Abstract
Oxidative stress leads to membrane lipid peroxidation, which yields products causing variable degrees of detrimental oxidative modifications in cells. Reactive oxygen species (ROS) are the key regulators in this process and induce lipid peroxidation in Escherichia coli. Application of nonthermal (cold) plasma is increasingly used for inactivation of surface contaminants. Recently, we reported a successful application of nonthermal plasma, using a floating-electrode dielectric-barrier discharge (FE-DBD) technique for rapid inactivation of bacterial contaminants in normal atmospheric air (S. G. Joshi et al., Am. J. Infect. Control 38:293-301, 2010). In the present report, we demonstrate that FE-DBD plasma-mediated inactivation involves membrane lipid peroxidation in E. coli. Dose-dependent ROS, such as singlet oxygen and hydrogen peroxide-like species generated during plasma-induced oxidative stress, were responsible for membrane lipid peroxidation, and ROS scavengers, such as α-tocopherol (vitamin E), were able to significantly inhibit the extent of lipid peroxidation and oxidative DNA damage. These findings indicate that this is a major mechanism involved in FE-DBD plasma-mediated inactivation of bacteria.
Nonthermal (cold) dielectric-barrier discharge (DBD) atmospheric-pressure plasma is widely under investigation for use as an alternative sterilization and disinfection method in the fields of biology and medicine. Most recently, we demonstrated that Escherichia coli, Staphylococcus aureus, and methicillin-resistant Staphylococcus aureus in both their planktonic form and in biofilms are rapidly inactivated by nonthermal DBD plasma using a floating-electrode technique (11). Complete inactivation of E. coli was seen in less than 120 s when E. coli was present in its planktonic form, and complete inactivation occurred in about 180 s when it was in the biofilm form, making this technique attractive for sterilization processes. E. coli is one of the most common Gram-negative bacterial contaminants responsible for hospital-acquired infections (HAI) and one of the most widely studied organisms in the laboratory and therefore is a good choice to track various oxidative-stress pathways.
A DBD plasma-generating probe is an apparatus that generates microsecond-long, high-voltage-pulsed cold plasma between the primary electrode covered with a quartz surface and the surface of the biological sample, which serves as a second electrode. The high-voltage electrode is completely covered with a dielectric barrier, which makes it safe for sterilization applications, and the nature of the applied microsecond pulses do not elevate the surface temperature above 28°C. In the floating-electrode DBD (FE-DBD) plasma setup, the second electrode (biological sample) is not grounded and remains at a floating potential. Discharge ignites when the powered electrode approaches the surface to be treated at a distance (discharge gap) less than about 3 mm, depending on the form, duration, and polarity of the driving voltage, and it is safe to apply to human or animal skin and delicate surfaces which are likely to be damaged by thermal (hot) plasma. In thermal plasma, argon is used, and in this instance, the local temperature is increased significantly. Other DBD plasmas can be generated either in special gas flow or vacuum, and the sample of interest is held between the two electrodes (5, 11) (see reference 5 for details). Thus, the FE-DBD plasma technique offers better potential for diversity, since it works in room air (normal atmospheric pressure) and is cold to touch.
Nonthermal plasma at normal atmospheric pressure (room air) generates many physical and chemical active species when applied to biological samples using the floating-electrode (FE) technique. Some of the species being characterized in our laboratories are ozone, hydrogen peroxide, singlet oxygen, superoxide, hydroxyl radical, nitric oxide, and UV (5). All or most of these species are capable of causing oxidative damage to bacteria, and if the damage is extensive (beyond the capacity of the cellular repair machinery), it will lead to cell death. There are many speculative reports supporting this mechanism, and although the exact changes occurring are not yet known, the involvement of reactive oxygen species (ROS) and oxidative damage specific to proteins, lipid layers, or DNA have been observed. Furthermore, the FE-DBD plasma application technique is relatively new, and plasma-generated species interact directly with the biological sample of interest. Therefore, it is anticipated that ROS generated during plasma treatment might be sufficient to activate lipid peroxidation and amplify oxidative damage to DNA, initiating cell death. In order to optimize plasma devices further to increase their efficacy for sterilization, a deeper understanding of this complex mechanism is required. Four major reactive oxygen species, superoxide, hydrogen peroxide, hydroxyl radical, and ozone, are possibly generated during nonthermal plasma discharge in air; all four are cytotoxic when they come in contact with bacterial cells or are internalized, and they display different kinetics and variable levels of severity (5, 11). We undertook this study to investigate ROS as the mechanism responsible for bacterial cell killing by FE-DBD plasma.
During preliminary experiments, we observed that E. coli mounts an oxidative stress response to FE-DBD plasma treatment and that antioxidants were able to inhibit it and prevent damage and cell death. In the present report, we demonstrate that the plasma-induced oxidative stress causes severe morphological changes in E. coli and leads to compromise of the cell membrane integrity and membrane lipid peroxidation. The lipid peroxidation was proportional to the amount of DNA damage during this oxidation, and the ROS scavenger, α-tocopherol (vitamin E), was able to significantly inhibit membrane lipid peroxidation and oxidative DNA damage in this cascade (2, 17, 28, 29).
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasma treatment.
A standard strain of E. coli, ATCC 25922, was used for all experiments. E. coli was grown aerobically at 37°C in Luria-Bertani (LB) medium to mid-exponential phase (optical density at 600 nm [OD600] of 0.2) and used for the experiments.
The microsecond-pulse plasma-generating device was used for nonthermal dielectric-barrier discharge (DBD) plasma generation in normal room air. The device was locally fabricated in our laboratory at the A. J. Drexel Plasma Institute of Drexel University. A floating-electrode (FE)-DBD technique was used to apply plasma (5, 6, 11). A detailed description of the plasma electrode configuration and operating system has been published previously (5, 6). The plasma generator was set at 15 kV and 500-Hz frequency (0.13-W/cm2 power) for all experiments, and the biological sample (cell suspension in phosphate-buffered saline [PBS]) of interest was directly exposed to plasma, which was generated at the gap (air) between the interface of the plasma probe surface and the upper surface of the sample. The distance between the probe and sample surface was kept at 2.0 mm. No special gas or airflow was used, and the plasma was created at normal atmospheric pressure. The samples were exposed to plasma for 0 (0), 3 (0.39), 6 (0.78), 12 (1.56), 15 (1.95), 24 (3.12), 30 (3.9), 60 (7.8), 90 (11.7), 120 (15.6), and 180 (23.4) seconds (the plasma energy in J/cm2 is shown in parentheses), depending upon the type of experiment.
Inactivation of E. coli and colony assay.
