Abstract
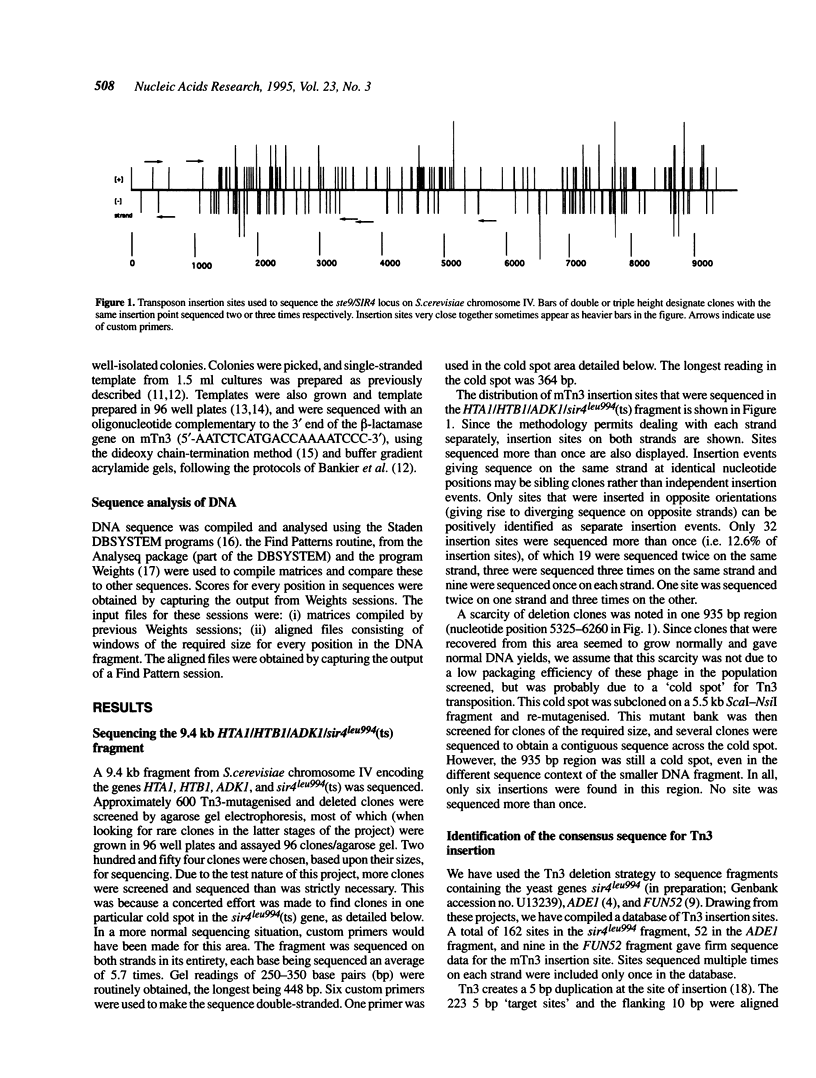
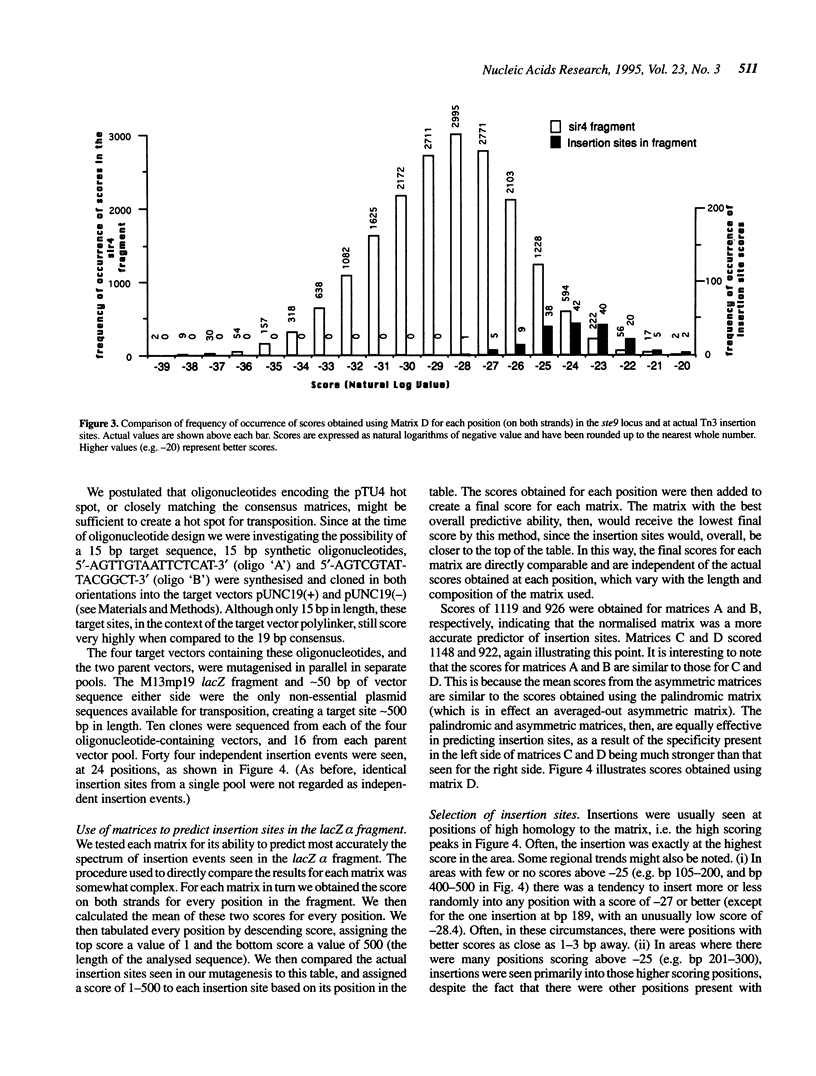
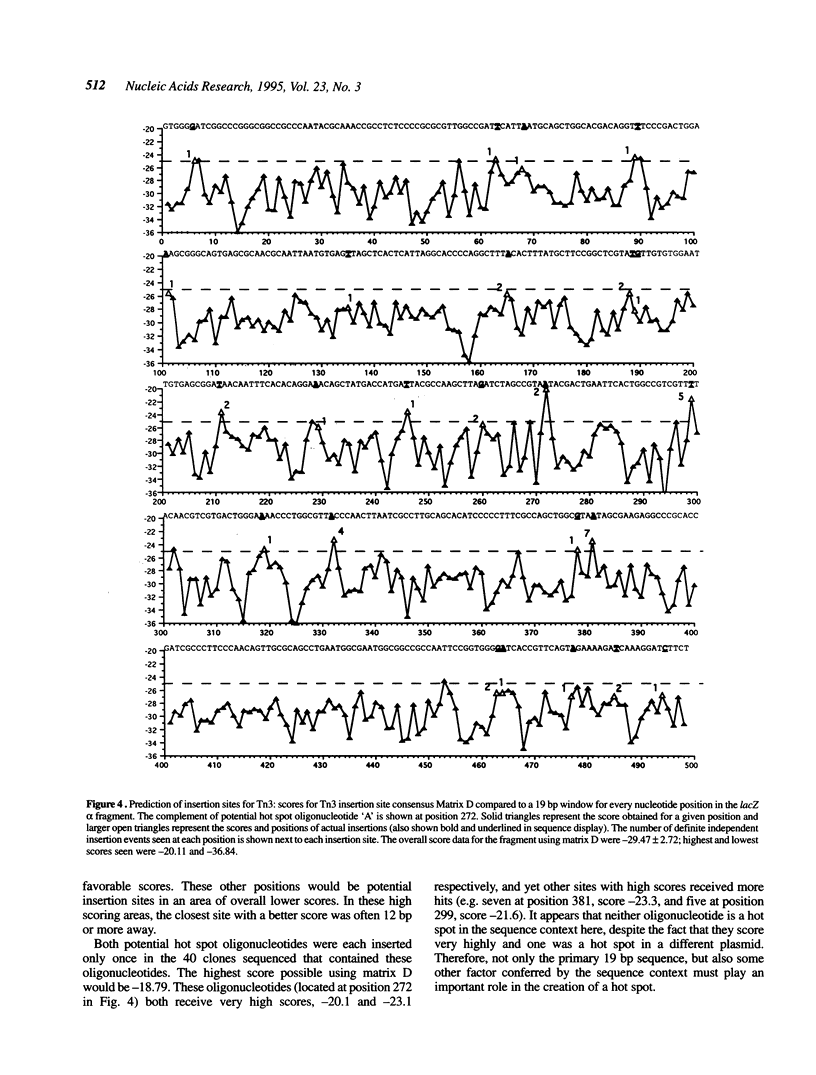
The Tn3-deletion method [Davies and Hutchison, Nucleic Acids Res. 19, 5731-5738, (1991)] was used to sequence a 9.4 kb DNA fragment. Transpositional 'warm' spots were not a limiting factor but a 935 bp 'cold' spot was completed using a synthetic oligonucleotide primer. Two hundred and twenty three miniTn3 insertion sites from three sequencing projects were aligned and a 19 bp asymmetric consensus site was identified. There is no absolute sequence requirement at any position in this consensus, so insertion occurs promiscuously (approximately 37% of sites are potential targets). In our sequencing projects, multiply targeted sites always closely matched the consensus, although not all close matches were targeted frequently. The 935 bp cold spot showed no unusual features when analysed with the consensus sequence. The consensus can be used to accurately predict likely insertion sites in a new sequence. Synthetic oligonucleotides based on the consensus and a known hot spot for Tn3 were mutagenised. These sequences were not hot spots in our vectors, suggesting that the primary sequence alone is not sufficient to create an insertional hot spot. We conclude that some other factor, such as DNA secondary structure, also plays an important role in target site selection for the transposon Tn3.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmed A. A rapid procedure for DNA sequencing using transposon-promoted deletions in Escherichia coli. Gene. 1985;39(2-3):305–310. doi: 10.1016/0378-1119(85)90328-2. [DOI] [PubMed] [Google Scholar]
- Bankier A. T., Weston K. M., Barrell B. G. Random cloning and sequencing by the M13/dideoxynucleotide chain termination method. Methods Enzymol. 1987;155:51–93. doi: 10.1016/0076-6879(87)55009-1. [DOI] [PubMed] [Google Scholar]
