted with the RNAzol method (Biotecx Laboratories, Houston). Total RNA was then fractionated on a 1% agarose-formaldehyde gel, transferred onto nylon membrane (ICN) in 20 × SSC for 20 h, and crosslinked with UV light. The cDNA inserts used to prepare probes were purified on agarose gels, labeled with 32P-deoxycytidine 5′-triphosphate with the Amersham multiprime DNA labeling system, and purified by using an Elutip (Schleicher & Schuell). RNA blots were hybridized at 65°C in 50 mM Pipes (pH 6.5)/100 mM NaCl/50 mM sodium phosphate/1 mM EDTA/5% SDS/100 μg/ml salmon sperm DNA for at least 24 h (4 × 106 cpm/ml). Filters were washed with 5% SDS/0.5 × SSC at room temperature for 15 min, twice at 65°C for 15 min, and then for 15 min with 0.1% SDS/0.2 × SSC at 65°C. Filters were exposed on Kodak X-Omat film with an intensifying screen at −70°C. To minimize variations between blots, analysis was carried out on multiple autoradiograms obtained from multiple electrophoretic gel runs of every sample. For all Northern blots using human RNA, a single sample from one normal individual was used as an internal control for normalization of data.
The cDNA probes for human EF 1-β (entire coding region) and EF 1-γ (complete coding sequence except the nucleotides coding for the four NH2-terminal amino acids) were kindly provided into the plasmid PUC120 by William Möller, Sylvius Laboratory, Leiden, The Netherlands (11, 12).
Results
Characterization by Subtraction Cloning of the Overexpression of EF-1α mRNA in Diabetic Human Muscle.
To identify changes in gene expression that might contribute to the pathogenesis of diabetes mellitus, subtraction libraries made from normal and Type 2 diabetic human skeletal muscle were screened by using subtractive probes (see Research Design and Methods) (4, 7). The presumptive differentially expressed clones were then used as probes in comparative Northern blot analysis of total RNA extracted from human skeletal muscle of controls, Types 1 and 2 diabetics. Among several diabetes-regulated mRNAs, we focused our attention on clone B10D6 (4), whose expression was markedly increased in diabetic muscle as compared with normal muscle. The cDNA insert of clone B10D6 was partially sequenced, and its sequence matched 99% to the sequence of human EF-1α mRNA by comparison in GenBank. This cDNA insert consisted of 1,254 nucleotides, starting at nucleotide 461 of EF-1α cDNA (13), including 989 nucleotides of coding region followed by 265 nucleotides of 3′-untranslated region. Thus, we concluded that clone B10D6 encoded human muscle EF-1α.
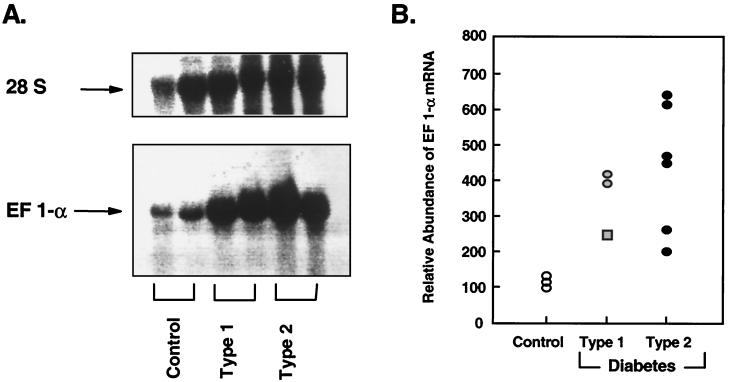
On Northern blot analysis of human skeletal muscle RNA, the major species of mRNA detected with the labeled insert B10D6 was 1.8 kb, consistent with the size of human EF-1α mRNA (13, 14). Furthermore, in this screening blot, EF-1α mRNA was markedly over-expressed in muscle samples of Types 1 and 2 diabetics compared with those from nondiabetic individuals (Fig 1A). Comparison of the levels of EF-1α mRNA in three nondiabetic, three Type 1, and six Type 2 diabetic muscle samples revealed a 2.5- to 4.2-fold increase in expression of EF-1α in Type 1 diabetes and a 2.0- to 6.4-fold increase in muscle of Type 2 diabetics (Fig. 1B). Interestingly, among the three Type 1 diabetics studied, we observed the lowest level of overexpression of EF-1α mRNA in the muscle sample from a patient who had pancreatic transplantation and subsequent near-normalization of blood glucose (Fig. 1B).
