Abstract
The mono-ADP-ribosyltransferase toxins are bacterial virulence factors that contribute to many disease states in plants, animals, and humans. These toxins function as enzymes that target various host proteins and covalently attach an ADP-ribose moiety that alters target protein function. We tested compounds from a virtual screen of commercially available compounds combined with a directed poly(ADP-ribose) polymerase (PARP) inhibitor library and found several compounds that bind tightly and inhibit toxins from Pseudomonas aeruginosa and Vibrio cholerae. The most efficacious compounds completely protected human lung epithelial cells against the cytotoxicity of these bacterial virulence factors. Moreover, we determined high-resolution crystal structures of the best inhibitors in complex with cholix toxin to reveal important criteria for inhibitor binding and mechanism of action. These results provide new insight into development of antivirulence compounds for treating many bacterial diseases.
Bacteria use virulence factors as tools to facilitate disease in plants, animals, and humans (14, 26, 30, 34); one strategy to combat infection is to inhibit these factors by small-molecule therapy, thereby helping to neutralize the offending microbe (5, 6, 12, 19, 22). It is now generally appreciated that an antivirulence approach is a powerful alternative strategy for antibacterial treatment and vaccine development (27) and that it may require multiple tactics to resolve the current drug resistance dilemma (6, 8). Antivirulence compounds offer significant advantages over conventional antibiotics since these inhibitors are directed toward specific mechanisms (targets) in the offending pathogen that promote infection rather than against an essential metabolic factor (12). Neutralizing the cytotoxic properties of virulence factors from microorganisms without threatening their survival offers reduced selection pressure, making the induction of drug resistance mutations less likely (6). Additionally, virulence-specific therapeutics avoid the undesirable effects on the host microbiota that are associated with current antibiotics.
The mono-ADP-ribosyltransferase (mART) family is a group of toxic bacterial enzymes, some of which possess a long history against human civilization. The best-characterized and well-known members of this lethal family are cholera toxin (CT) from Vibrio cholerae, diphtheria toxin (DT) produced by Corynebacterium diphtheriae, pertussis toxin (PT) from Bordella pertussis, heat-labile enterotoxin from Escherichia coli, C3-like exoenzyme produced by Clostridium botulinum and Clostridium limosum, and exotoxin A (ExoA) from Pseudomonas aeruginosa. These enzymes act on NAD+ and facilitate the scission of the glycosidic bond (C-N) between nicotinamide and its conjugated ribose followed by the transfer of the ADP-ribose group to a nucleophilic residue on a target macromolecule (35). This family can be divided into the CT and DT groups. The CT group consists of an ExoS-like subgroup (enzymatic A domain alone or paired with another domain), which targets the RAS family of G proteins; the C2-like subgroup (A/B motif, where B is the translocation domain), which targets actin; the C3-like subgroup (A only), which targets the Rho G-protein family; and the CT-PT-like subgroup (A/B5), targeting the Gα family of G proteins. The three characterized members of the DT group consist of three-domain A/B toxins that target the ribosomal translocase, eukaryotic elongation factor 2 (eEF2) (16). The mART family is characterized by low primary sequence identity, but the catalytic domain is structurally conserved. We recently developed an in silico approach based on fold recognition methods to identify prospective new mART members from bacterial genomes (13). These newly discovered toxins can now be exploited as targets in the development of new antivirulence therapeutics for treating bacterial diseases and infections (9, 29).
Here, we focus on two DT-group mARTs targeting elongation factor 2—ExoA, a well-characterized factor produced by P. aeruginosa, and cholix, a new mART toxin recently identified with our in silico approach from V. cholerae (16)—which, along with diphtheria toxin, show nearly identical enzyme activities and inhibitor specificities (2, 16, 31, 35, 36). Using the 1.25-Å cocrystal structure of cholix toxin with PJ34 [N-(6-oxo-5,6-dihydrophenanthridin-2-yl)-(N,N-dimethylamino)acetamide hydrochloride] inhibitor (Protein Data Bank [PDB] accession number 2Q6M) as a template, a virtual screen of over 500,000 commercial compounds identified 72 prospective inhibitors. After these inhibitors were filtered for chemical stability and redundancy, 31 compounds were then tested experimentally (see Tables S1 and S2 in the supplemental material). We also tested a small, directed poly(ADP-ribose) polymerase (PARP) inhibitor library of 12 compounds and found that several of these PARP inhibitors showed potent mART inhibition both in vitro and in cell-based assays. The resulting library of mART inhibitors includes eight compounds that showed nearly 100% protection of mammalian cells against high doses of bacterial toxin, six compounds that showed moderate protection, and 11 compounds that showed weak protection. In vitro kinetic studies correlate these levels of protection with the 50% inhibitory concentration (IC50) and dissociation constant (Kd) for each compound. Crystal structures of 9 novel inhibitors in complex with cholix toxin clearly demonstrate their binding within the toxin active site.
MATERIALS AND METHODS
Strains and media.
Saccharomyces cerevisiae W303 (MATa his3 ade2 leu2 trp1 ura3 can1), ERG6− (MATa his3 leu2 met15 ura3 erg6::KanMX), MTID:2955 (MATa leu2 trp1 can1 ura3 ade2 his3 pdr1D::NAT pdr3D::URA3), 2775 (MATa his3 leu2 lys2 ura3 MNN6::KanMX), and 7034 (MATa his3 leu2 lys2 ura3 MNN4::KanMX) were grown on yeast-peptone-dextrose or synthetic dextrose (SD) dropout medium. Human lung epithelial cells (C38) were cultured as previously described (37) in LHC-8 supplemented with 5% fetal bovine serum.
PARP inhibitor library.
A small, directed poly(ADP-ribose) polymerase (PARP) library of 8 compounds was a gift from Guilford Pharmaceuticals (Baltimore, MD). They were numbered P1 to P8 to indicate that these compounds originated from this series.