Colony count assays were used to demonstrate kinetic killing curves and the antimicrobial effects of a given compound or device. In brief, 1-ml samples of the culture were centrifuged at 6,000 rpm for 10 min to remove interference from extracellular protein and washed three times with cold PBS. The cell suspension in PBS was used for inactivation studies. Aseptically, 65 μl of cell suspension was transferred to a monoconcavity glass slide and spread in the cavity. The cavity slide was put on the mechanical stage, and a plasma probe holding a clamp was set over the sample at a fixed distance of 2 mm. The sample was either unexposed or exposed to plasma for a given time, and then it was immediately diluted appropriately (with PBS [pH 7.2]) and spread over Trypticase soy agar (TSA) and incubated at 37°C for 18 to 24 h. The plates with no growth were incubated for up to 72 h and checked for the presence of colonies every 24 h. The percentage of surviving cells was calculated from the total count of CFU. For percent calculations, the starting bacterial cell concentration was set at 100%. The bacterial cell density-dependent response to plasma was studied in a similar fashion; only the starting cell density was varied.
Plasma-induced morphological changes in E. coli cells.
To determine whether plasma treatment induces cellular stress and leads to cell morphological changes, the cells were either untreated or treated with plasma and then studied using two different techniques. In the first technique, smears were prepared after plasma treatments, Gram stained, and examined by bright-field microscopy (1). In the second technique, to ascertain whether morphologically compromised cells were dead or dormant, we performed an XTT [2,3-bis-(methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide] assay, which measures respiration (9, 11, 16).
Quantification of viable cells by the XTT assay.
XTT reagent solutions were prepared, and the XTT assay was performed as described previously (11, 16) after appropriate plasma treatment or no treatment. XTT metabolic products were measured in the wells of a 96-well plate by reading absorption at 492 nm using a Synergy Mx multimode microtiter plate reader (BioTek, Winooski, VT). The readings were normalized, and the percentages of surviving cells were calculated by comparison to untreated samples.
Influence of antioxidants on cells exposed to plasma-induced stress.
A panel of known antioxidants in bacteriology was used to demonstrate their protective effect against plasma-mediated stress. Glutathione, ascorbic acid, histidine, mannitol, sodium pyruvate (all from Sigma Chemical Co.), and a water-soluble formulation of vitamin E (α-tocopherol derivative) (Eastman Chemical Co., United Kingdom) were freshly prepared in PBS (pH 7.2), filter sterilized (0.22-μm filter), stored, and used at a final concentration of 10 mM. The experiments were carried out using three techniques. In the first technique, the E. coli cells were washed with PBS three times and suspended in antioxidant during the plasma treatment. In the second technique, the cells were exposed first to plasma, and then the antioxidant was added. In the third technique, the cells were preincubated with the antioxidant for 30 min at room temperature and then subsequently treated with plasma. After the plasma treatments, the cells were appropriately diluted, and colony assays were performed.
Live/Dead BacLight bacterial viability assay.
The BacLight bacterial viability (Live/Dead) assay kit is routinely used to detect the efficacy of bactericidal agents, and the Live/Dead BacLight bacterial viability assay was performed as described previously (11, 19). The sample tubes were incubated in the dark at room temperature for 2 h and centrifuged at 6,000 rpm for 8 min. The supernatant was collected separately, and the cell pellet was loosened to smear on a clean glass slide and viewed using a Leica DMRX fluorescence microscope with attached Leica DG300FX digital camera system, using fluorescein and Texas Red band-pass filters (for corresponding SYTO9 green dye and propidium iodide). The images were captured from five randomly selected areas for three different sets of experiments, saved as TIFF files, and edited using Adobe Photoshop CS3 for analysis.
Detection of reactive oxygen species during plasma treatment.
To test for reactive oxygen species in solutions and bacterial cell suspensions, we used three different techniques. In the first technique, a commercially available singlet oxygen sensor green reagent (SOSGR) (Molecular Probes) was used. Unlike other fluorescent and chemiluminescent singlet oxygen reagents available, this reagent is highly selective for 1O2 and does not show any appreciable response to hydroxyl radical (OH) or superoxide (O2−) (3, 4, 18). A 1 mM stock solution was freshly prepared in methanol (aliquoted and stored at −20°C in amber-colored microtubes), and a final working concentration of 1 μM (i.e., 1:1,000 dilutions) was used in reaction solution after optimization. In the sample where this was omitted, an equal volume of methanol in PBS was added. A cell suspension of known density was treated with plasma over time, and this reagent was immediately added to reaction solution and held for 15 min in dark at room temperature. Non-plasma-treated cells and cells treated with H2O2 were used as negative and positive controls, respectively. The suspension was then centrifuged at 8,000 rpm for 8 min, and the supernatant was transferred to the wells of a black (bottom opaque) 96-well plate for fluorescent multimode plate reading (BioTek) and read at excitation/emission maxima of 485/530 nm. In the presence of singlet oxygen, it emits a bright green fluorescence similar to that of fluorescein. In a parallel experiment, the cell pellet was suspended in 20 μl of PBS and smeared on a clean glass slide and examined with a fluorescence microscope (Leica microscope, as mentioned above) using the appropriate fluorescein band-pass filters. The graphs were generated after background subtractions.
In the second technique, the experiment was carried out as described above, but a highly specific ultrasensitive fluorescent probe, namely, Amplex UltraRed reagent (AUR) (Molecular Probes) was used. AUR is brightly fluorescent (emission/excitation spectra of 530 nm/590 nm) and specifically sensitive to assay H2O2 (23, 27). After incubation and centrifugation as described above, the supernatant and pellet cells were assayed separately, using excitation/emission spectra of 530/590 nm in the fluorescence microtiter plate reader, and the signals were calculated as arbitrary fluorescence units (AFU) after the readings were normalized. In the third technique, in some of the experimental conditions, bovine catalase, which is used to decay hydrogen peroxide (catalog no. C1345; Sigma) was preincubated with E. coli cells for 15 min at room temperature, and then the cells were exposed to plasma treatments. Based on optimization from the stock of 6 mg ml−1 of catalase solution, a working solution diluted 1:2,000 (final) was used. After plasma treatments, the treated cells were appropriately diluted with PBS, and the colony count assay was performed as described above. In a parallel experiment, non-plasma-treated or hydrogen peroxide-treated samples were run as negative and positive controls.
Measurement of membrane potential.
To determine whether plasma treatment induces a drop in the membrane potential, we exposed cells suspended in PBS to plasma treatments over time essentially as described above and immediately added a freshly prepared slow-response potential-sensitive probe, DiSBAC2(3) [bis-(1,3-diethylthiobarbituric acid)trimethine oxonol] (Molecular Probes) (final concentration of 200 μM) following the manufacturer's directions. Nontreated cells or cells treated with H2O2 and 70% ethanol were included as negative and positive controls, respectively. Potential-dependent fluorescence changes are typically ∼1% per mV when measured at 530/560 nm. This fluorescent probe is highly specific for decreased membrane potential. The probe enters depolarized cells and binds to intracellular proteins or the inner membrane and exhibits enhanced fluorescence and a red spectral shift (20, 21).