- Barton A. B., Davies C. J., Hutchison C. A., 3rd, Kaback D. B. Cloning of chromosome I DNA from Saccharomyces cerevisiae: analysis of the FUN52 gene, whose product has homology to protein kinases. Gene. 1992 Aug 1;117(1):137–140. doi: 10.1016/0378-1119(92)90502-g. [DOI] [PubMed] [Google Scholar]
- Bender J., Kleckner N. Tn10 insertion specificity is strongly dependent upon sequences immediately adjacent to the target-site consensus sequence. Proc Natl Acad Sci U S A. 1992 Sep 1;89(17):7996–8000. doi: 10.1073/pnas.89.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Casadaban M. J., Chou J., Tu C. P. Studies of the specificity and control of transposition of the Tn3 element. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1247–1255. doi: 10.1101/sqb.1979.043.01.141. [DOI] [PubMed] [Google Scholar]
- Craig N. L. Tn7: a target site-specific transposon. Mol Microbiol. 1991 Nov;5(11):2569–2573. doi: 10.1111/j.1365-2958.1991.tb01964.x. [DOI] [PubMed] [Google Scholar]
- Davies C. J., Hutchison C. A., 3rd A directed DNA sequencing strategy based upon Tn3 transposon mutagenesis: application to the ADE1 locus on Saccharomyces cerevisiae chromosome I. Nucleic Acids Res. 1991 Oct 25;19(20):5731–5738. doi: 10.1093/nar/19.20.5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon L. P., Graham I. R., Griffiths A. D., Eperon I. C. Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell. 1988 Jul 29;54(3):393–401. doi: 10.1016/0092-8674(88)90202-4. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H. Mutants of Saccharomyces cerevisiae unresponsive to cell division control by polypeptide mating hormone. J Cell Biol. 1980 Jun;85(3):811–822. doi: 10.1083/jcb.85.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C. J., Heffron F., Twu J. S., Schloemer R. H., Lee C. H. Analysis of Tn3 sequences required for transposition and immunity. Gene. 1986;41(1):23–31. doi: 10.1016/0378-1119(86)90263-5. [DOI] [PubMed] [Google Scholar]