P. aeruginosa drug sensitivity assay.
Overnight cultures of P. aeruginosa strain PA103 were streaked onto LB plates and grown overnight at 37°C. A single colony was selected from the plate and grown overnight in 5 ml of LB broth at 37°C. One hundred microliters of the overnight culture was used to inoculate 250 ml of fresh LB broth. The inoculated medium was added to sterile culture tubes in 5-ml aliquots. Inhibitors were added to each aliquot at a final concentration of 50 μM and incubated overnight at 37°C. Growth was deemed positive if the optical density at 600 nm (OD600) was comparable (within 0.2 absorbance [Abs] units) to that of the untreated P. aeruginosa culture. Dimethyl sulfoxide (DMSO) (0.5%) was used as the negative control, and tetracycline (5 μg/ml) and gentamicin (10 μg/ml) were used as positive controls.
Protein purification of cholix toxin.
The full-length form and the catalytic fragment (cholixc) of cholix toxin were purified as previously described (17). Nonrecombinant, full-length exotoxin A was purchased from Sigma (St. Louis, MO).
IC50 determination for inhibitory compounds.
The concentration of inhibitor that reduced enzymatic activity by 50%, the IC50, was measured as previously described (36), with some modifications. The reaction mixtures (70-μl total volume) consisted of 300 μM ɛ-NAD, 10 μM eEF2, and a range of inhibitor concentrations in 20 mM Tris, 85 mM KCl (pH 7.9) buffer. The reaction mixtures were incubated at 25°C for 5 min in disposable Ultra-micro-UV cuvettes (Brand Scientific, Essex, CT), and the reactions were initiated by the addition of 5 nM cholix toxin. The IC50 was determined by fitting the data to the Boltzman sigmoidal function by nonlinear regression with Origin 6.1 (OriginLab, Northampton, MA).
Calculated Ki values.
Since the IC50 is not a direct indicator of affinity, these values were converted to Ki values according to the Cheng-Prusoff equation (7): Ki = IC50/(1 + [S]/Km), where [S] is the NAD+ concentration and Km is for the NAD+ substrate.
Calculated logP and logD values.
logP is log10(partition coefficient), and partition coefficient is [compound]octanol/[compound]water and was calculated (clogP) using an online clogP calculator (http://intro.bio.umb.edu/111-112/OLLM/111F98/newclogp.html) by ChemAxon Ltd. The logD pH profiles were generated by the same software and represent the calculated log distribution coefficient, logD = log10(distribution coefficient), where distribution coefficient D = Σ[microspecies]octanol/Σ[microspecies]water. Calculated logD values (pH 5.5 and 7.4) are shown for the most active inhibitor compounds in Table 2 and for all tested compounds in Tables S1 and S2 in the supplemental material.
Virtual screen.
We performed a virtual screen using GLIDE 5.0 (virtual docking program; Schrodinger, New York, NY) against our cholixc structure (PDB code 2Q6M). The approach was divided into the following steps. First, the Chembridge, Chemdiv, and Enamine libraries of 450,000, 750,000, and 850,000 compounds, respectively, were merged into a nonredundant data set of approximately 1,500,000 unique molecules. Molecular descriptors were calculated, and compounds that did not satisfy the Lipinski rules or have calculated logS values lower than −6.5 (indicative of poor aqueous solubility) were filtered out. The resulting library of about 500,000 drug-like molecules with favorable development potential constituted our virtual screening library. Second, each compound within the assembled library was docked to the active site of the enzyme using the GLIDE program and assigned a score (a predicted pseudo-binding energy) that reflects the quality of the fit and accounts for shape complementarity, Coulomb and continuum electrostatics, hydrogen bonding networks, and entropic penalty upon ligand binding.
Yeast-based compound screen.
Saccharomyces cerevisiae cells (strains W303, ERG6−, MTID:2955, 2775, and 7034) expressing the catalytic domain of P. aeruginosa ExoA (ExoAc) were cultured in the presence of 50 μM (each) compound and 1% dimethyl sulfoxide (DMSO) in 96-well plates for 48 h as previously described (3, 31).
Mammalian C38 cell drug sensitivity assay.
The C38 cells were cultured in 25-cm2 culture flasks containing LHC-8 medium with 5% fetal bovine serum at 37°C in 5% CO2 until the cells reached 90 to 100% confluence. The cells were washed with phosphate-buffered saline (PBS) and then detached from the culture flask by treating the cells with Puck's saline solution containing 0.02% EDTA. The cell suspension was diluted to a final density of 1.0 × 105 cells/ml, and 100-μl aliquots were used to seed a 96-well plate. The cells were then grown in the presence of 50 μM inhibitor (nonpolar compounds required 0.5% DMSO for solubility). If the inhibitor compound reduced cell viability (using the MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assay) to less than 70%, then the compound was considered toxic.
LD50 for ExoA against C38 cells.
The LD50 is the lethal dose of toxin that kills 50% of the C38 cells. The C38 cells were cultured as described above, except that full-length ExoA was added to the wells at 1 to 2,000 ng/ml and the cells were allowed to grow for 96 h, after which cell viability was determined using an MTT dye-based colorimetric assay (31). The LD50 values were determined by extrapolation from each corresponding line-of-best-fit (sigmoidal function in Origin 6.1), and the average values were reported ± standard deviation.
Inhibitor cytoprotection in mammalian cells.
C38 cells were cultured in the presence of various concentrations of ExoA as previously described (31) with the addition of each compound at 50 μM. Compounds V1 to V31, 1,8-naphthalimide (NAP), and PJ97A were assayed with 0.5% DMSO in the culture medium, whereas compounds PJ34 and P1 to P8 were assayed without DMSO. Cell viability was determined using an MTT dye-based colorimetric assay as previously described (31).
Determination of EC50.
C38 cells were cultured in the presence of 650 ng/ml of ExoA (approximately 10 times the LD50) in 96-well plates for 96 h. Each active inhibitor compound (NAP, V30, P1, P4, P5, and P6) was added to the growth medium to give final concentrations varying from 0.5 to 100 μM. Cell viability was determined using the same method as that in the cytoprotection assay. The half-maximal effective concentrations (EC50s) were extrapolated from each corresponding line-of-best-fit (sigmoidal function in Origin 6.1), and the average values were reported ± the standard deviation.
Crystallization, data collection, and refinement of cholixc-inhibitor complexes.
