Abstract
This article provides an analysis of the in vitro effect of qnrA1, qnrB1, and qnrS1 genes, combined with quinolone-resistant Ser83Leu substitutions in GyrA and/or Ser80Arg in ParC, on fluoroquinolone (FQ) resistance in isogenic Escherichia coli strains. The association of Ser83Leu substitution in GyrA, Ser80Arg substitution in ParC, and qnr gene expression increased the MIC of ciprofloxacin to 2 μg/ml. qnr genes present in E. coli that harbored a Ser83Leu substitution in GyrA increased mutant prevention concentration (MPC) values to 8 to 32 μg/ml. qnr gene expression in E. coli may play an important role in selecting for one-step FQ-resistant mutants.
Fluoroquinolone (FQ) resistance occurs mainly as a result of mutations in chromosomal genes encoding quinolone targets, DNA gyrase and topoisomerase IV (5). More recently, plasmid-mediated mechanisms, such as those mediated by the qnr, aac(6′)-Ib-cr, and qepA genes, have been reported (11, 18). In the absence of other mechanisms, the presence of any qnr gene increased the MIC of FQ between 4- and 128-fold, although MIC values remained below CLSI breakpoints (9, 18).
It has been suggested that Qnr proteins facilitate the selection of higher-level quinolone-resistant mutants. In spite of this, the therapeutic relevance of the acquisition of qnr genes on FQ bactericidal activity remains unclear (10, 13, 16). Since spontaneous bacterial mutants usually arise at a low frequency of 10−6 to 10−8, the prevention of mutant bacterial populations may help to restrict the development of antimicrobial resistance. To avoid selecting for resistance, drug concentrations should be kept above the mutant prevention concentration (MPC) (4, 20). In vivo studies have shown that the presence of qnr genes in association with additional quinolone resistance mechanisms might be relevant in the activity of these antimicrobial agents (1, 15).
In a recent study (12), the combined effect of topoisomerase mutations on FQ resistance in isogenic Escherichia coli strains showed that at least three mutations—two of which had to be in gyrA—were necessary to exceed CLSI resistance breakpoints. Plasmid-mediated quinolone resistance (PMQR) genes confer low levels of quinolone resistance, and their precise effect on selecting for quinolone resistance in association with other mechanisms is not well known. In addition, recent studies have shown that the qnrA gene increased the MPC against FQ (16). The aim of this study was to evaluate the effect of qnrA, qnrB, and qnrS genes on the development of quinolone resistance in wild-type E. coli strains compared to isogenic E. coli strains harboring mutated gyrA and/or parC genes.
Ser83Leu and Ser80Arg mutations, located in GyrA and ParC, respectively, were obtained by gene replacement, as described by Posfai et al. (14). The qnr genes carried on the pBK-CMV cloning vector were transformed by electroporation into E. coli ATCC 25922 and its isogenic mutant strains E. coli ATCC 25922-S83L, E. coli ATCC 25922-S80R, and E. coli ATCC 25922-S83L-S80R (Table 1). The primers used to obtain the different isogenic strains are indicated in Table 2.
TABLE 1.