Figure 1.
Northern blot analysis of human skeletal muscle EF-1α mRNA. (A) Representative Northern blot of human skeletal muscle EF 1-α expression. Twenty micrograms of total RNA was extracted from two nondiabetic, two Type 1, and two Type 2 diabetic muscle samples, hybridized with labeled 1.2 kb cDNA insert of clone B10D6 (corresponding to human EF-1α cDNA), and autoradiographed. This blot shows a typical experiment. Upper shows that the amount of RNA loaded on the gels was equivalent in the different individuals based on reprobing of the blots with an oligonucleotide probe to human 28S RNA. (B) Relative abundance of the EF1-α mRNA. Twenty micrograms of total RNA was extracted from three nondiabetic, three Type 1, and six Type 2 diabetic muscle samples and hybridized with 1.2 kb cDNA insert of clone B10D6. Autoradiograms from several Northern blot analyses were quantified by scanning densitometry. The data were normalized to the integrated intensities of signals obtained from one nondiabetic RNA sample used as an internal standard. Each data point represents the mean of at least three determinations per RNA sample, except for the square data point, which represents one determination on a muscle sample of a Type 1 diabetic patient who had a pancreatic transplant several months before this study.
Effect of Diabetes on EF-1α, -β, and -γ mRNA Expression in Human Skeletal Muscle.
EF-1α is a GTP-binding protein that plays a critical role in the early steps of protein synthesis and ribosome formation (9). At each round of elongation, EF-1α is responsible for the binding of aminoacyl-tRNA to the ribosome acceptor site, GTP is hydrolyzed, and EF-1α recycles via a process in which EF-1βγ catalyzes the exchange of GDP for GTP. Modification of either the guanine nucleotide-binding subunit EF-1α or the guanine exchange subunits EF-1β/γ could affect the rate of formation of ternary EF-1α-GTP-aminoacyl-tRNA complexes and thereby alter the rate of elongation (15). We therefore measured the effect of diabetes on EF-1α, -β, and -γ mRNA expression in human skeletal muscle.
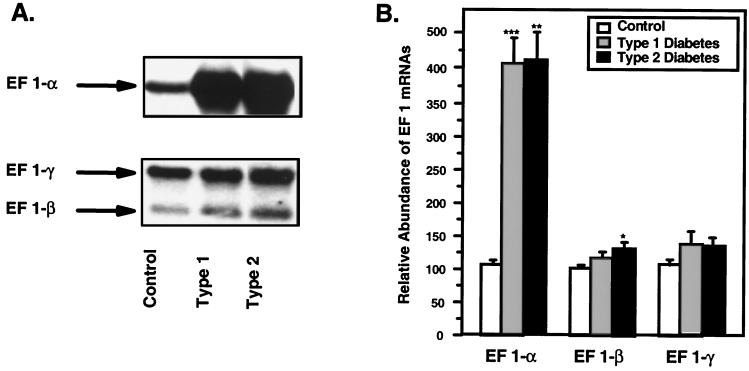
As shown in Fig. 2A, a single hybridizing mRNA species specific to each cDNA was expressed in human muscle, and the size of the mRNA species was in accordance with that in the literature (12–14). With the same Northern blots that revealed a ≈4-fold increase in EF-1α mRNA in Types 1 and 2 diabetic muscle samples, there was no significant change in EF-1γ mRNA in either Type 1 or 2 diabetics (Fig. 2B) and only a very small (32%), marginally significant increase in EF-1β in Type 2 diabetics. Thus, the effect of diabetes was to produce a relatively selective increase in EF-1α mRNA.