The purified cholixc (6.6 mg/ml, final concentration) was cocrystallized with the inhibitor compounds (15 mM, final concentration) by the sitting drop vapor diffusion method against reservoirs containing 15 to 18% polyethylene glycol 8000 (PEG 8000) and 10 to 20 mM KH2PO4 at 19°C. Before flash freezing in liquid N2, the crystals were transferred to either paratone-N (Hampton Research) or 25% glycerol, 16% PEG 10000, and 0.2 M KH2PO4 for cryoprotection. All data sets were collected at beamline ID-08 (Canadian Light Source, Saskatoon, Canada) except for cholixc-V30, which was collected at I911-2 (Max-Lab, Lund, Sweden). The data were processed and reduced with XDS and XSCALE (18). All structures were solved by molecular replacement using either Molrep (32) or Phaser (23) with the cholixc structure in complex with PJ34 (PDB entry 2Q6M) as a search model. Finally, the structure was iteratively rebuilt in Coot (11) and refined using Refmac5 (25). Structures with a resolution of 1.32 Å or higher were refined anisotropically, whereas all other structures were refined isotropically, including translation/libration/screw (TLS) motion determination with the cholixc structure divided into 10 groups.
Protein structure accession numbers.
The coordinates and structure factors are deposited in the RCSB Protein Data Bank as follows: cholix-P1, 3KIO; cholix-P2, 3KI1; cholix-P3, 3KI2; cholix-P4, 3KI3; cholix-P5, 3KI7; cholix-P6, 3KI6; cholix-P7, 3KI5; cholix-P8, 3KI4; cholix-V30, 3NY6.
RESULTS AND DISCUSSION
Inhibitor libraries.
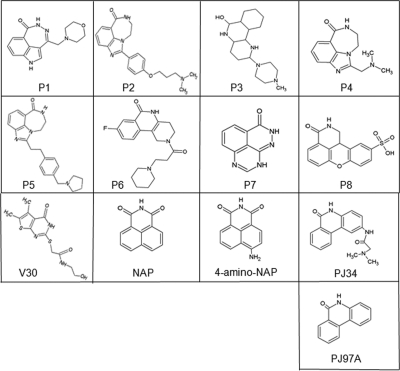
In this study, there were two libraries of compounds that were used. The first, designated the P series, was a small library of known PARP inhibitors (Fig. 1), and the second, designated the V series, was a library of commercially available compounds generated from a virtual screen conducted based on the crystal structure of cholix toxin with PJ34 inhibitor (PDB code 2Q6M). In addition, two well-characterized mART inhibitors, 1,8-naphthalimide (NAP) (31) and N-(6-oxo-5,6-dihydrophenanthridin-2-yl)-(N,N-dimethylamino)acetamide hydrochloride (PJ34) (36), were included as controls for the in vitro and cell-based studies (Fig. 1). Also, a polar derivative of NAP, 4-amino-NAP, and the nonpolar parent of PJ34, PJ97A, were also tested (Fig. 1). It was previously demonstrated that the P-series compounds compete with the NAD+ substrate in ExoA/cholix by binding within the nicotinamide binding site within these enzymes (2, 36). The virtual screen approach identified 72 hits (prospective inhibitors), which were filtered based upon chemical stability and redundancy, and 31 compounds were then tested experimentally (Table 1; see also Table S1 in the supplemental material). A small, in-house, poly(ADP-ribose) polymerase (PARP) inhibitor library of 8 compounds was also tested, and it was found that several of these PARP inhibitors showed potent mART inhibition both in vitro and in cell-based assays.
FIG. 1.
Chemical structures of several mART inhibitors. The P-series (P1 to P8) inhibitors are shown along with the most active V-series inhibitor, V30, and previously characterized mART inhibitors, 1,8-naphthalimide (NAP) and N-(6-oxo-5,6-dihydrophenanthridin-2-yl)-(N,N-dimethylamino)acetamide hydrochloride (PJ34). Also shown are the structures of a NAP derivative, 4-amino-NAP, and the parent compound for PJ34, PJ97A.
TABLE 1.
Overview of the cell-based results for potential ExoA inhibitorsa
| Compound | Protection in C38 cells | Protection in yeast | Reference or source |
|---|---|---|---|
| NAP | ++++ | ++++ | 31 |
| 4-NAP | +++ | ++ | 31 |
| PJ97A | +++ | +++ | 15 |
| PJ34 | − | − | 15 |
| V1 | − | − | This study |
| V2 | + | − | This study |
| V3 | Toxic | − | This study |
| V4 | Toxic | − | This study |
| V5 | Toxic | Toxic | This study |
| V6 | − | − | This study |
| V7 | Toxic | Toxic | This study |
| V8 | − | − | This study |
| V9 | ++ | − | This study |
| V10 | + | − | This study |
| V11 | + | − | This study |
| V12 | ++ | − | This study |
| V13 | − | − | This study |
| V14 | − | − | This study |
| V15 | ++ | + | This study |
| V16 | + | − | This study |
| V17 | ++ | − | This study |
| V18 | − | − | This study |
| V19 | Toxic | − | This study |
| V20 | − | − | This study |
| V21 | + | − | This study |
| V22 | + | − | This study |
| V23 | ++ | ++ | This study |
| V24 | ++ | − | This study |
| V25 | + | − | This study |
| V26 | + | − | This study |
| V27 | − | − | This study |
| V28 | Toxic | Toxic | This study |
| V29 | + | + | This study |
| V30 | ++++ | − | This study |
| V31 | + | + | This study |
| P1 | ++++ | − | This study |
| P2 | Toxic | − | This study |
| P3 | Toxic | − | This study |
| P4 | ++++ | − | This study |
| P5 | ++++ | − | This study |
| P6 | ++++ | − | This study |
| P7 | Toxic | − | This study |
| P8 | + | − | This study |
All compounds were screened in yeast expressing the catalytic domain of ExoA (31) as well as against C38 cells dosed with the toxin. In column 2, compounds capable of protecting C38 cells from ExoA concentrations of greater than 1,000, 500 to 1,000, 250 to 500, and 100 to 250 ng/ml were scored as ++++, +++, ++, and +, respectively. Compounds showing protection at less than 100 ng/ml toxin were scored as −. In column 3, compounds found to restore growth to 75 to 100%, 50 to 75%, 25 to 50%, and 5 to 25% were scored as ++++, +++, ++, and +, respectively. Compounds showing less than 5% growth were scored as −. Protection was assessed by determining the ability of each inhibitor to increase the LD50 of ExoA. “Toxic” indicates that the inhibitor was cytotoxic to the cells, and a toxic compound is defined as any compound that reduced cell viability to less than 30%. All compounds were screened at a final concentration of 50 μM. Yeast strains W303, ERG6−, MTDID:2955, 2775, and 7034 were screened in duplicate, and no significant differences in protection were observed.