Bacterial strains and fluoroquinolone MIC and MBCs
| E. coli strain/plasmid gene | Relevant features | Plasmid containing PMQRa gene | Ciprofloxacin susceptibilityb | MIC (μg/ml)c |
MBC (μg/ml) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CIP | LVX | MXF | NFX | CIP | LVX | MXF | NFX | ||||
| ATCC 25922 | Wild type | None | S | 0.002 | 0.008 | 0.008 | 0.015 | 0.015 | 0.03 | 0.015 | 0.03 |
| ATCC/qnrA | Wild type | pBK-QnrA1 | S | 0.125 | 0.5 | 0.25 | 0.5 | 0.125 | 0.5 | 0.5 | 1 |
| ATCC/qnrB | Wild type | pBK-QnrB1 | S | 0.125 | 0.125 | 0.25 | 0.25 | 0.125 | 0.25 | 1 | 0.5 |
| ATCC/qnrS | Wild type | pBK-QnrS1 | S | 0.125 | 0.5 | 0.25 | 0.5 | 0.25 | 0.5 | 0.25 | 1 |
| ATCC 25922-S83L | GyrA Ser83Leu | None | S | 0.125 | 0.125 | 0.06 | 0.125 | 0.125 | 0.25 | 0.125 | 0.5 |
| ATCC-S83L/qnrA | GyrA Ser83Leu | pBK-QnrA1 | S | 0.5 | 0.5 | 0.5 | 2 | 0.5 | 0.5 | 0.5 | 2 |
| ATCC-S83L/qnrB | GyrA Ser83Leu | pBK-QnrB1 | S | 0.5 | 0.25 | 0.5 | 1 | 0.5 | 0.5 | 0.5 | 1 |
| ATCC-S83L/qnrS | GyrA Ser83Leu | pBK-QnrS1 | S | 1 | 1 | 1 | 2 | 1 | 2 | 2 | 4 |
| ATCC 25922-S80R | ParC Ser80Arg | None | S | 0.004 | 0.008 | 0.008 | 0.03 | 0.004 | 0.008 | 0.008 | 0.03 |
| ATCC-S80R/qnrA | ParC Ser80Arg | pBK-QnrA1 | S | 0.25 | 0.25 | 0.5 | 0.5 | 0.25 | 0.5 | 0.5 | 2 |
| ATCC-S80R/qnrB | ParC Ser80Arg | pBK-QnrB1 | S | 0.125 | 0.25 | 0.5 | 0.5 | 0.25 | 0.5 | 0.5 | 0.5 |
| ATCC-S80R/qnrS | ParC Ser80Arg | pBK-QnrS1 | S | 0.125 | 0.25 | 0.25 | 0.5 | 0.25 | 0.25 | 0.5 | 0.5 |
| ATCC 25922-S83L-S80R | GyrA Ser83Leu, ParC Ser80Arg | None | S | 0.25 | 0.25 | 0.25 | 2 | 2 | 0.25 | 0.25 | 2 |
| ATCC-S83L-S80R/qnrA | GyrA Ser83Leu, ParC Ser80Arg | pBK-QnrA1 | I | 2 | 2 | 2 | 8 | 4 | 4 | 4 | 64 |
| ATCC-S83L-S80R/qnrB | GyrA Ser83Leu, ParC Ser80Arg | pBK-QnrB1 | S | 1 | 1 | 1 | 4 | 2 | 1 | 2 | 4 |
| ATCC-S83L-S80R/qnrS | GyrA Ser83Leu, ParC Ser80Arg | pBK-QnrS1 | I | 2 | 4 | 2 | 8 | 4 | 4 | 4 | 16 |
PMQR, plasmid-mediated quinolone resistance.
S and I, susceptible and intermediate susceptibility, respectively, according to CLSI guidelines (2).
MICs determined by microdilution for ciprofloxacin (CIP), levofloxacin (LVX), moxifloxacin (MXF), and norfloxacin (NFX).
TABLE 2.
Oligonucleotides and plasmids used in this study
| Primer or plasmid | Sequencea | Use in this study | Source or reference |
|---|---|---|---|
| qnr cloning | |||
| Pre-QnrA1 | 5′-CGGGATCCCGCGGCAGTTAAAATTGGGGCT-3′ | Cloning of qnrA1 | This study |
| Post-QnrA1 | 5′-CGGGATCCCGACGCCGAGTCCCGACCAGACTGC-3′ | Cloning of qnrA1 | This study |
| Pre-QnrB1 | 5′-CGGGATCCCGCTTGGTCGCCCTGGCCAACC-3′ | Cloning of qnrB1 | This study |
| Post-QnrB1 | 5′-CGGGATCCCGGCAAACCAGCTTACAGCAGGC-3′ | Cloning of qnrB1 | This study |
| Pre-QnrS1 | 5′-CGGGATCCCGCCACTTAAAACAGGTAAATTG-3′ | Cloning of qnrS1 | This study |
| Post-QnrS1 | 5′-CGGGATCCCGTACATGGTTGTCCCTATGTC-3′ | Cloning of qnrS1 | This study |
| Gene replacement | |||
| gyrAS83L-Fw | 5′-CCATGGTGACCTGGCGGTCTATG-3′ | Mutagenesis of gyrA | This study |
| gyrAS83L-Rv | 5′-CATAGACCGCCAGGTCACCATGG-3′ | Mutagenesis of gyrA | This study |
| parCS80R-Fw | 5′-CCGCACGGCGATCGCGCCTGTTATGAAGC-3′ | Mutagenesis of parC | This study |
| parCS80R-Rv | 5′-GCTTCATAACAGGCGCGATCGCCGTGCGG-3′ | Mutagenesis of parC | This study |
| Pre-gyrAS83 | 5′-CGGGATCCCGAGCGATCTCTTCGTGGTCTACG-3′ | Partial gyrA amplification | This study |
| Post-gyrAS83 | 5′-CGGGATCCCGCCTGATACGGAATTTCGTGGAC-3′ | Partial gyrA amplification | This study |
| Pre-parCS80 | 5′-CGGGATCCCGGACCGCGATAGCGTTGTCTTCCG-3′ | Partial parC amplification | This study |