Detection of involvement of membrane lipid peroxidation.
Lipid peroxides are the unstable indicators of oxidative stress in animal, plant, and microbial cells that decompose to form fairly complex and reactive compounds, such as malondialdehyde (MDA) and 4-hydroxynonenal (4-HNE), and the process of lipid peroxidation is well defined and reported to cause cellular damage (17, 29). The thiobarbituric acid-reactive substance (TBARS) assay is a well-established assay for monitoring and screening lipid peroxidation. MDA forms a 1:2 adduct with thiobarituric acid (TBA), and can be measured colorimetrically or fluorometrically using an MDA equivalent standard. We detected the appearance of MDA in E. coli cells and isolated membrane-rich fraction of E. coli cells during plasma treatment over time using the OxiSelect TBARS assay kit (Cell Biolabs, San Diego, CA) following the manufacturer's instructions. A nonqualitative method of membrane fraction collection was used, wherein E. coli cells were subjected to sonication in PBS as described previously (32) and centrifuged at 6,000 rpm at 4°C for 15 min in order to separate unbroken cells, and the supernatant was centrifuged again at a high speed of 20,000 rpm at 4°C for 120 min to obtain a membrane-rich fraction at the bottom of the tube. The supernatant was discarded, and the membrane-rich deposit was resuspended in PBS to treat with plasma as described above.
Detection of DNA damage in E. coli cells.
DNA damage was detected by two different methods. In the first method, agarose gel electrophoresis was used to demonstrate physical disintegration of DNA (29, 33), and in the second method, an enzyme-linked immunosorbent assay (ELISA) technique was utilized to demonstrate oxidative DNA damage markers during plasma treatment. E. coli cell suspensions were treated with plasma as described above, and the samples were processed by established methods as described above. Equal amounts of DNA samples were subjected to agarose electrophoresis, and ethidium bromide-stained DNA gels were photographed using a UV transillluminator Gel Doc system.
The oxidative DNA damage ELISA kit (Cell Biolabs) is a competitive enzyme immunoassay available for rapid detection and quantification of 8-hydroxydeoxyguanosine (8-OHdG), a ubiquitous marker of oxidative stress and a by-product of oxidative DNA damage from cellular DNA samples (17, 29). The quantity of 8-OHdG in an unknown sample is determined by comparing its absorption with that of a known standard curve. The DNA was isolated and purified from 1 ml of plasma-treated or untreated sample using a bacterial DNA isolation kit (Promega Corporation, Madison, WI).
Statistical analysis.
Data sets were analyzed using Microsoft Excel and verified using GraphPad Prism 4 and GraphPad InStat 3 (San Diego, CA). The P values are derived against corresponding untreated conditions, unless and otherwise stated, and a P value of <0.05 is considered significant. The experiments are repeated a minimum of three times unless stated otherwise, and data are means ± standard deviations (SDs).
RESULTS
The floating-electrode dielectric-barrier discharge (FE-DBD) technique rapidly inactivates E. coli in a concentration-dependent manner.
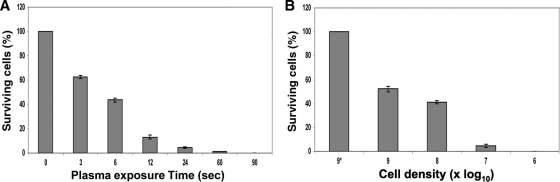
The bacterial colony count assay is the gold standard in determining antibacterial efficacies. Figure 1 is a graphical representation of the percentages of surviving cells and shows a rapid inactivation of E. coli around 60 s (and <90 s) of exposure time to the plasma treatment, and this phenomenon is plasma dose dependent (Fig. 1A). To see the effect of biomass on plasma's antimicrobial efficacy, we conducted E. coli cell density-dependent experiments. Figure 1B shows that 30 s of plasma treatment inactivates ∼95% of 107 E. coli cells and causes complete inactivation (sterilization) when 106 cells are treated.
FIG. 1.
Colony count assay, the gold standard in bacteriology, shows that plasma inactivates E. coli rapidly upon exposure (at 7 log10), and the antimicrobial effect is plasma dose (energy) dependent (A) and bacterial cell density dependent (B). In panel B, the bar labeled 9* shows the percentage of surviving cells in cells not treated with plasma treatment (for 9 log10). The values are means ± standard deviations (SDs) (error bars) for 3 separate experiments.
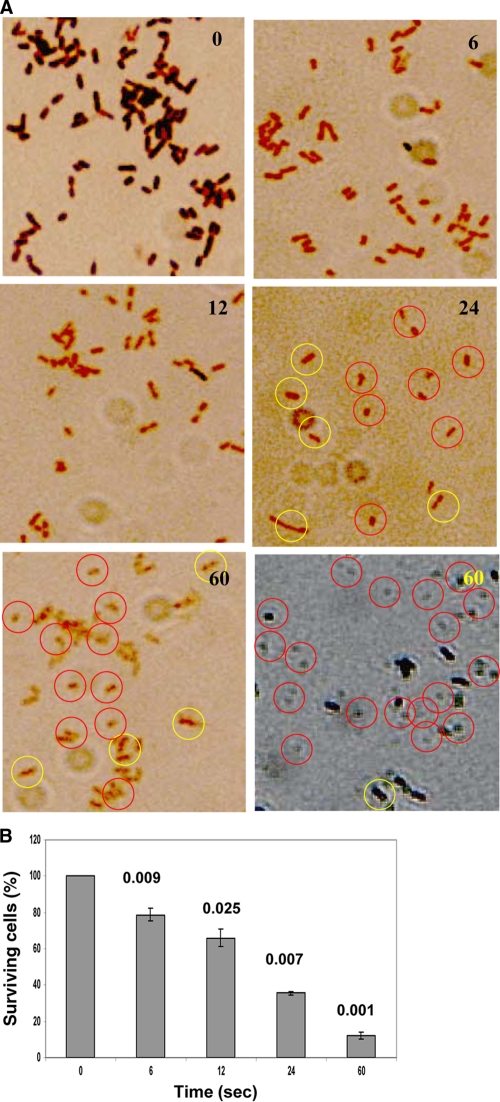
Plasma treatment causes morphological changes in E. coli that are peculiar to oxidative stress and eventual death.