Inhibitor activities in yeast.
All potential inhibitors (both the P and the V series) were screened in five different yeast strains, previously developed to facilitate compound delivery to the yeast cytoplasm, expressing the ExoAc. However, there were no significant differences in the effects of inhibitors in these yeast strains, and so only the results of screens in the W303 strain are shown (Table 1). We previously demonstrated that intracellular expression of the ExoAc gene in yeast caused a growth defect phenotype which can be used as a cell-based measure of mART toxicity in this model system (31). Effective inhibitors of mART (ExoAc) activity can be screened in this yeast-based system, where such agents abrogate the growth defect phenotype (33). The yeast strains employed for initial screening were W303 (wild type), ERG6− (which lacks the Δ24-sterol C-methyltransferase), MTID:2955 (which lacks two master regulators in the expression of pleiotropic drug response elements), and 2775 and 7034 (deficient in mannosylphosphate transferase); however, no differences between the various yeast strains were observed, and so W303 was used as the tester strain for this study (3). In yeast, generally compounds from both the P and V series that were able to at least partially restore yeast growth (although V23 and V30 are exceptions) were nonpolar, lacked solubility in aqueous buffer, and were not ionizable (log values; see Tables S1 and S2 in the supplemental material), suggesting that the cell wall acts as a physical barrier, particularly to polar molecules. This was most obvious in the structure/activity relationships of NAP and its more polar derivative, 4-amino-NAP, and between PJ97A and the more polar PJ34, where the nonpolar parent compound was active against ExoAc in yeast, but the more polar derivative was considerably less active (Table 1; Fig. 1; see Tables S1 and S2) (1, 28). None of the polar P-series compounds (P1 to P8) showed inhibitor efficacy in yeast against ExoAc, and in the V series only compound V23 showed modest protection in yeast, with weak protection afforded by compounds V15, V29, and V31. Yeasts grow on less complex media; are genetically simpler, lacking many redundant systems of mammalian cells; and provide more rapid feedback as a screening tool; however, yeasts are limited for identifying polar/ionizable inhibitors because their cell wall presents a barrier to the permeation of these compounds (1, 28).
ExoA cytotoxicity in mammalian cells and inhibitor protection.
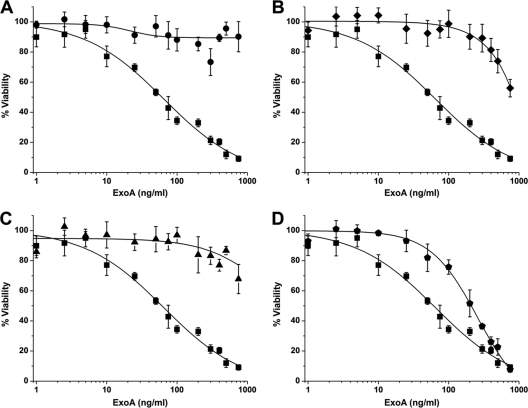
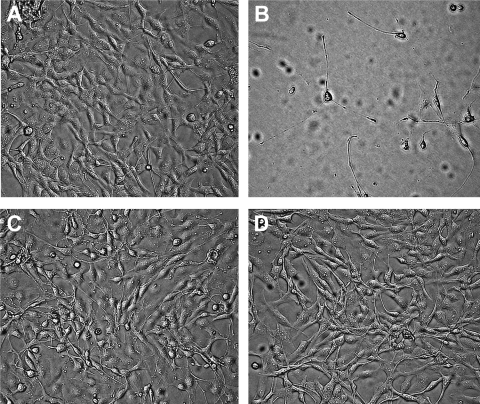
ExoA invades target mammalian cells through the recognition of the low-density lipoprotein receptor (LRP) in the plasma membrane (20), and it kills the host cell by arresting protein synthesis and inducing apoptosis (10). ExoA showed an LD50 of 65 ± 12 ng/ml for the C38 cells (Fig. 2 A to C, control curve for ExoA with no inhibitor). NAP and V30 provided excellent protection of C38 cells against ExoA toxicity, even at very high toxin doses (Fig. 1 and Fig. 2A and C). At high doses (500 ng/ml), ExoA was highly toxic to the C38 cells (Fig. 3 B) compared with the healthy control cells (Fig. 3A). Remarkably, compounds V30 and P1 completely protected the C38 cells from the lethal action of high doses of ExoA (Fig. 3C and D).
FIG. 2.
Cytoprotective effects of nonpolar inhibitors in ExoA-intoxicated C38 cells. (A) NAP protects cells from the toxic effects of ExoA (circles) compared to a 0.5% DMSO control (squares). (B to D) Compounds PJ97A (diamonds) (B), V30 (triangles) (C), and V23 (polygons) (D) all show various levels of protection in C38 cells. The ExoA dose-response curve is shown in each panel as filled squares. All compounds were assayed at a final concentration of 50 μM in the presence of 0.5% DMSO. The LD50 of ExoA in C38 cells with 0.5% DMSO was 65 ± 3 ng/ml. The LD50 of ExoA in the presence of NAP and V30 was >1,000 ng/ml, that in the presence of PJ97A was >750 ng/ml, and that in the presence of V23 was 199 ± 30 ng/ml. Experiments were performed in quadruplicate, in three independent determinations. Error bars represent the mean ± standard deviation for each set of replicates.
FIG. 3.