| Post-parCS80 | 5′-CGGGATCCCGCAGATCGGTGGTAGCGAAGAGGTG-3′ | Partial parC amplification | This study |
| QRDRb sequencing | |||
| gyrA-1 | 5′-AAATCTGCCCGTGTCGTTGGT-3′ | Sequencing | 17 |
| gyrA-2 | 5′-GCCATACCTACGGCGATACC-3′ | Sequencing | 17 |
| parC-A | 5′-CTGAATGCCAGCGCCAAATT-3′ | Sequencing | 17 |
| parC-B | 5′-GCGAACGATTTCGGATCGTC-3′ | Sequencing | 17 |
| Plasmids | |||
| pBK-CMV | Cloning vector | ||
| pST76C | Gene replacement/suicide vector | ||
| pUC19RP12 | Gene replacement/resolution vector |
Underlined nucleotides correspond to the BamHI site used for cloning.
QRDR, quinolone resistance-determining region.
Susceptibility tests were performed in duplicate for each bacterial strain by the broth microdilution method according to CLSI reference methods (2). The presence of any qnr gene increased MIC levels in all E. coli genotypes. The ciprofloxacin (CIP) MIC for E. coli ATCC 25922 harboring any qnr gene was 0.125 μg/ml, which is more than 62-fold higher than that for the empty wild-type strain (Table 1). The expression of qnr genes in the E. coli ATCC-Ser83Leu strain gave a less marked increase, with CIP MICs of 0.5, 0.5, and 1 μg/ml, meaning 4-, 4-, and 8-fold increases in expression for qnrA1, qnrB1, and qnrS1, respectively. The Ser80Arg substitution in ParC played a secondary role in FQ resistance (Table 1), as previously described (7, 12). In E. coli ATCC-Ser83Leu-Ser80Arg, the presence and expression of qnrA1 or qnrS1 genes increased the CIP MIC to 2 μg/ml (or intermediate susceptibility according to CLSI guidelines) (Table 1) (2). Isogenic strains containing the qnrB1 gene were always susceptible to FQ according to CLSI breakpoints, including the double-topoisomerase mutant (Table 1). qnrB1 seems to be slightly less efficient than qnrA1 and qnrS1 in terms of MIC values. Minimal bactericidal concentrations (MBCs) were similar to the corresponding MIC values.
MPC was determined as described previously by Marcusson et al. (8, 16). The presence of qnr genes increased MPC values of FQ in all E. coli genotypes. The MPC values of CIP in the wild-type E. coli ATCC 25922 strain coding for Qnr proteins as the only quinolone resistance mechanism increased 8-, 2-, and 4-fold for qnrA1, qnrB1, and qnrS1, respectively, compared to E. coli ATCC 25922 (Table 3). A similar increase was observed for the other quinolones. The presence of a single Ser83Leu mutation in the gyrA gene raised MPC values 2- to 4-fold, compared to those for wild-type E. coli ATCC 25922. The additional presence of qnr genes in E. coli ATCC-Ser83Leu increased MPC values to 8 to 32 μg/ml, depending on the FQ (Table 3). E. coli ATCC-Ser83Leu-Ser80Arg MPC values ranged from 4 to 32 μg/ml. E. coli ATCC-Ser83Leu-Ser80Arg expressing qnr genes showed MPC values ranging from 32 to 128 μg/ml (except for qnrB1, for which values ranged from 8 to 32 μg/ml), well in excess of the breakpoint concentrations of CLSI guidelines (data not shown). The effect of qnr genes on MPC was similar to the presence of a Ser83Leu substitution in GyrA as the single quinolone resistance mechanism. MPC concentrations were clearly higher than the maximum serum concentrations obtained when using drugs in antimicrobial therapy for E. coli ATCC 25922-Ser83Leu expressing qnr genes (6, 19). The presence of quinolone resistance mechanisms produced a reduction in the mutant selection window (MSW). It is therefore difficult to predict MPC from the MIC values, and on this basis, the MPC will vary according to FQ and the specific resistance mechanism involved (Table 3).