By using bright-field microscopy, we observed two major changes. First, a change in the morphological features of E. coli from the typical bacillus form to the coccoid form; second, upon plasma treatments, E. coli showed loss of Gram stain-retaining capabilities over the length of time of exposure to plasma treatment. Both these features were plasma dose dependent. The bacillary cells became coccoid and smaller (shrunken) after 24 s and almost lost their classical Gram-staining features (Fig. 2A). These changes were consistent when the experiments were repeated and were obvious at longer treatment times. Therefore, in order to make the changes stand out, we overstained with Gram stain and viewed the cells with ×1,000 magnification, which revealed small, shrunken coccoid cells, which were not seen during normal staining (may be due to loss of stain) after 60 s of plasma treatment. In order to determine whether plasma treatment inactivates E. coli cells or merely causes transient changes in their morphology which can be repaired by cellular machinery, we performed the XTT assay concurrent with Gram staining, which demonstrated that the cells were being inactivated as we increased the plasma dose and not respiring and not dormant (Fig. 2B). A mere 30 s of plasma treatment resulted in about 90% inactivation of 108 E. coli cells at 500 Hz of plasma frequency (15 kV), and the P value was highly significant compared to untreated control (P < 0.001) (data not shown).
FIG. 2.
(A) Representative images showing the appearance of morphological changes peculiar to the Gram-staining response and E. coli cells upon increasing moderate plasma (500-Hz) exposure over time. The Gram-staining property of E. coli cells changes over time upon plasma exposure, indicative of membrane-associated and/or cytoplasmic pH changes, and the cells lose their stain-retaining property. This indicates chemical and/or physical changes in the cells (including the membrane), one of the features of oxidative changes. Cells shrink to reduce surface area and tend to become coccoid (black-and-white image in the left bottom corner of panel A has increased contrast to show the changes). The yellow and red circles show the healthy bacillary cells and damaged cells with coccoid/spherical appearance, respectively. Bright-field microscopy was done at a magnification of ×1,000. The numbers in the top right corners of the images in panel A show the time (in seconds) of exposure to plasma. (B) The XTT assay for E. coli (8 log10) was performed to determine whether the cells die in a dose-dependent manner or whether the cells are still respiring or are dormant. The treatment time (in seconds) is displayed on the x axis, and P values were calculated for the values compared to the values for the untreated condition (0 s). Unlike the colony assay, the XTT assay gives instant results for their survival status. The numbers above the bars indicate the P values comparing the values to the values for nontreated cells (0 s). Error bars shows the SDs.
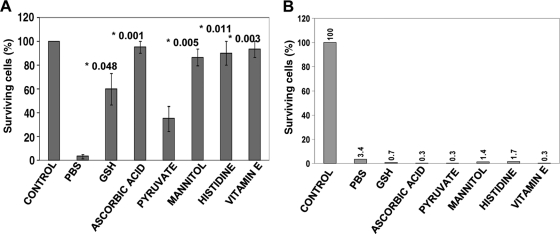
Antioxidants prevent plasma-induced E. coli inactivation.
Antioxidants are known to mediate protection from oxidative stress in bacteria. We used a battery of known antioxidants, including glutathione, ascorbic acid (vitamin C), sodium pyruvate, mannitol, histidine, and α-tocopherol (vitamin E). Pretreatment with these agents (final concentration of 10 mM) of 107 E. coli showed protection from plasma-induced oxidation in variable but significant proportions (Fig. 3A). Though all antioxidants performed well, ascorbic acid (vitamin C) and water-soluble α-tocopherol (vitamin E) could significantly protect E. coli from oxidation-mediated death by scavenging reactive oxygen species (ROS) (P values of <0.001 and <0.003, respectively, versus control without antioxidant [PBS]). In the colony count assay, the E. coli cells exposed to antioxidant after plasma treatment demonstrated no protective effect (Fig. 3B), possibly due to severe cellular damage. This showed that the cells may be inactivated quickly by oxidative stress beyond the capabilities of the cell's repair machinery.
FIG. 3.
Colony assays demonstrating the effects of antioxidants on E. coli survival during plasma treatment. (A) Effects of antioxidants on E. coli survival during plasma treatment in the presence of antioxidants (in PBS). The plasma settings were 500 Hz (3.12 J/cm2) for 24 s. The control was not treated with plasma. All other bars show 24 s of exposure in PBS or PBS with antioxidant. The numbers above the bars and preceded by an asterisk are the P values comparing the value to the value for plasma treatment (in PBS) without antioxidant (n = 3). (B) Exposure to antioxidants after plasma treatment does not protect E. coli cells from death, and the results show that plasma causes severe and nonrepairable damage to bacterial cells. The settings and parameters are the same as in panel A.
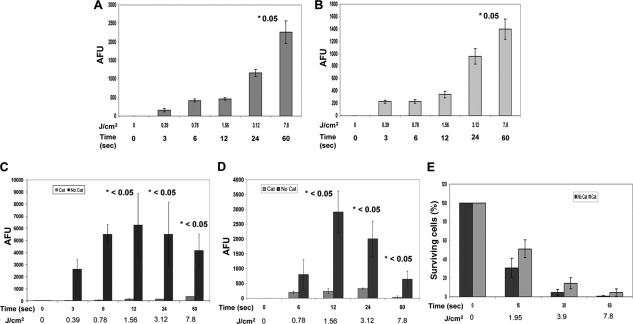
Plasma stress generates reactive oxygen species.
Plasma treatment generated ROS in PBS in a dose-dependent manner. Figure 4A demonstrates that when the fluid (PBS) medium is treated with plasma, ROS are generated as detected with a specific fluorescent probe for singlet oxygen (1O2). The plasma treatment-generated 1O2-like species in PBS was also detected in E. coli cells (Fig. 4B) when a cell suspension was treated and then centrifuged to separate the fluid component (PBS) from the cell pellet. The initial fluorescing units were subtracted from all treatment conditions to normalize the values, and no treatment (0 J/cm2 and 0 s) is shown for comparison. In order to verify whether ROS generation also involves other widely reported species, such as hydrogen peroxide (H2O2), we conducted specific-enzyme-mediated scavenging experiments and probed with another specific hydrogen peroxide-detecting fluorescent probe (Amplex UltraRed). In a separate set of experiments, the catalase enzyme was added to PBS or cell suspensions after plasma treatment, which decreased the concentration of hydrogen peroxide significantly at each treatment time, compared to controls without catalase, demonstrating that hydrogen peroxide is generated by plasma treatment (Fig. 4C and D). In parallel experiments, colony count assays were performed on E. coli cells that were preincubated with catalase before plasma treatment; there were significantly more colonies from the catalase-containing tubes than from the tubes without catalase during plasma treatment (Fig. 4E). In both experiments, appropriate negative [no plasma treatment, but with enzyme(s)] or positive (H2O2) controls were used. Collectively, these findings suggest that singlet oxygen is generated in significant amounts and that the hydrogen peroxide is one of the major reactive species generated during plasma treatment.
FIG. 4.