Effect of inhibitor treatment on the morphology of ExoA-intoxicated C38 cells. Human C38 cells were grown for 72 h in the absence of ExoA and inhibitor (control) (A), 500 ng/ml ExoA (B), 500 ng/ml ExoA and 50 μM compound V30 (C), and 500 ng/ml ExoA and 50 μM compound P1 (D). Pictures were taken at ×200 magnification with a Zeiss inverted microscope (Invertoskop 40C).
V-series inhibitor activities in mammalian cells.
In C38 cells, virtual screen compound V30 showed excellent protection against ExoA with moderate activity exhibited by V9, V12, V15, V17, V23, and V24 (Table 1). Weak protection was shown by V10, V11, V16, V21, V22, V25, V26, V29, and V31 (Table 1). Therefore, V30, which was the only V-series inhibitor to show excellent protection of C38 from toxin, was chosen for further testing and quantification for in vitro inhibitory (IC50) activity. NAP, a previously characterized, competitive inhibitor of ExoA (2), served as a known toxin inhibitor control; both V30 and NAP were remarkably similar in being able to protect C38 cells and increased the LD50 values for ExoA by 30- to 40-fold (Fig. 2A and C). Compound PJ97A, a derivative of the well-characterized ExoA/cholix inhibitor, PJ34 (36), also showed strong protection against ExoA but not as much as NAP and V30 (LD50 increased nearly 12-fold for PJ97A) (Table 2; Fig. 2B). Compound V23 is an example of a modest ExoA inhibitor in C38 cells, and it was, in fact, the weakest in this active inhibitor group (Fig. 2D). Modest V-series inhibitors such as V12 increased the toxin LD50 by about only 3-fold (Table 1; see Fig. S1 in the supplemental material).
TABLE 2.
Comparison of Kd, Tm, IC50, and EC50s for inhibitors of Pseudomonas aeruginosa ExoA
| Compound | Kd (nM)a | Ki (nM)b | IC50 (nM)c | EC50 (μM)d | clogPf | logD pH 5.5f | logD pH 7.4f |
|---|---|---|---|---|---|---|---|
| P1 | 10 ± 2; 650 ± 50 | 22 ± 4 | 170 ± 30 | 2.9 ± 0.8 | −0.08 | 0.48 | 1.59 |
| P2 | 260 ± 10 | 63 ± 5 | 480 ± 40 | NDe | 2.25 | −1.09 | 0.4 |
| P3 | 1,470 ± 30 | 90 ± 17 | 690 ± 130 | ND | 1.01 | −4.67 | −1.18 |
| P4 | 1,380 ± 30 | 132 ± 7 | 960 ± 50 | 12.6 ± 3.3 | 0.4 | −1.49 | 0.1 |
| P5 | 750 ± 10 | 582 ± 124 | 4,460 ± 950 | 16.7 ± 1.9 | 3.14 | −0.42 | 1.06 |
| P6 | 1,100 ± 20 | 80 ± 18 | 610 ± 140 | 3.4 ± 1.6 | 1.13 | −2.21 | −0.81 |
| P7 | 680 ± 40 | 118 ± 7 | 908 ± 118 | ND | 1.82 | 0.26 | 1.24 |
| P8 | 160 ± 30; 5,210 ± 1,780 | 136 ± 3 | 1,040 ± 136 | ND | 1.29 | −1.08 | −1.08 |
| V30 | 931 ± 74 | 367 ± 3 | 2,815 ± 22 | 8.8 ± 0.5 | 1.66 | 1.15 | 1.13 |
| NAP | 950 ± 30 | 12 ± 1 | 90 ± 10 | 3.8 ± 0.9 | 1.68 | 1.68 | 1.67 |
| PJ34 | 820 ± 54 | 37 ± 9 | 280 ± 70 | ND | 1.91 | 0.35 | 1.75 |
| PJ97A | 393 ± 64 | 610 ± 175 | 4,674 ± 175 | ND | 2.74 | 2.74 | 2.74 |
The binding affinity of inhibitors to wild-type ExoAc was measured by the quenching of the intrinsic Trp fluorescence caused by the binding of the ligand to the enzyme active site.
The absolute inhibition constant (Ki) was calculated from the experimentally determined IC50s according to the following relationship: Ki = IC50/(1 + {[SNAD]/KM(NAD)}) (7); see Materials and Methods for details. The [NAD+] was 300 μM, and the KM(NAD) was 45 μM.
The IC50s were determined by fitting each dose-response curve to a Boltzmann sigmoidal function in Origin 6.1.
The EC50s were determined for inhibitors added to C38 cells in the presence of 650 ng/ml of ExoA in 96-well plates for 96 h (see Materials and Methods for details).
ND, not determined.
The logP and logD values were calculated as described in Materials and Methods.
P-series inhibitor activities in mammalian cells.
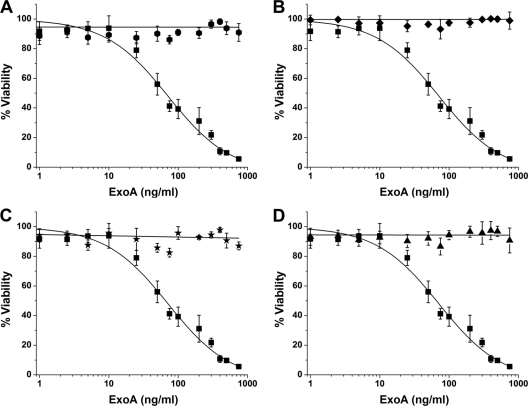
In the P-series library (Fig. 1), compounds P1, P4, P5, and P6 showed excellent protection (over 90%) at ExoA doses near 1,000 ng/ml (ExoA LD50 = 65 ng/ml) (Fig. 4 A to D), which was even greater than the protection provided by both NAP and V30 inhibitors. Notably, compounds P2, P3, and P7 showed good IC50s against purified cholix (Table 2) but were toxic to C38 cells (Table 1, listed as toxic) and thus were excluded from cell-based testing. The C38-toxic compounds reduced viability to less than 10%, and the nontoxic inhibitors did not reduce viability below 95% at the 50 μM dose (Table 1). Compound P8 also showed a good in vitro IC50 but exhibited only a modest ability to protect C38 cells from ExoA (Fig. 1; Tables 1 and 2).