Figure 2.
Northern blot analysis of human skeletal muscle EF-1α, EF-1β and -γ mRNAs. (A) Representative Northern blot of human skeletal muscle EF 1-α, -β, and -γ expression. Twenty micrograms of total RNA was extracted from nondiabetic, Type 1, and Type 2 diabetic muscle samples, hybridized respectively with the cDNA insert from clone B10D6, EF 1-β, and -γ, and autoradiographed. (B) Relative abundance of the EF-1α, -β, and -γ mRNAs. Twenty micrograms of total RNA was extracted from three nondiabetic, two Type 1, and six Type 2 diabetic muscle samples and hybridized with EF-1α (B10D6), -β, and -γ cDNA probes. Autoradiograms from several Northern blot analyses were quantified separately by scanning densitometry, and the data were normalized to the integrated intensities of signals obtained from the same nondiabetic RNA sample used on all blots. The histograms represent the mean of the normalized values ± SEM. Statistical comparisons were made (against control values in each case) by using the unpaired Student's t test (***, P < 0.0001; **, P < 0.002; *, P < 0.05).
EF-1α, -β, and -γ mRNA Expression in Animal Models of Diabetes.
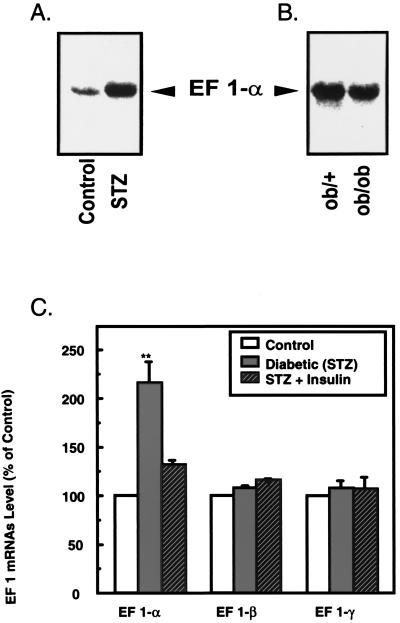
To further examine the regulation of EF-1α mRNA in diabetes, we studied the expression of EF-1 in three different animal models of diabetes: the STZ-induced diabetic rat, the nonobese diabetic mouse, and the ob/ob mouse (16). Table 1 summarizes the body weight, plasma glucose, and serum insulin levels of the STZ rats and ob/ob mice used in this study. As shown in Fig. 3 A and C, EF-1α mRNA level was increased 2.2-fold in STZ-induced diabetic rats, whereas there was no significant change in EF-1β and -γ mRNA expression compared with nondiabetic muscle. When this well characterized model of insulin deficiency Type 1 diabetes was treated with insulin, the amount of EF-1α mRNA decreased to values just above that in controls, whereas EF-1β and -γ mRNA expression was still unchanged (Fig. 3C).
Table 1.
Characteristics of STZ and ob/ob diabetic animals: Effect of diabetes mellitus and insulin therapy on body weight, plasma glucose, and serum insulin concentrations of rats and mice studied
| Body weight, g | Plasma glucose, mg/dl | Serum insulin, microunits/ml | |
|---|---|---|---|
| Control rats | 193 ± 4 | 182 ± 13 | 27 ± 5 |
| STZ-induced diabetic rats | 151 ± 7 | 513 ± 23 | 5.3 ± 1.5 |
| STZ rats insulin-treated | 183 ± 10 | 66 ± 13 | >2,000 |
| Lean mice ob/+ | 18.2 ± 0.4 | 117 ± 17 | 39 ± 7 |
| Lean mice ob/ob | 34.2 ± 0.4 | 390 ± 35 | 388 ± 89 |
Male Sprague-Dawley rats were either studied as controls or injected with streptozotocin, without or with subsequent insulin treatment, as described in Research Design and Methods. All three groups of animals were then killed, and total RNA was extracted from hindlimb muscles as described in Research Design and Methods and analyzed by Northern blot as described in Fig. 3. All plasma measurements were made just before death. The data represent means ± SEM from three separate experiments, each with at least two animals per group.