Plasma treatment generates detectable amounts of reactive oxygen species (ROS). (A and B) Generation of singlet oxygen (1O2)-like ROS upon exposure to plasma treatments, as detected by specific fluorescence from molecular probes. (A to D) Plasma treatments produced ROS in both the medium (PBS) (A and C) and the E. coli cells (B and D). The plasma treatment time (in seconds) and the corresponding amount of energy (in J/cm2) are shown on the x axes. Arbitrary fluorescence units (AFU), an indicator of the corresponding ROS, are shown on the y axes. (For details, see Materials and Methods.) (C) The generation of hydrogen peroxide (H2O2)-like species upon exposure to plasma treatments is detected using a specific fluorescence molecular probe, and catalase-mediated scavenging is observed. Plasma exposure produced hydrogen peroxide in both the medium (PBS) (C) and the E. coli cells (D). Cat, catalase. (E) A colony assay demonstrating catalase-mediated protection to E. coli during plasma treatment is shown. The numbers above the bars and preceded by an asterisk are P values comparing the value with either the value for the corresponding untreated sample (A and B) or the relevant corresponding condition (with catalase or without catalase) (C and D). A statistically nonsignificant but appreciable amount of protection by catalase was seen in panel E. The values are means ± SDs (error bars) for 3 experiments.
Plasma-induced oxidants attack the cell membrane and depolarize it in E. coli.
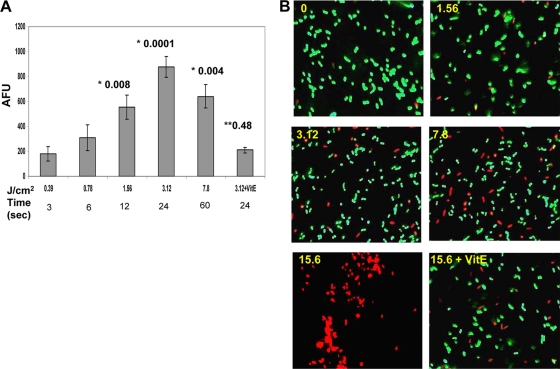
A membrane-interactive slow-response potential-sensitive probe, DiSBAC2(3), which can enter depolarized cells was used. DiSBAC2(3) typically binds to intracellular proteins or membrane and enhances fluorescence drastically and causes a red spectral shift. Figure 5A demonstrates a significant increase in fluorescence by this probe, proportionate to the amount of plasma energy (see the P values for 12 s and 24 s of plasma treatment). A saturation point is achieved after 24 s of plasma treatment. When the E. coli cells were plasma treated in the presence of vitamin E, a protective effect was seen. The amount of plasma energy corresponding to plasma treatment time (in seconds) is also shown (in Fig. 5A) for quick reference. To demonstrate that this change is responsible for eventual leakage of the cell membrane, the cells were stained with a Live/Dead BacLight staining kit. Figure 5B depicts the findings of fluorescence microscopy, showing gradual leakage/breach in the cell membrane, allowing the cell-impermeable dye propidium iodine to enter cells and to bind to DNA, imparting an increase in red fluorescence. Thus, the plasma treatment-dependent increase in fluorescence was reduced upon treatment in the presence of vitamin E (Fig. 5B).
FIG. 5.
Plasma-induced stress attacks the membrane and leads to the loss of membrane potential in E. coli cells in a dose-dependent manner. (A) A fluorescence molecular probe [DiSBAC2(3)] detects any membrane damage which leads to compromise in membrane integrity and thus membrane potential. The plasma treatment time (in seconds) and the corresponding amount of energy (in J/cm2) are shown on the x axis. An increase in arbitrary fluorescence unit (AFU) is taken as an indicator of a drop in membrane potential and is shown on the y axis. Values are means ± SDs (error bars). The numbers above the bars are P values. The P values preceded by one asterisk compare the value to the value for nontreated control. The P value preceded by two asterisks compare that value to the value for the corresponding treatment without antioxidant (vitamin E [VitE]). (B) A BacLight viability test essentially tests the compromised and leaky cell membrane in E. coli, and thus propidium iodide (PI) stain is taken up by membrane-damaged cells where it interacts with cellular DNA. These PI-positive cells are often taken as an indicator for dying or dead cells. The yellow numbers in the top right corners of the images are the amount of energy of the plasma treatment (in J/cm2).
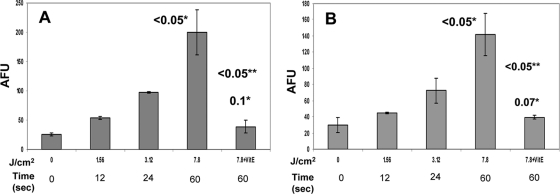
Plasma-mediated oxidation leads to membrane lipid peroxidation.
ROS is one of the major causes of membrane lipid peroxidation. We demonstrate here that intact E. coli cells and an isolated membrane-rich fraction undergo lipid peroxidation in a manner that is proportional to the amount of plasma energy. Maximum peroxidation is observed at 60 s both by intact cells and the membrane fraction. Comparatively, the membrane fraction exhibited more formation of the lipid peroxidation product malondialdehyde (MDA) (P < 0.05) (Fig. 6A and B). This change was inhibited significantly when the treatment was carried out in the presence of the antioxidant α-tocopherol (P ≤ 0.05), and the fluorescence signals were close to the baseline level (0 J/cm2 and 0 s) (Fig. 6).
FIG. 6.
The malondialdehyde (MDA) assay shows that plasma treatment leads to lipid peroxidation in E. coli. (A and B) Both an isolated membrane-rich fraction of E. coli (A) and intact E. coli cells (B) show a dose-dependent generation of MDA, the lipid peroxidation product. This peroxidation is inhibited by the antioxidant α-tocopherol (a form of vitamin E [VitE]), suggesting the involvement of oxidative stress-mediated damage. The values are means ± SDs (error bars) for 3 experiments. The numbers above the bars followed by one asterisk are P values comparing the value to the value for the untreated control. The numbers followed by two asterisks are P values comparing the value to the value for the corresponding plasma treatment condition without vitamin E.
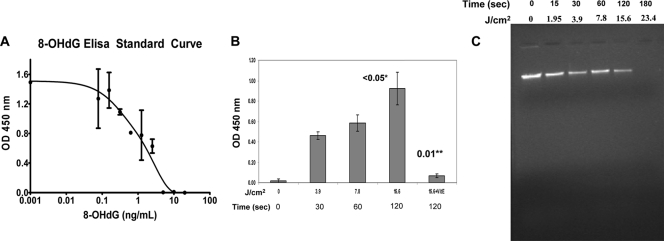
Detection of the markers of oxidative DNA damage (e.g., 8-OHdG) and DNA damage.