TABLE 3.
Fluoroquinolone MPC, MSW, MPC time window, and MIC or MIC range for mutants of the eight isogenic strains used in this study
| E. coli strain/plasmid gene | MPC (μg/ml)a |
MSWb (MPC/MIC [μg/ml]) |
MPC time window (h)c |
MIC or MIC range for mutant (μg/ml)d |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CIP | LVX | MXF | NFX | CIP | LVX | MXF | NFX | CIP | LVX | MXF | NFX | CIP | LVX | MXF | NFX | |
| ATCC 25922 | 1 | 2 | 2 | 4 | 500 | 250 | 250 | 266.7 | 48 | 48 | 48 | 48 | 0.06 | 0.125 | 0.06 | 0.25 |
| ATCC/qnrA | 8 | 8 | 8 | 8 | 64 | 16 | 32 | 16 | 48 | 24 | 24 | 24 | 0.5 | 1-2 | 0.5-1 | 1-2 |
| ATCC/qnrB | 2 | 4 | 4 | 8 | 16 | 32 | 16 | 32 | 72 | 48 | 24 | 48 | 0.125 | 0.5 | 0.5-1 | 0.5-1 |
| ATCC/qnrS | 4 | 4 | 4 | 8 | 32 | 8 | 8 | 16 | 24 | 24 | 24 | 24 | 0.5-1 | 1-2 | 0.5-1 | 1-2 |
| ATCC 25922-S83L | 4 | 4 | 4 | 8 | 32 | 32 | 66.7 | 32 | 24 | 24 | 24 | 24 | 0.5 | 2 | 1 | 2 |
| ATCC-S83L/qnrA | 16 | 32 | 16 | 16 | 32 | 64 | 64 | 16 | 24 | 24 | 24 | 24 | 1-4 | 2-4 | 2-4 | 8-16 |
| ATCC-S83L/qnrB | 8 | 16 | 32 | 16 | 16 | 64 | 64 | 16 | 24 | 72 | 24 | 24 | 1-2 | 1-4 | 1-4 | 2-16 |
| ATCC-S83L/qnrS | 8 | 16 | 32 | 32 | 8 | 16 | 32 | 16 | 24 | 96 | 24 | 24 | 1-2 | 4 | 1-2 | 2-16 |
MPC values were determined on Mueller-Hinton plates for ciprofloxacin (CIP), levofloxacin (LVX), moxifloxacin (MXF), and norfloxacin (NFX); MPC was defined as the lowest antibiotic concentration (in the range of concentration steps analyzed) at which resistant colonies do not form.
MSW, mutant selection window (i.e., the antibiotic concentration found between the MIC and MPC).
Earliest time (in hours) at which resistant colonies were visible one step below the MPC.
MICs for resistant colonies were recovered on Mueller-Hinton plates one step below the MPC value.
Mutants were recovered from the plated concentrations closest to the MPC value at a very low frequency. The quinolone resistance-determining region (QRDR) of target genes gyrA and parC was analyzed. All of the characterized mutants of the E. coli ATCC 25922 strain had just a Ser83Leu substitution in the QRDR of gyrA, supporting the view that this is the most frequent modification in E. coli. On the other hand, most colonies of the E. coli ATCC-Ser83Leu strain selected in the MSW showed additional modifications in the QRDR of the parC gene (Gly78Asp or Ser80Ile substitutions), also previously associated with quinolone resistance. Clinical FQ resistance according to CLSI guidelines (2) (MIC of ≥4 μg/ml for CIP) was observed for some of these mutants (Table 3). The PMQR might enable mutant bacteria with low levels of FQ resistance to survive long enough for them to grow and emerge during FQ treatment. The detection of mutations in type II topoisomerase genes reflects the ability of this mechanism to select for mutants with higher quinolone resistance. In vivo selection of FQ-resistant Enterobacteriaceae expressing qnr genes has been reported (3, 13). With respect to bacterial survival, although some bacteria did survive the MPC for a 96-h extended period, no quinolone-resistant mutants were selected, these being a persistent phenotype and indicating that the MPC parameter was working as specified (Table 3) (8).