Figure 3.
Comparative Northern blot analysis of EF-1α, -β, and -γ expression in hindlimb muscle from animal models of diabetes mellitus. (A) Representative analysis of EF-1α mRNA level in skeletal muscle of control and STZ-induced diabetic rats. (B) Representative analysis of EF-1α mRNA level in lean and genetically obese (ob/ob) mice. (C) Effect of diabetes mellitus and insulin therapy on EF-1 subunit expression in rat skeletal muscle. Twenty micrograms of total RNA was extracted from groups of control, diabetic, and insulin-treated diabetic rats (described in Table 1) and hybridized with random prime labeled cDNA probes for EF-1α (B10D6), -β, and -γ. Analysis of ribosomal RNA indicated that equal amounts of RNA were loaded onto each lane (not shown). Specific mRNAs were quantified from autoradiograms of several Northern blot analysis by laser-scanning densitometry relative to the amount in controls, which was assigned a value of 100. Data represent means ± SD from two or three separate experiments, each with at least two animals per group. Statistical comparisons were made by using the unpaired Student's t test (**, P < 0.002).
We also observed a 2.4-fold overexpression of EF-1α in muscle of nonobese diabetic mice, another model of Type 1 diabetes, compared with muscle of their nondiabetic controls (data not shown). Not surprisingly, skeletal muscle from ob/ob mice, a model of obesity associated with diabetes, did not show an increase in EF-1α mRNA expression relative to control ob/+ mice (Fig. 3B). In comparison to the other rodent model of diabetes, the ob/ob mouse is much less hyperglycemic and more hyperinsulinemic, as well as being obese and insulin-resistant; moreover, this model is because of a specific gene defect and may not be reflective of a more general diabetic condition (16).
These results suggest that poorly controlled diabetes results in specific increases in skeletal muscle EF-1α mRNA that are not observed in mild hyperglycemic states or when treatment of diabetes restores this level to approximately that of nondiabetic muscle (i.e., after insulin treatment or pancreatic transplant).
Discussion
By screening subtraction libraries prepared from skeletal muscle of normal and diabetic humans, we have begun to identify diabetes-associated changes in gene expression and to examine which of these changes may be secondary to the metabolic abnormalities of diabetes (i.e., present in both Types 1 and 2 diabetes) versus primary or unique defects related to the insulin resistance of Type 2 diabetes (4–7). Using this approach, we demonstrated increased expression of four different mitochondrially encoded genes in diabetes [cytochrome oxidase I, cytochrome oxidase III, NADH dehydrogenase IV, and 12s rRNA] (5), glycogen phosphorylase (6), and a Ras-related protein, Rad (7, 17). We demonstrated that Rad may participate in insulin resistance by serving as a negative regulator of glucose transport (18), whereas the others could play a role in some of the other altered glucose metabolic responses observed in skeletal muscle of diabetic patients. In the present work, we have identified a marked increase in expression of EF-1α and an imbalance with other subunits of the EF-1 complex in diabetic skeletal muscle that may contribute to the altered protein synthesis observed in this disease.
The superfamily of guanine nucleotide-binding proteins consists of three major families, including Ras and Ras-related small GTP-binding proteins, the heterotrimeric GTP-binding proteins (G proteins) involved in signaling (19), the initiation and elongation factors involved in protein synthesis, and the tubulins (20). Considerable evidence suggests that Ras and Ras-related proteins are involved in the insulin-signaling pathway (18, 21). We (22) and others (23) have suggested a functional “crosstalk” between the insulin receptor and the inhibitory guanine nucleotide-binding protein (Gi), and abnormal expression and/or function of G protein subunits have been described in diabetic states by several groups (24, 25). Moreover, Moxham and Malbon (26) have shown that Gi(α2) deficiency in transgenic mice creates a model for the insulin resistance characteristic of non-insulin-dependent diabetes mellitus. The possibility that the elongation factors could also participate in insulin signaling or contribute to the pathogenesis of diabetes now also needs to be added to this list of GTP-binding proteins linked to these processes.