Cellular DNA in intact E. coli cells is protected from degradation by the cell's antioxidation mechanisms, and degradation usually occurs only when there is an imbalance between the oxidation and antioxidation regulatory pathways. The oxidatively damaged DNA can be identified using specific biochemical markers such as 8-hydroxydeoxyguanosine (8-OHdG). Figure 7A shows a standard 8-OHdG concentration-dependent curve for comparison, whereas Fig. 7B demonstrates plasma energy-dependent oxidation responses as detected by the amount of 8-OHdG. It reached a saturation point after 120 s of plasma treatment (plasma generator set at 1,250 Hz and 0.31 W/cm2) and thus required about 2.5 times more energy for such a change. The total amount of bacterial suspension used for this experiment was 1 ml (∼10-fold more than earlier experiments), mainly to enable isolation and purification of an appreciable amount of DNA. When the antioxidant α-tocopherol was added prior to plasma treatment, it inhibited DNA damage mediated by oxidative stress. Figure 7C depicts an agarose gel after electrophoresis showing damaged genomic DNA upon plasma treatment. Figure 7B shows an appreciable amount of inhibition when the cells were plasma treated in the presence of vitamin E.
FIG. 7.
The oxidative DNA damage marker 8-hydroxydeoxyguanosine (8-OHdG) appears as the oxidative stress increases. (A) A graph showing detection of 8-OHdG with serially diluted solutions of 8-OHdG by ELISA technique. This standard curve was used for comparison with experimental conditions. (B) Plasma dose (energy)-dependent response showing increasing appearance of 8-OHdG, which was is inhibited significantly upon pretreatment and the treatment in the presence of an antioxidant, vitamin E (VitE). Error bars show SDs. The number above the bar followed by one asterisk is the P value comparing the value to the value for the untreated control. The number followed by two asterisks is the P value comparing the value to the value for corresponding plasma condition without antioxidant (VitE). (C) The ethidium bromide-stained agarose gel, showing fragmentation of genomic DNA of E. coli cells as the amount of plasma energy application increases. The length of time of plasma treatment of the cells (in seconds) and corresponding energy of plasma treatment (in J/cm2) are shown above the gel.
DISCUSSION
The present study demonstrates that plasma treatment induced oxidative stress and generation of reactive oxygen species and that these agents affect cell morphology and membrane and DNA-associated changes in E. coli. In previous studies through a series of experiments, we demonstrated that the floating-electrode dielectric-barrier discharge (FE-DBD) technique of applying nonthermal plasma is efficient in inactivating bacteria (11, 22). This phenomenon can be seen in Fig. 1. Considering the speed of these inactivation events, we decided to study plasma's mechanism of interaction with E. coli cells. As plasma created in normal atmospheric room air generates various physical and chemical species (5) and is responsible for oxidation, we hypothesized that plasma-induced oxidative stress may be responsible for bacterial inactivation. Involvement of oxidative stress versus specific site targeting in bacterial inactivation is an emerging area of interest (7, 25).
The bacilli became coccoid and smaller (shrunken) with longer plasma treatment times (especially after 24 s of treatment). This property of transformation of the bacillary form to the coccoid form under adverse environmental conditions (especially under oxidative stress) has been observed by many other investigators (12, 14, 30). This is a typical behavior wherein an organism tries to reduce its surface area to minimize or reduce further injury, energy requirement, and metabolic activities and has been observed during severe oxidative stress (30). This type of oxidative stress response is organism specific, and so is the reversal of changes, and it is largely dependent upon the type and intensity of stress/stimulus (12). The observations like those presented in Fig. 2A are also noted by Nakamura et al., who reported the changes as irreversible and who reported that the changes were associated with the quality and quantity of ROS (14). It is also postulated that in some of the bacterial species, the coccoid form represents a dormant stage, whereas others proposed that this change occurs irreversibly (14). These types of cell are typically dying or dead cells and have been documented elsewhere (30). The experiments were carried out using E. coli cells in their exponential growth phase, and cellular morphology was verified each time. During the exponential phase, the organisms under normal conditions do not become coccoid and tend to show peculiar normal bacillary forms. Therefore, the changes observed remain associated with the plasma-induced stress. We exposed E. coli to sublethal doses of various plasma energy levels (and thus treatments in seconds), and the time of delay required for sample processing (∼45 min) was considered a constant throughout these experiments. It is likely that the 24 s and onwards of plasma exposure together with the delay in times in sample processing is critical and would be responsible for such changes, as we did not observe such changes with shorter treatment times (lower energy levels). The results of our XTT assay confirmed that the proposed changes are indicative of dying/dead cells (Fig. 2B). The inability of the cells to retain Gram stain indicates changes (associated with membrane and/or cytoplasmic pH or proteins) of a physical or chemical nature. It is possible that an interaction of plasma-generated species may have damaged the cell membrane by physical or chemical modifications. Lipid peroxidation of membrane often leads to leaky cells, and these cells are likely to lose dye (Gram stain).
There are a variety of antioxidants recommended for bacterial studies; they reportedly protect bacteria from oxidative stress (7), and the protection largely depends upon the type, quantity, and exposure time of the bacterial cell to generated ROS. In the present study, ascorbic acid (vitamin C) and water-soluble α-tocopherol (vitamin E) stood out as the best in scavenging ROS and protecting cells from oxidation effects of plasma treatment (Fig. 3A). However, these agents failed to revive bacteria after plasma treatment (Fig. 3B), which certainly shows the severity of plasma-induced damage and the amount of ROS in play. Although histidine and mannitol are reported as 1O2 and .OH scavengers (7, 8), mostly they are regarded as nonspecific ROS scavengers (by virtue of their relatively low redox potentials). The major ROS generated by FE-DBD nonthermal plasma are likely singlet oxygen, hydrogen peroxide, ozone, superoxide, and hydroxyl radical, but most of these ROS are interconvertible and hard to estimate or quantify. For this purpose, we selected two different probes that detect two of the major reactive species, 1O2 and H2O2, and used a strategy in which catalase was added prior to plasma treatment to determine whether catalase gives protection to E. coli cells against plasma-generated hydrogen peroxide. We used a widely used chemical trapping method for 1O2 detection following plasma treatment of fluid and E. coli. SOSGR is relatively stable in acidic pH (plasma causes slightly acidic pH), where it fluoresces bright green upon activation/trap (3, 4, 18). Singlet oxygen sensor green reagent (SOSGR) has been successfully applied to animal cells, plant cells, and bacterial cells, such as Staphylococcus aureus (13), and to plasma engineering of 1O2 production (31). In our studies, significant amounts of 1O2 were generated in the fluid component (Fig. 4A) and inside E. coli cells (Fig. 4B) in a dose-dependent manner by plasma treatments. To complement the generation of ROS, we detected another important species, H2O2, quantitatively using Amplex UltraRed reagent (AUR). We demonstrate here that H2O2 was generated in large amounts and that this plasma product was converted by the catalase enzyme into H2O and O2 after plasma treatment, decreasing the net amount of H2O2 with a concurrent sharp reduction in AUR fluorescent product. Both findings together suggest that ROS were the products of plasma-induced oxidative stress, that the hydrogen peroxide generated by plasma failed to exert its effect, and that catalase was able to protect E. coli cells.