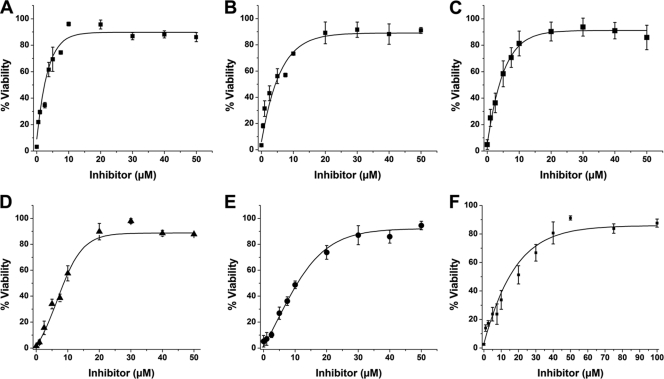
FIG. 4.
Cytoprotective effects of water-soluble inhibitors in ExoA-intoxicated C38 cells. Compounds P1 (hexagons) (A), P4 (diamonds) (B), P5 (stars) (C), and P6 (triangles) (D) protect C38 from high doses of ExoA. The ExoA dose-response curve is shown in each panel as filled squares. All compounds were assayed at a final concentration of 50 μM. The LD50 of ExoA in C38 was 61 ± 9 ng/ml. The LD50 of ExoA in the presence of compounds P1, P4, P5, and P6 was >1,000 ng/ml. Experiments were performed in quadruplicate, in three independent determinations. Error bars represent the mean ± standard deviation for each set of replicates.
Correlation of cell-based and in vitro inhibitor characteristics.
Several compounds from both the V and P series showed good inhibitor efficacy with EC50s that ranged from 2.9 to 16.7 μM, including P1, P6, NAP, V30, P4, and P5 (Fig. 5A to F; Table 2). IC50s and binding affinities (Kd) were measured for these compounds in vitro for comparison with the cell-based results. NAP, P1, and P6 compounds showed low- to midnanomolar IC50s whereas P4, V30, and P5, gave higher values (960 nM, 2,815 nM, and 4,460 nM, respectively) (Table 2). The higher IC50s for these latter compounds were reflected in the corresponding Ki values (inhibitor binding constants) and represent weaker binding to the toxin active site. However, this weaker affinity was not a factor in the ability of these compounds to block toxin activity in C38 cells (Tables 1 and 2) at the 50 μM experimental dose. Interestingly, the inhibitor efficacy (EC50s) did not correlate with the inhibition constant (Ki) of these compounds with the toxin target enzyme, indicating that the minimum requirement for a good inhibitor is competitive binding to the active site at 1 μM affinity or better (Table 2, Ki values). Furthermore, EC50s reflect the pharmacokinetic properties of these compounds that involve a number of processes within the target cell such as the ability to cross a membrane, the ability to diffuse to the target protein within the cell milieu, and stability within the cell. It is notable that the Kd values of cholix with inhibitors also did not correlate with their corresponding Ki values; this is likely because the Ki reflects the binding constant of the inhibitor in complex with its protein target, eEF2 (taken from kinetic inhibition data), whereas the Kd is the binding constant for the free enzyme with inhibitor. Two compounds, P1 and P8, showed dual binding constants, indicating two distinct binding sites with approximately a 50-fold difference in affinity for cholix. One explanation for this phenomenon is that the high-affinity site is the active site and the lower-affinity site involves a region of the enzyme that is not the active site but is located elsewhere on the protein surface. This has been observed for a number of enzymes during crystallization with substrate analogues/inhibitors (24).
FIG. 5.
Inhibitor potency in C38 cells as determined by half-maximal effective concentration (EC50). C38 cells were exposed to lethal doses of ExoA (650 ng/ml, 10 times the LD50) at various concentrations of each inhibitor. (A and B) NAP protected with an EC50 of 3.8 ± 0.9 μM (A) and V30 had an EC50 of 8.8 ± 0.5 μM (B), both in the presence of 0.5% DMSO. (C to F) Compounds P1 (C), P4 (D), P5 (E), and P6 (F) had EC50s of 2.9 ± 0.8 μM, 12.6 ± 3.3 μM, 16.7 ± 1.9 μM, and 3.4 ± 1.6 μM, respectively. EC50s represent the average value obtained from four independent determinations performed in quadruplicate. Error bars represent the mean ± standard deviation for each set of replicates.
The resulting library of mART inhibitors (Table 1) includes eight compounds that showed nearly 100% protection of mammalian cells against high doses of bacterial toxin (++++ or +++), six compounds that showed moderate protection (++), and 11 compounds that showed weak protection (+). Two compounds from our combined libraries showed good protection of yeast, two showed moderate protection, and three showed only weak protection from bacterial toxin (Table 1). Importantly, none of the inhibitors tested from both the virtual screen and the PARP library were toxic to P. aeruginosa at the doses administered to yeast and mammalian cells (see P. aeruginosa drug sensitivity assay in Materials and Methods). Although the mART toxin family shares a common NAD+ binding site with many mammalian enzymes, including dehydrogenases, it is not likely that competitive inhibitors specific for the NAD+ binding site within these toxins will cause mammalian cell toxicity. This is because the toxin NAD+ binding site coordinates NAD+ in a unique, twisted horseshoe configuration (4) that is very different from the extended NAD+ conformation associated with dehydrogenases, for instance.
Physicochemical properties of potential toxin inhibitors.
The log partition coefficient (logP) is a measure of the differential solubility of a compound in immiscible solvents such as octanol and water, and in drug discovery it is a useful parameter to understand the behavior of drug molecules in cell culture and in the human body. According to Lipinski's rule of 5, the logP (clogP [calculated logP]) for a drug candidate should be <5 and can be applied to neutral compounds (21); however, logD pH profiles should be used to estimate the bioavailability of ionizable compounds. The best inhibitors in C38 cells (Tables 1 and 2) are ionizable compounds that were ambivalent toward aqueous or organic solvent (logD = −1.49 to 1.68) (Table 2; see Tables S1 and S2 in the supplemental material); however, NAP and PJ97A are neutral compounds—the latter compound suffered from lower aqueous solubility (clogPPJ97A = 2.74) (Table 2; see also Table S2), likely compromising its bioavailability compared with that of NAP (clogPNAP = 1.68) (Table 2; see also Table S2).