One of the principal end points of insulin action is the stimulation of protein synthesis. Insulin induces both a general increase in the rate of mRNA transcription and preferential increases in the transcription of specific mRNAs. Insulin also appears to regulate both the initiation and elongation phases of translation (3, 27). Insulin deficiency has been shown to inhibit protein synthesis in adipose tissue, liver, heart, and skeletal muscle, whereas insulin administration to diabetic animals can readily reverse inhibition in these tissues. The exact nature of these changes depends on the duration of the hormonal change. In rats, short-term (2-day) diabetes results in reduction in the level of RNA in all muscles and impairment in peptide-chain initiation. Long-term (7-day) diabetes causes no further reduction in RNA but results in the development of an additional impairment to protein synthesis (28, 29). The decrease in protein synthesis appears to be caused by a decreased rate of mRNA translation mediated primarily by impairment of peptide-chain elongation and termination (2, 27, 30). Thus, elongation factors are potential targets for the mechanism(s) by which insulin alters translational activity.
In mammalian cells, peptide-chain elongation requires two elongation factors, multimeric EF-1 and monomeric EF-2, both phosphoproteins, which are the primary sites of regulation of protein translation (27). It is not currently clear to what extent the rate of elongation limits the overall rate of protein synthesis, because few data on the control of elongation, especially in tissues that are physiological targets of insulin, are available. Levenson et al. have shown that insulin can rapidly stimulate the biosynthesis of eukaryotic EF-2 in NIH 3T3 cells overexpressing human insulin receptors (31), and this insulin-induced enhanced EF-2 synthesis could conceivably affect overall rates of peptide chain elongation. In addition, insulin stimulates the dephosphorylation of EF-2 probably via inhibition of EF-2 kinase in a rapamycin-sensitive fashion (32, 33).
In the case of EF-1, the entire complex of EF-1αβγ is required for protein synthesis, because the α subunit binds to both GTP and aminoacyl-tRNA, whereas the β and γ subunits exchange the GDP for GTP (15). EF-1α expression is subject to regulation at a number of different levels that may involve several mechanisms (34). It has been reported that mammalian cells treated with phorbol esters and vitamin A have a several-fold increase in the levels of EF-1α expression that may render them more susceptible to transformation (35, 36). On the other hand, expression of the EF-1α gene decreases as mouse and human fibroblasts in culture become senescent, whereas forced expression of EF-1α in Drosophila melanogaster prolongs the life span of the fly (34). Thus, this component of the protein synthesis apparatus seems to be involved in the control of cell proliferation, and it is of critical importance to delineate the regulatory mechanisms that control its level of expression.
In cultured cells, mRNA levels for EF-1α, -β, and -γ increase in parallel, suggesting coordinate regulation of the expression of these three genes (12), and recently coordinate transcriptional induction of the three EF-1 polypeptide subunits has been observed in response to pathophysiologic concentrations of homocysteine (37). Insulin has also been shown to stimulate the phosphorylation of EF-1α in 3T3-L1 cells in culture, probably via S6 kinase, which is associated with enhanced elongation activity (38). The present study shows, to our knowledge for the first time, unbalanced regulation of the expression of mRNAs encoding for EF-1 subunits as a result of the diabetic state. Thus, in skeletal muscle of two animal models of diabetes as well as muscle of diabetic humans, the expression level of EF-1α mRNA is dramatically increased, whereas the levels of EF-1β and -γ mRNAs are unchanged. This dysregulation of subunit expression and balance, coupled with altered states of phosphorylation, may be of significance, because EF-1α is a multifunctional protein that affects a wide variety of cellular processes in addition to its critical role in the elongation step of protein synthesis.