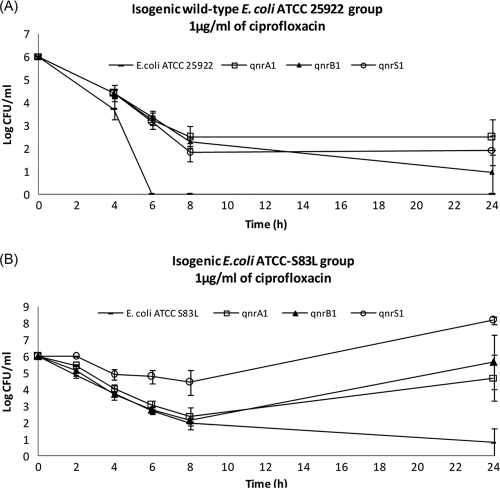
Killing-curve assays showed a selective advantage for survival at 1 μg/ml of CIP in strains expressing qnr genes, both with and without the Ser83Leu substitution in GyrA in E. coli (Fig. 1). This CIP concentration defines the limit for establishing susceptibility or intermediate susceptibility (according to CLSI guidelines) in Enterobacteriaceae (2). CIP at 1 μg/ml in the isogenic wild-type E. coli strain ATCC 25922 (with or without qnr gene expression [Fig. 1A]) caused a marked reduction in viable bacteria after 8 h of incubation. After 6 h, no viable bacteria were recovered for the wild-type E. coli ATCC 25922 strain without qnr genes, while E. coli ATCC 25922 with qnr gene expression maintained levels of 102 to 103 CFU/ml for up to 24 h. At 4× the MIC of CIP, all qnr gene expression in wild-type E. coli ATCC 25922 maintained a viable CFU/ml at least 100-fold higher at 24 h compared to empty wild-type strains (data not shown). CIP at 1 μg/ml in the isogenic E. coli ATCC-Ser83Leu strain (with and without qnr gene expression [Fig. 1B]), caused a marked reduction in viable bacteria during the first 8 h, except for strains expressing qnrS1. After 8 h, bacterial regrowth was noted for strains expressing qnrA1, qnrB1, and qnrS1, and this continued up to 24 h, although not for E. coli ATCC-Ser83Leu, demonstrating the impact of qnr genes in terms of bacterial viability.
FIG. 1.
Viable bacterial counts in time-kill curve assays with ciprofloxacin (CIP) 1 μg/ml. (A) Isogenic wild-type E. coli ATCC 25922, with and without qnrA1, qnrB1, or qnrS1 gene expression; (B) isogenic mutant E. coli ATCC 25922-S83L, with and without qnrA1, qnrB1, or qnrS1 gene expression.
Finally, we evaluated 16 isogenic E. coli strains harboring different QRDR modifications, and with and without qnr-expressing genes such as qnrA1, qnrB1, and qnrS1. This study showed that these mechanisms, implicated in low-level plasmid-mediated FQ resistance, may play a significant role in the acquisition of clinical resistance to FQ and, therefore, therapeutic failure. Animal models are necessary to confirm these in vitro results.
Acknowledgments
This work was supported by the Ministerio de Sanidad y Consumo, Instituto de Salud Carlos III (project PI060580) and the Consejería de Innovación Ciencia y Empresa, Junta de Andalucía (P07-CTS-02908), Spain. It was partly supported by the Ministerio de Sanidad y Consumo, Instituto de Salud Carlos III-FEDER, Spanish Network for Research in Infectious Diseases (REIPI RD06/0008). A.B. was funded by a predoctoral grant from the Instituto de Salud Carlos III (PFIS), Spain.
Footnotes
Published ahead of print on 20 December 2010.
REFERENCES
- 1.Allou, N., E. Cambau, L. Massias, F. Chau, and B. Fantin. 2009. Impact of low-level resistance to fluoroquinolones due to qnrA1 and qnrS1 genes or a gyrA mutation on ciprofloxacin bactericidal activity in a murine model of Escherichia coli urinary tract infection. Antimicrob. Agents Chemother. 53:4292-4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clinical and Laboratory Standards Institute. 2010. Performance standards for antimicrobial susceptibility testing: nineteenth informational supplement M100-S20. CLSI, Wayne, PA.