Structures of inhibitors with cholix toxin.
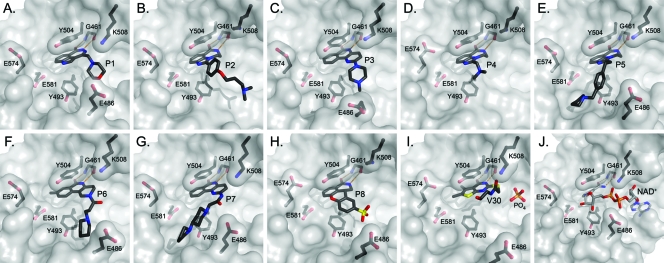
ExoA and cholix are closely related mART toxins that possess nearly identical biological activities (16). However, cholixc produces superior crystal structures and was used as our model for structure determination of inhibitor complexes (31). The high-resolution crystal structures (1.27 to 1.68 Å) of cholixc with inhibitors P1 to P8 and V30 were solved by molecular replacement and provided strong support to the in vitro and cell-based inhibition data (Tables 1 and 2; see also Table S3 in the supplemental material). These structures unequivocally demonstrated that these compounds are competitive inhibitors of NAD+ and bind within the nicotinamide binding domain of cholix (Fig. 6 A to I compared with 6J). The cocrystal structures of cholixc with NAP and PJ34 were previously solved by our group (16, 31) and serve as useful references for understanding the binding of inhibitors P1 to P8 and V30 within the cholixc active site (Fig. 1 and 6). The key features for these active-site inhibitors of ExoA include a benzamido group fused into a hetero-ring structure, which acts to mimic the nicotinamide moiety of NAD+ (36) by intercalating deeply within the nonpolar pocket while forming aromatic ring stacking interactions with the two conserved Tyr residues of cholixc (Tyr 493 and Tyr 504) (Fig. 1 and 6J). These two Tyr residues flank the core of the inhibitor structures and provide a hydrophobic cradle for nonpolar, planar aromatic ligands such as NAP and compounds P1 to P8 and V30 (Fig. 1 and 6; see also Fig. S2). Furthermore, in all compounds, a key interaction includes H bonding between the inhibitor imide and the backbone groups of a conserved Gly residue (Gly461) within the Y-H-G motif of the active-site core as seen for the NAD+ substrate (Fig. 6J) (31). Two-dimensional drawings of the interactions of each inhibitor with the cholix active-site residues can be seen in Fig. S3, which help to illustrate the common mode of interaction of these inhibitory compounds within the toxin active site and help to explain their competitive inhibition kinetics (Table 2).
FIG. 6.
Crystal structures of the catalytic fragment of cholix bound to inhibitor compounds. Cholix-P1 (A), cholix-P2 (B), cholix-P3 (C), cholix-P4 (D), cholix-P5 (E), cholix-P6 (F), cholix-P7 (G), cholix-P8 (H), cholix-V30 (I), and model of cholix-NAD+ complex (J). The inhibitors and the NAD+ substrate are shown with standard atom colors, and nearby residues are shown as black sticks. Hydrogen bonds are shown in orange dashed lines. The model of the cholix-NAD+ complex is based upon the ExoA-NAD+ complex (PDB code 3B78).
We previously developed a yeast-based approach for the identification and characterization of mART toxins from pathogenic bacteria (31). This screening method provides great promise for the discovery and characterization of novel therapeutics against these toxins. Importantly, this yeast cell-based approach may have a more general appeal as a method to identify and characterize any protein that is toxic to eukaryotic cells, including bacterial virulence factors, as well as eukaryotic proteins involved in apoptosis, necrosis, and cancer. In addition, we demonstrated the utility of a virtual chemical screen in combination with our new screening method to identify lead compounds against mART toxins that show efficacy in cell-based systems. One of these compounds, V30, shows great potential as a lead for further development as an antivirulence compound against this family of bacterial toxins. We also tested a small, directed PARP library of inhibitor compounds against ExoA and found that all of the compounds bound to the cholix active site and provided some protection in C38 cells (except for toxic compounds P2, P3, and P7), whereas only nonpolar compounds were effective in yeast cells (Table 1). We clearly demonstrated that the ExoA/cholix inhibitors dock within the nicotinamide binding pocket of the active site of these toxins and inhibit the mART enzymatic activity of purified toxin. Furthermore, we conclusively showed that the most efficacious compounds function as DT-group mART toxin inhibitors with prophylactic properties for human lung epithelial cells. We also showed that the best compounds are not toxic to either the lung cells or the producing bacterial pathogen, in this case, P. aeruginosa, and hence have great potential as antivirulence agents for treating many bacterial infections and diseases. Finally, we plan to use a rational drug design approach to improve the binding affinity of our best lead compounds, V30 and P1, to the cholix/ExoA target enzymes through modern drug discovery and combinatorial chemistry methods.
Supplementary Material
Acknowledgments
This research was funded by the Canadian Institutes of Health Research and the Canadian Cystic Fibrosis Foundation to A.R.M.
We thank Dawn White for excellent technical support during this study.