As a result of oxidative stress, the membrane potential is likely to be reduced, and depending upon the severity and amount of ROS generated, it may lead to a breach in the membrane and eventually cytosolic leakage. In the present studies, plasma-induced oxidative stress was responsible for the loss of membrane potential in E. coli (Fig. 5A), and led to leaky membranes permitting a nuclear dye (propidium iodide) to enter the cell and bind to DNA (Fig. 5B). Hyperpolarization is indicated by a decrease in fluorescence. A drop in membrane potential was inhibited by the antioxidant vitamin E, which also prevented membrane damage (Fig. 5), indicating a positive relationship between ROS and membrane potential in the present study. He and Chen (10) reported conversion of over 99% Campylobacter jejuni from the spiral form to the coccoid form upon exposure to stress, and membrane integrity was not compromised in ∼96% of the coccoid cells, in contrast to the present studies wherein a concurrent quantitative loss of membrane integrity was observed in E. coli. This observation also suggests that these changes may be organism specific and depend upon the quality and quantity of stress stimulus. A slow-response potential-sensitive probe DiSBAC2(3) is the gold standard for measuring membrane potential (20, 21), and the BacLight bacterial viability test is known to detect the loss of membrane integrity (10, 15).
In biological membrane systems, lipid peroxidation is a frequent consequence of free radical and ROS attack and yields products that often react with DNA and proteins, leading to oxidative modifications. Peroxidation of lipid further generates reactive species, such as free radicals, carbonyl compounds, and hydroperoxides; the latter often decompose via chemical degradation or catalytic transition metal ions to give rise to more-toxic breakdown products, such as alkoxy radicals, peroxy radicals, hydroxyl radicals, and reactive aldehydes like 2-alkenals, 2-propenal (acrolein), 4-hydroxynonenal, and malondialdehyde (MDA) (17, 29). MDA is a major lipid peroxidation product. ROS scavengers, such as vitamin E, reportedly inhibit such peroxidation mediated through ROS (24, 26). In the present study, a significant accumulation of MDA following plasma treatment was noted (P ≤ 0.05), and vitamin E was able to prevent a significant amount of lipid peroxidation (Fig. 6). It is most likely that vitamin E scavenged plasma-induced reactive species and thus prevented subsequent changes of lipid peroxidation. Yamada et al. reported a positive correlation between ROS and membrane lipid peroxidation and a scavenging effect of vitamin E in E. coli (26). Reactive aldehydes can also generate DNA adducts that are responsible for an increased rate of mutagenesis in E. coli and Salmonella or extensive damage to DNA, leading to death (17, 28).
DNA is probably the most biologically significant target of oxidative attacks. It is thought that continuous oxidative damage of DNA is a significant contributor to all major cancers. Among numerous types of oxidative DNA damage, the formation of 8-hydroxydeoxyguanosine (8-OHdG) is a ubiquitous marker of oxidative stress and a by-product of oxidative stress. An oxidative modification of guanine residues generates 8-OHdG. 8-OHdG is also a major marker of oxidative DNA damage in prokaryotic cells, such as E. coli (29). Kinetic analysis demonstrated a significant increase in 8-OHdG formation both qualitatively and quantitatively when the cells were exposed to plasma treatment (Fig. 7). Nakamura et al. demonstrated a marked decrease in genomic DNA during the transformation of Helicobacter pylori from the bacillary form to the coccoid form, with a progressive increase in 8-OHdG under oxidative stress (14). In the present studies, a drastic decrease in the formation of 8-OHdG when vitamin E was preincubated or present during treatment indicates that the ROS scavenger prevented the cells from progressing to the prooxidant state and eventually DNA damage.
Lipid peroxidation is an ongoing process in living aerobic cells, but at a low level, and the process is prevented from entering the autocatalytic phase by defense enzymes, such as superoxide dismutase (SOD) and catalase, and other antioxidants. Although the observations on lipid peroxidation-induced DNA damage are limited, it is proposed that this is a major mode of membrane-mediated DNA damage (2, 29) in the cascade of free radical- and ROS-induced genetic damage. The electron transport chain provides a constant supply of ROS, and .OH, .O2, and H2O2 are typical side products of aerobic metabolism. Furthermore, ROS can also form during treatment of cells to redox-cycling chemicals, but in aerobes, such as E. coli, the synthesis of a variety of antioxidant enzymes is induced as surveillance strategy (17). In conclusion, the results of the present study indicate that upon plasma treatment, E. coli cells undergo substantial oxidative stress, generate reactive species, such as 1O2 and H2O2, which cause a drop in the membrane potential and cause membrane lipid peroxidation and a breach in membrane integrity. These signals are amplified sufficiently to lead to oxidative DNA damage and eventual cellular death in E. coli.
Acknowledgments
This work was supported in part by a Drexel University MRI grant and funds from the Beukenkamp Endowment of the Department of Surgery, Drexel University College of Medicine, Philadelphia, PA.
Footnotes
Published ahead of print on 3 January 2011.
REFERENCES
- 1.Baron, E. J., S. Mix, and W. Moradi. 2010. Clinical utility of an automated instrument for Gram staining single slides. J. Clin. Microbiol. 48:2014-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cerutti, P. A. 1985. Prooxidant states and tumor promotion. Science 227:375-381. [DOI] [PubMed] [Google Scholar]
- 3.Flors, C., et al. 2006. Imaging the production of singlet oxygen in vivo using a new fluorescent sensor, Singlet Oxygen Sensor Green. J. Exp. Bot. 57:1725-1734. [DOI] [PubMed] [Google Scholar]
- 4.Flors, C., and S. Nonell. 2006. Light and singlet oxygen in plant defense against pathogens: phototoxic phenalenone phytoalexins. Acc. Chem. Res. 39:293-300. [DOI] [PubMed] [Google Scholar]
- 5.Fridman, A. 2008. Plasma chemistry. Cambridge University Press, New York, NY.