EF-1α is a component of the valyl–tRNA synthase complex (39), is associated with the mRNA ribonuclease protein complex (40), and is a participant in protein degradation (15). Through these reactions, EF-1α can modify the aminoacyl-tRNA content and regulate cellular proteins by modifying rates of RNA processing and protein turnover. EF-1α also functions in the reorganization of the cell cytoskeleton, has been identified as an actin-binding protein in Dictyostelium (41), and is a component of the spindle-organizing center in mammalian cells (42). Recent studies have described an interaction between EF-1α and calmodulin (43) and between EF-1α and microtubules (44). It has been proposed that the association with GTP may serve as a switch that regulates the aminoacyl-tRNA addition to the ribosomes, whereas association with calmodulin in response to calcium may determine which of the other activities are activated (43). Because most cytoplasmic mRNA is anchored to either microtubules or actin filaments in eukaryotic cells, it has been suggested that the ability of EF-1α to alter the assembly and geometry of these polymers may be a crucial property of EF-1α in regulating the efficiency of polypeptide elongation (45). EF-1α may also help integrate signal transduction with the cytoskeleton and is an activator of phosphatidylinositol-4 kinase (46). Recently, Edmonds et al. have shown that EF-1α is an overexpressed actin-binding protein in metastatic rat mammary adenocarcinoma and have also confirmed in their study that increased EF-1α mRNA translated to increased EF-1α protein (47). This broad diversity of functions may also explain why EF-1α is such a well-conserved and abundant protein, constituting around 3% of the total cytoplasmic protein content in some eukaryotic cells and therefore present in excess compared with other translational factors, including EF-1βγ subunits and ribosomes.
What is not clear from our studies is why EF-1α is increased, whereas the other subunits are not. One possibility is that the up-regulation of EF-1 α mRNA is somehow a compensatory mechanism in an attempt to reverse the decreased protein synthesis observed in diabetes. Alternatively, the increase could reflect common regulatory elements in the promoters of several GTP-binding proteins that are activated in the diabetic state. Further study will be required to dissect these possibilities. In any case, this study indicates that EF-1α is a diabetes-regulated gene that may coordinate protein synthetic activity with other cellular events and thus may represent an interesting target for the search for defective mechanisms common to both Types 1 and 2 diabetes.
Acknowledgments
We are grateful to Dr. M. Saad for assistance with experiments using animal muscle tissue and to M. Petruzzelli for technical assistance with animal care. We thank Dr. W. Möller and J. Sanders for EF-1β and -γ cDNAs. This work was supported by grants from the Juvenile Diabetes Foundation (C.R.), the Association de Langue Française pour l'Etude du Diabete et des Maladies Metaboliques (C.R.), the Joslin National Institutes of Health (NIH) Diabetes Endocrinology Research Care P30 DK 36836, and NIH Grant DK 33201 (C.R.K.).
Abbreviations
- EF
elongation factor
- STZ