- Hutchison C. A., 3rd, Swanstrom R., Loeb D. D. Complete mutagenesis of protein coding domains. Methods Enzymol. 1991;202:356–390. doi: 10.1016/0076-6879(91)02019-6. [DOI] [PubMed] [Google Scholar]
- Lodge J. K., Weston-Hafer K., Berg D. E. Transposon Tn5 target specificity: preference for insertion at G/C pairs. Genetics. 1988 Nov;120(3):645–650. doi: 10.1093/genetics/120.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T., Amemura-Maekawa J., Ohtsubo E. DNA binding domains in Tn3 transposase. Mol Gen Genet. 1993 Jan;236(2-3):267–274. doi: 10.1007/BF00277122. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Automation of the computer handling of gel reading data produced by the shotgun method of DNA sequencing. Nucleic Acids Res. 1982 Aug 11;10(15):4731–4751. doi: 10.1093/nar/10.15.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Methods to define and locate patterns of motifs in sequences. Comput Appl Biosci. 1988 Mar;4(1):53–60. doi: 10.1093/bioinformatics/4.1.53. [DOI] [PubMed] [Google Scholar]
- Strathmann M., Hamilton B. A., Mayeda C. A., Simon M. I., Meyerowitz E. M., Palazzolo M. J. Transposon-facilitated DNA sequencing. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1247–1250. doi: 10.1073/pnas.88.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausbaugh L. D., Bourke M. T., Sommer M. T., Coon M. E., Berg C. M. Probe mapping to facilitate transposon-based DNA sequencing. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6213–6217. doi: 10.1073/pnas.87.16.6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu C. P., Cohen S. N. Translocation specificity of the Tn3 element: characterization of sites of multiple insertions. Cell. 1980 Jan;19(1):151–160. doi: 10.1016/0092-8674(80)90396-7. [DOI] [PubMed] [Google Scholar]