- 3.de Toro, M., et al. 2010. In vivo selection of aac(6′)-Ib-cr and mutations in the gyrA gene in a clinical qnrS1-positive Salmonella enterica serovar Typhimurium DT104B strain recovered after fluoroquinolone treatment. J. Antimicrob. Chemother. 65:1945-1949. [DOI] [PubMed] [Google Scholar]
- 4.Drlica, K., and X. Zhao. 2007. Mutant selection window hypothesis updated. Clin. Infect. Dis. 44:681-688. [DOI] [PubMed] [Google Scholar]
- 5.Hooper, D. C. 2001. Emerging mechanisms of fluoroquinolone resistance. Emerg. Infect. Dis. 7:337-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lipman, J., J. Scribante, A. G. Gous, H. Hon, and S. Tshukutsoane. 1998. Pharmacokinetic profiles of high-dose intravenous ciprofloxacin in severe sepsis. The Baragwanath Ciprofloxacin Study Group. Antimicrob. Agents Chemother. 42:2235-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marcusson, L. L., N. Frimodt-Moller, and D. Hughes. 2009. Interplay in the selection of fluoroquinolone resistance and bacterial fitness. PLoS Pathog. 5:e1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcusson, L. L., S. K. Olofsson, L. P. Komp, O. Cars, and D. Hughes. 2005. Mutant prevention concentrations of ciprofloxacin for urinary tract infection isolates of Escherichia coli. J. Antimicrob. Chemother. 55:938-943. [DOI] [PubMed] [Google Scholar]
- 9.Martinez-Martinez, L., C. M. Eliecer, J. Manuel Rodriguez-Martinez, J. Calvo, and A. Pascual. 2008. Plasmid-mediated quinolone resistance. Expert Rev. Anti Infect. Ther. 6:685-711. [DOI] [PubMed] [Google Scholar]
- 10.Martinez-Martinez, L., A. Pascual, I. Garcia, J. Tran, and G. A. Jacoby. 2003. Interaction of plasmid and host quinolone resistance. J. Antimicrob. Chemother. 51:1037-1039. [DOI] [PubMed] [Google Scholar]
- 11.Martinez-Martinez, L., A. Pascual, and G. A. Jacoby. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797-799. [DOI] [PubMed] [Google Scholar]
- 12.Morgan-Linnell, S. K., and L. Zechiedrich. 2007. Contributions of the combined effects of topoisomerase mutations toward fluoroquinolone resistance in Escherichia coli. Antimicrob. Agents Chemother. 51:4205-4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poirel, L., et al. 2006. In vivo selection of fluoroquinolone-resistant Escherichia coli isolates expressing plasmid-mediated quinolone resistance and expanded-spectrum beta-lactamase. Antimicrob. Agents Chemother. 50:1525-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Posfai, G., V. Kolisnychenko, Z. Bereczki, and F. R. Blattner. 1999. Markerless gene replacement in Escherichia coli stimulated by a double-strand break in the chromosome. Nucleic Acids Res. 27:4409-4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez-Martinez, J. M., et al. 2008. Activity of ciprofloxacin and levofloxacin in experimental pneumonia caused by Klebsiella pneumoniae deficient in porins, expressing active efflux and producing QnrA1. Clin. Microbiol. Infect. 14:691-697. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez-Martinez, J. M., et al. 2007. Mutant prevention concentrations of fluoroquinolones for Enterobacteriaceae expressing the plasmid-carried quinolone resistance determinant qnrA1. Antimicrob. Agents Chemother. 51:2236-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez-Martinez, J. M., C. Velasco, A. Pascual, I. Garcia, and L. Martinez-Martinez. 2006. Correlation of quinolone resistance levels and differences in basal and quinolone-induced expression from three qnrA-containing plasmids. Clin. Microbiol. Infect. 12:440-445. [DOI] [PubMed] [Google Scholar]
- 18.Strahilevitz, J., G. A. Jacoby, D. C. Hooper, and A. Robicsek. 2009. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin. Microbiol. Rev. 22:664-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turnidge, J. 1999. Pharmacokinetics and pharmacodynamics of fluoroquinolones. Drugs 58(Suppl. 2):29-36. [DOI] [PubMed] [Google Scholar]
- 20.Zhao, X., and K. Drlica. 2008. A unified anti-mutant dosing strategy. J. Antimicrob. Chemother. 62:434-436. [DOI] [PMC free article] [PubMed] [Google Scholar]