streptozotocin
References
- 1.Miers W R, Barrett E J. J Basic Clin Physiol Pharmacol. 1998;9:235–253. doi: 10.1515/jbcpp.1998.9.2-4.235. [DOI] [PubMed] [Google Scholar]
- 2.Dillmann W H. Diabetes Metab Rev. 1988;4:789–797. doi: 10.1002/dmr.5610040807. [DOI] [PubMed] [Google Scholar]
- 3.Kimball S R, Vary T C, Jefferson L S. Annu Rev Physiol. 1994;56:321–348. doi: 10.1146/annurev.ph.56.030194.001541. [DOI] [PubMed] [Google Scholar]
- 4.Kahn C R, Reynet C. In: Molecular Biology of Diabetes. Draznin B, LeRoith D, editors. II. Totowa, NJ: Humana; 1994. pp. 51–77. [Google Scholar]
- 5.Antonetti D A, Reynet C, Kahn C R. J Clin Invest. 1995;95:1383–1388. doi: 10.1172/JCI117790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reynet C, Kahn C R, Loeken M R. Diabetologia. 1996;39:183–189. doi: 10.1007/BF00403961. [DOI] [PubMed] [Google Scholar]
- 7.Reynet C, Kahn C R. Science. 1993;262:1441–1444. doi: 10.1126/science.8248782. [DOI] [PubMed] [Google Scholar]
- 8.Merrick W C, Hershey J W B. In: Translational Control. Hershey J B W, Mathews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 31–69. [Google Scholar]
- 9.Ryazanv A G, Rudkin B B, Spirin A S. FEBS Lett. 1991;285:170–175. doi: 10.1016/0014-5793(91)80798-8. [DOI] [PubMed] [Google Scholar]
- 10.Saad M J A, Araki E, Miralpeix M, Rothenberg P L, White M F, Kahn C R. J Clin Invest. 1992;90:1839–1849. doi: 10.1172/JCI116060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanders J, Maassen J A, Amons R, Möller W. Nucleic Acids Res. 1991;19:45. doi: 10.1093/nar/19.16.4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanders J, Maassen J A, Möller W. Nucleic Acids Res. 1992;20:5907–5910. doi: 10.1093/nar/20.22.5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uetsuki T, Naito A, Nagata S, Kaziro Y. J Biol Chem. 1988;264:5791–5798. [PubMed] [Google Scholar]
- 14.Opdenakker G, Cazeba-Arvelaiz Y, Fiten P, Dijkmans R, Van Damme J, Volckaert G, Billiau A, Van Elsen A, Van der Schueren B, Van den Berghe H, et al. Hum Genet. 1987;75:339–344. doi: 10.1007/BF00284104. [DOI] [PubMed] [Google Scholar]
- 15.Merrick W C. Microbiol Rev. 1992;56:291–315. doi: 10.1128/mr.56.2.291-315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mordes J P, Rossini A A. In: Joslin's Diabetes Mellitus. Marble A, Krall L P, Bradley R F, Christlieb H R, Soeldner J S, editors. Philadelphia: Lea & Febiger; 1985. pp. 110–137. [Google Scholar]
- 17.Andreelli F, Vidal H, Khalfallah Y, Riou J P, Laville M. Diabetes Suppl. 1. 1998. , 1701. [Google Scholar]
- 18.Moyers J S, Bilan P J, Reynet C, Kahn C R. J Biol Chem. 1996;271:23111–23116. doi: 10.1074/jbc.271.38.23111. [DOI] [PubMed] [Google Scholar]
- 19.Bourne H R, Sanders D A, McCormick F. Nature (London) 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 20.Kaziro Y, Itoh H, Kozasa T, Nakafuku M, Satoh T. Annu Rev Biochem. 1991;60:349–400. doi: 10.1146/annurev.bi.60.070191.002025. [DOI] [PubMed] [Google Scholar]
- 21.Cheatham B, Kahn C R. Endocr Rev. 1995;16:117–142. doi: 10.1210/edrv-16-2-117. [DOI] [PubMed] [Google Scholar]
- 22.Rothenberg P L, Kahn C R. J Biol Chem. 1988;263:15546–15552. [PubMed] [Google Scholar]
- 23.Caro J F, Raju M S, Caro M, Lynch C J, Poulos J, Exton J H, Thakkar J K. J Cell Biochem. 1994;54:309–319. doi: 10.1002/jcb.240540307. [DOI] [PubMed] [Google Scholar]