Footnotes
Published ahead of print on 6 December 2010.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Aouida, M., O. Tounekti, O. Belhadj, and L. M. Mir. 2003. Comparative roles of the cell wall and cell membrane in limiting uptake of xenobiotic molecules by Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 47:2012-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong, S., J. H. Li, J. Zhang, and A. R. Merrill. 2002. Characterization of competitive inhibitors for the transferase activity of Pseudomonas aeruginosa exotoxin A. J. Enzyme Inhib. Med. Chem. 17:235-246. [DOI] [PubMed] [Google Scholar]
- 3.Arnoldo, A., et al. 2008. Identification of small molecule inhibitors of Pseudomonas aeruginosa exoenzyme S using a yeast phenotypic screen. PLoS Genet. 4:e1000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell, C. E., T. O. Yeates, and D. Eisenberg. 1997. Unusual conformation of nicotinamide adenine dinucleotide (NAD) bound to diphtheria toxin: a comparison with NAD bound to the oxidoreductase enzymes. Protein Sci. 6:2084-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benghezal, M., et al. 2007. Inhibitors of bacterial virulence identified in a surrogate host model. Cell. Microbiol. 9:1336-1342. [DOI] [PubMed] [Google Scholar]
- 6.Cegelski, L., G. R. Marshall, G. R. Eldridge, and S. J. Hultgren. 2008. The biology and future prospects of antivirulence therapies. Nat. Rev. Microbiol. 6:17-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, Y., and W. H. Prusoff. 1973. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 22:3099-3108. [DOI] [PubMed] [Google Scholar]
- 8.Clatworthy, A. E., E. Pierson, and D. T. Hung. 2007. Targeting virulence: a new paradigm for antimicrobial therapy. Nat. Chem. Biol. 3:541-548. [DOI] [PubMed] [Google Scholar]
- 9.Diep, B. A., and M. Otto. 2008. The role of virulence determinants in community-associated MRSA pathogenesis. Trends Microbiol. 16:361-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du, X., R. J. Youle, D. J. FitzGerald, and I. Pastan. 2010. Pseudomonas exotoxin A-mediated apoptosis is Bak dependent and preceded by the degradation of Mcl-1. Mol. Cell. Biol. 30:3444-3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 12.Escaich, S. 2008. Antivirulence as a new antibacterial approach for chemotherapy. Curr. Opin. Chem. Biol. 12:400-408. [DOI] [PubMed] [Google Scholar]
- 13.Fieldhouse, R. J., and A. R. Merrill. 2008. Needle in the haystack: structure-based toxin discovery. Trends Biochem. Sci. 33:546-556. [DOI] [PubMed] [Google Scholar]
- 14.Higgins, D. A., et al. 2007. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature 450:883-886. [DOI] [PubMed] [Google Scholar]
- 15.Jagtap, P., et al. 2002. Novel phenanthridinone inhibitors of poly(adenosine 5′-diphosphateribose) synthetase: potent cytoprotective and antishock agents. Crit. Care Med. 30:1071-1082. [DOI] [PubMed] [Google Scholar]
- 16.Jorgensen, R., et al. 2008. Cholix toxin, a novel ADP-ribosylating factor from Vibrio cholerae. J. Biol. Chem. 283:10671-10678. [DOI] [PubMed] [Google Scholar]
- 17.Jorgensen, R., Y. Wang, D. Visschedyk, and A. R. Merrill. 2008. The nature and character of the transition state for the ADP-ribosyltransferase reaction. EMBO Rep. 9:802-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kabsch, W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26:795-800. [Google Scholar]
- 19.Kollef, M. H. 2008. Broad-spectrum antimicrobials and the treatment of serious bacterial infections: getting it right up front. Clin. Infect. Dis. 47(Suppl. 1):S3-S13. [DOI] [PubMed] [Google Scholar]
- 20.Kounnas, M. Z., et al. 1992. The alpha 2-macroglobulin receptor/low density lipoprotein receptor-related protein binds and internalizes Pseudomonas exotoxin A. J. Biol. Chem. 267:12420-12423. [PubMed] [Google Scholar]
- 21.Lipinski, C. A., F. Lombardo, B. W. Dominy, and P. J. Feeney. 2001. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46:3-26. [DOI] [PubMed] [Google Scholar]
- 22.Maresso, A. W., and O. Schneewind. 2008. Sortase as a target of anti-infective therapy. Pharmacol. Rev. 60:128-141. [DOI] [PubMed] [Google Scholar]
- 23.McCoy, A. J., et al. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McPherson, A. 2004. Introduction to protein crystallization. Methods 34:254-265. [DOI] [PubMed] [Google Scholar]
- 25.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 26.Raghavan, S., P. Manzanillo, K. Chan, C. Dovey, and J. S. Cox. 2008. Secreted transcription factor controls Mycobacterium tuberculosis virulence. Nature 454:717-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rappuoli, R. 2007. Bridging the knowledge gaps in vaccine design. Nat. Biotechnol. 25:1361-1366. [DOI] [PubMed] [Google Scholar]
- 28.Scherrer, R., L. Louden, and P. Gerhardt. 1974. Porosity of the yeast cell wall and membrane. J. Bacteriol. 118:534-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sleno, L., and A. Emili. 2008. Proteomic methods for drug target discovery. Curr. Opin. Chem. Biol. 12:46-54. [DOI] [PubMed] [Google Scholar]
- 30.Tsigrelis, C., K. V. Singh, T. D. Coutinho, B. E. Murray, and L. M. Baddour. 2007. Vancomycin-resistant Enterococcus faecalis endocarditis: linezolid failure and strain characterization of virulence factors. J. Clin. Microbiol. 45:631-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turgeon, Z., et al. 2009. Yeast as a tool for characterizing mono-ADP-ribosyltransferase toxins. FEMS Microbiol. Lett. 300:97-106. [DOI] [PubMed] [Google Scholar]
- 32.Vagin, A., and A. Teplyakov. 1997. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30:1022-1025. [Google Scholar]
- 33.Visschedyk, D. D., et al. 2010. Photox, a novel actin-targeting mono-ADP-ribosyltransferase from Photorhabdus luminescens. J. Biol. Chem. 285:13525-13534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yahr, T. L., and M. C. Wolfgang. 2006. Transcriptional regulation of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol. 62:631-640. [DOI] [PubMed] [Google Scholar]
- 35.Yates, S. P., R. Jorgensen, G. R. Andersen, and A. R. Merrill. 2006. Stealth and mimicry by deadly bacterial toxins. Trends Biochem. Sci. 31:123-133. [DOI] [PubMed] [Google Scholar]
- 36.Yates, S. P., et al. 2005. Structure-function analysis of water-soluble inhibitors of the catalytic domain of exotoxin A from Pseudomonas aeruginosa. Biochem. J. 385:667-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeitlin, P. L., et al. 1991. A cystic fibrosis bronchial epithelial cell line: immortalization by adeno-12-SV40 infection. Am. J. Respir. Cell Mol. Biol. 4:313-319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.