- 6.Fridman, G., et al. 2007. Floating electrode dielectric barrier discharge plasma in air promoting apoptotic behavior in melanoma skin cancer cell lines. Plasma Chem. Plasma Processing 27:163-176. [Google Scholar]
- 7.Goswami, M., S. H. Mangoli, and N. Jawali. 2006. Involvement of reactive oxygen species in the action of ciprofloxacin against Escherichia coli. Antimicrob. Agents Chemother. 50:949-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartman, P. E., Z. Hartman, and K. T. Ault. 1990. Scavenging of singlet molecular oxygen by imidazole compounds: high and sustained activities of carboxy terminal histidine dipeptides and exceptional activity of imidazole-4-acetic acid. Photochem. Photobiol. 51:59-66. [DOI] [PubMed] [Google Scholar]
- 9.Hatzinger, P. B., P. Palmer, R. L. Smith, C. T. Penarrieta, and T. Yoshinari. 2003. Applicability of tetrazolium salts for the measurement of respiratory activity and viability of groundwater bacteria. J. Microbiol. Methods 52:47-58. [DOI] [PubMed] [Google Scholar]
- 10.He, Y., and C. Y. Chen. 2010. Quantitative analysis of viable, stressed and dead cells of Campylobacter jejuni strain 81-176. Food Microbiol. 27:439-446. [DOI] [PubMed] [Google Scholar]
- 11.Joshi, S. G., et al. 2010. Control of methicillin-resistant Staphylococcus aureus in planktonic form and biofilms: a biocidal efficacy study of nonthermal dielectric-barrier discharge plasma. Am. J. Infect. Control 38:293-301. [DOI] [PubMed] [Google Scholar]
- 12.Lu, Z., H. Min, and Y. Xia. 2004. The response of Escherichia coli, Bacillus subtilis, and Burkholderia cepacia WZ1 to oxidative stress of exposure to quinclorac. J. Environ. Sci. Health B 39:431-441. [DOI] [PubMed] [Google Scholar]
- 13.Maisch, T., et al. 2007. The role of singlet oxygen and oxygen concentration in photodynamic inactivation of bacteria. Proc. Natl. Acad. Sci. U. S. A. 104:7223-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakamura, A., et al. 2000. Oxidative cellular damage associated with transformation of Helicobacter pylori from a bacillary to a coccoid form. Free Radic. Biol. Med. 28:1611-1618. [DOI] [PubMed] [Google Scholar]
- 15.Ooi, N., et al. 2009. XF-73, a novel antistaphylococcal membrane-active agent with rapid bactericidal activity. J. Antimicrob. Chemother. 64:735-740. [DOI] [PubMed] [Google Scholar]
- 16.Peeters, E., H. J. Nelis, and T. Coenye. 2008. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J. Microbiol. Methods 72:157-165. [DOI] [PubMed] [Google Scholar]
- 17.Perez, J. M., F. A. Arenas, G. A. Pradenas, J. M. Sandoval, and C. C. Vasquez. 2008. Escherichia coli YqhD exhibits aldehyde reductase activity and protects from the harmful effect of lipid peroxidation-derived aldehydes. J. Biol. Chem. 283:7346-7353. [DOI] [PubMed] [Google Scholar]
- 18.Ragas, X., A. Jimenez-Banzo, D. Sanchez-Garcia, X. Batllori, and S. Nonell. 2009. Singlet oxygen photosensitisation by the fluorescent probe Singlet Oxygen Sensor Green. Chem. Commun. (Camb.) 2009:2920-2922. [DOI] [PubMed] [Google Scholar]
- 19.Smith, K., and I. S. Hunter. 2008. Efficacy of common hospital biocides with biofilms of multi-drug resistant clinical isolates. J. Med. Microbiol. 57:966-973. [DOI] [PubMed] [Google Scholar]
- 20.Solly, K., et al. 2008. Miniaturization and HTS of a FRET-based membrane potential assay for K(ir) channel inhibitors. Assay Drug Dev. Technol. 6:225-234. [DOI] [PubMed] [Google Scholar]
- 21.Tang, Y., X. Li, J. He, J. Lu, and Z. Diwu. 2006. Real-time and high throughput monitoring of cAMP in live cells using a fluorescent membrane potential-sensitive dye. Assay Drug Dev. Technol. 4:461-471. [DOI] [PubMed] [Google Scholar]
- 22.Taylor, J., et al. 2008. In-vitro susceptibilities of E. coli and S. aureus in fluid to FE-DBD plasma, abstr. 21, p. 9. Spirit of Entrepreneurship in Life Saving Solutions: Biomedical Talent and Technology Showcase Poster Abstracts. School of Biomedical Engineering, Science and Health Systems, Drexel University, Philadelphia, PA. http://www.biomed.drexel.edu/new04/Content/entrepreneurship/Abstracts__ALL_Nov19.pdf.
- 23.Van Gough, D., A. Wolosiuk, and P. V. Braun. 2009. Mesoporous ZnS nanorattles: programmed size selected access to encapsulated enzymes. Nano. Lett. 9:1994-1998. [DOI] [PubMed] [Google Scholar]
- 24.Venditti, P., A. Bari, L. Di Stefano, and S. Di Meo. 2007. Vitamin E attenuates cold-induced rat liver oxidative damage reducing H2O2 mitochondrial release. Int. J. Biochem. Cell Biol. 39:1731-1742. [DOI] [PubMed] [Google Scholar]
- 25.Wang, X., and X. Zhao. 2009. Contribution of oxidative damage to antimicrobial lethality. Antimicrob. Agents Chemother. 53:1395-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamada, T., Y. Shimomura, Y. Hiraoka, and K. Kimbara. 2006. Oxidative stress by biphenyl metabolites induces inhibition of bacterial cell separation. Appl. Microbiol. Biotechnol. 73:452-457. [DOI] [PubMed] [Google Scholar]
- 27.Yan, F., et al. 2005. Near-real-time determination of hydrogen peroxide generated from cigarette smoke. J. Environ. Monit. 7:681-687. [DOI] [PubMed] [Google Scholar]
- 28.Yang, I. Y., et al. 2001. Responses to the major acrolein-derived deoxyguanosine adduct in Escherichia coli. J. Biol. Chem. 276:9071-9076. [DOI] [PubMed] [Google Scholar]
- 29.Yoon, S. J., J. E. Park, J. H. Yang, and J. W. Park. 2002. OxyR regulon controls lipid peroxidation-mediated oxidative stress in Escherichia coli. J. Biochem. Mol. Biol. 35:297-301. [DOI] [PubMed] [Google Scholar]
- 30.Zeng, H., G. Guo, X. H. Mao, W. D. Tong, and Q. M. Zou. 2008. Proteomic insights into Helicobacter pylori coccoid forms under oxidative stress. Curr. Microbiol. 57:281-286. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, Y., K. Aslan, M. J. Previte, and C. D. Geddes. 2008. Plasmonic engineering of singlet oxygen generation. Proc. Natl. Acad. Sci. U. S. A. 105:1798-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zia, M., M. S. Nazir, T. Ismail, and M. H. Qureshi. 2007. Isolation and purification of membrane-bound cytochrome c from Proteus mirabilis. Afr. J. Biotechnol. 6:1132-1135. [Google Scholar]
- 33.Zirkle, R. E., and N. R. Krieg. 1996. Development of a method based on alkaline gel electrophoresis for estimation of oxidative damage to DNA in Escherichia coli. J. Appl. Bacteriol. 81:133-138. [DOI] [PubMed] [Google Scholar]