- 24.Gawler D, Milligan G, Spiegel A S, Unson C G, Houslay M D. Nature (London) 1987;327:229–232. doi: 10.1038/327229a0. [DOI] [PubMed] [Google Scholar]
- 25.Spiegel A M, Shenker A, Weinstein L S. Endocr Rev. 1992;13:536–565. doi: 10.1210/edrv-13-3-536. [DOI] [PubMed] [Google Scholar]
- 26.Moxham C M, Malbon C C. Nature (London) 1996;379:840–844. doi: 10.1038/379840a0. [DOI] [PubMed] [Google Scholar]
- 27.Proud C G, Denton R M. Biochem J. 1997;328:329–341. doi: 10.1042/bj3280329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flaim K E, Copenhaver M E, Jefferson L S. Am J Physiol. 1980;239:E88–E95. doi: 10.1152/ajpendo.1980.239.1.E88. [DOI] [PubMed] [Google Scholar]
- 29.Kent J D, Kimball S R, Jefferson L S. Am J Physiol. 1991;260:C409–C416. doi: 10.1152/ajpcell.1991.260.3.C409. [DOI] [PubMed] [Google Scholar]
- 30.Yoshizawa F, Tonouchi A, Miura Y, Yagasaki K, Funabiki R. Biosci Biotechnol Biochem. 1995;59:348–349. doi: 10.1271/bbb.59.348. [DOI] [PubMed] [Google Scholar]
- 31.Levenson R M, Nairn A C, Blackshear P J. J Biol Chem. 1989;264:11904–11911. [PubMed] [Google Scholar]
- 32.Wang L, Wang X, Proud C G. Am J Physiol. 2000;278:H1056–H1068. doi: 10.1152/ajpheart.2000.278.4.H1056. [DOI] [PubMed] [Google Scholar]
- 33.Diggle T A, Redpath N T, Heesom K J, Denton R M. Biochem J. 1998;336:525–529. doi: 10.1042/bj3360525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riis B, Rattan S I, Clark B F, Merrick W C. Trends Biochem Sci. 1990;15:420–424. doi: 10.1016/0968-0004(90)90279-k. [DOI] [PubMed] [Google Scholar]
- 35.Ann D K, Wu M, M J, Huang T, Carlson D M, Wu R. J Biol Chem. 1988;263:3546–3549. [PubMed] [Google Scholar]
- 36.Tatsuka M, Mitsui H, Wada M, Nagata A, Nojima H, Okayama H. Nature (London) 1992;359:333–336. doi: 10.1038/359333a0. [DOI] [PubMed] [Google Scholar]
- 37.Chacko G, Ling Q, Hajjar K A. J Biol Chem. 1998;273:19840–19846. doi: 10.1074/jbc.273.31.19840. [DOI] [PubMed] [Google Scholar]
- 38.Chang Y W, Traugh J A. Eur J Biochem. 1998;251:201–207. doi: 10.1046/j.1432-1327.1998.2510201.x. [DOI] [PubMed] [Google Scholar]
- 39.Bec G, Kerjan P, Zha X D, Waller J P. J Biol Chem. 1989;264:21131–21137. [PubMed] [Google Scholar]
- 40.Slobin L I. In: Translation in Eukaryotes. Trachsel H, editor. Boca Raton, FL: CRC; 1991. pp. 149–175. [Google Scholar]
- 41.Yang F, Demma M, Warren V, Dharmawardhane S, Condeelis J. Nature (London) 1990;347:494–496. doi: 10.1038/347494a0. [DOI] [PubMed] [Google Scholar]
- 42.Marchesi V T, Ngo N. Proc Natl Acad Sci USA. 1993;90:3028–3032. doi: 10.1073/pnas.90.7.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaur K J, Ruben L. J Biol Chem. 1994;269:23045–23050. [PubMed] [Google Scholar]
- 44.Shiina N, Gotoh Y, Kubomura N, Iwamatsu A, Nishida E. Science. 1994;266:282–285. doi: 10.1126/science.7939665. [DOI] [PubMed] [Google Scholar]
- 45.Condeelis J. Trends Biochem Sci. 1995;20:169–170. doi: 10.1016/s0968-0004(00)88998-7. [DOI] [PubMed] [Google Scholar]
- 46.Yang W, Boss W F. J Biol Chem. 1994;269:3852–3857. [PubMed] [Google Scholar]
- 47.Edmonds B T, Wyckoff J, Yeung Y G, Wang Y, Stanley E R, Jones J, Segall J, Condeelis J. J Cell Sci. 1996;109:2705–2714. doi: 10.1242/jcs.109.11.2705. [DOI] [PubMed] [Google Scholar]