Abstract
Other than cleavage site mutations, there is little data on specific positions within Gag that impact on HIV protease inhibitor susceptibility. We have recently shown that non-cleavage site mutations in gag, particularly within matrix protein can restore replication capacity and further reduce protease inhibitor drug susceptibility when coexpressed with a drug-resistant (mutant) protease. The matrix protein of this patient-derived virus was studied in order to identify specific changes responsible for this phenotype. Three amino acid changes in matrix (R76K, Y79F, and T81A) had an impact on replication capacity as well as drug susceptibility. Introduction of these three changes into wild-type (WT) matrix resulted in an increase in the replication capacity of the protease mutant virus to a level similar to that achieved by all the changes within the mutant matrix and part of the capsid protein. Pairs of changes to wild-type matrix led to an increased replication capacity of the protease mutant (although less than with all three changes). Having only these three changes to matrix in a wild-type virus (with wild-type protease) resulted in a 5- to 7-fold change in protease inhibitor 50% effective concentration (EC50). Individual changes did not have as great an effect on replication capacity or drug susceptibility, demonstrating an interaction between these positions, also confirmed by sequence covariation analysis. Molecular modeling predicts that each of the three mutations would result in a loss of hydrogen bonds within α-helix-4 of matrix, leading to the hypothesis that more flexibility within this region or altered matrix structure would account for our findings.
Current British HIV Association (BHIVA) and other guidelines for highly active antiretroviral therapy (HAART) in the treatment of HIV and AIDS recommend first-line therapy with three active drugs: two nucleoside reverse transcriptase (RT) inhibitors and a nonnucleoside RT inhibitor. Protease inhibitors (PIs) are used with two active RT inhibitors in second-line therapy after the failure of first-line therapy (11). PIs are some of the most potent of the antiretroviral drugs in HIV clinical practice. Resistance to PIs develops by the accumulation of mutations within the protease gene that change amino acids within the enzymatic active site, reducing the binding of the inhibitor. Many of these primary PI resistance mutations have a negative effect on virus fitness or replication capacity, resulting in further secondary mutations that do not cause resistance themselves but instead increase the replication capacity of the resistant virus (2-5, 20, 22, 26). HIV protease cleaves Gag and Gag-Pol polyproteins, resulting in viral maturation after cellular release. Mutations within the Gag protein, particularly at the cleavage sites (cleavage site mutations [CSMs]) have also been associated with the recovery of replication capacity (9, 10, 24, 31, 35) as well as with PI resistance without protease mutations (27). Structural analysis showed that the A431V CSM has increased contact between the cleavage site and the mutated protease enzyme active site (29).
More recently, preexisting CSMs have been shown to have an impact on PI therapy in patients taking part in a clinical trial (ANRS 127) to determine the use of two protease inhibitors with or without other antiretrovirals. In this study, by 16 weeks of treatment, 26 patients did not have viral load below 50 copies per ml and were therefore defined as failing therapy. Nucleotide sequence analysis of the HIV protease from these patients did not reveal any known PI resistance mutations, suggesting that determinants of PI therapy failure can lie outside of the protease gene (17). Another clinical trial of PI monotherapy (MONARK) also suggests that determinants of PI therapy failure are not fully understood, since of 33 patients failing PI monotherapy, only 5 had known major PI resistance mutations. The cause of PI therapy failure in the remaining 28 patients is therefore unclear (8).
Phenotypic assays have shown that Gag, when expressed with a wild-type (WT) protease, can confer reduced susceptibility to PIs, although these gag genes were from patients who had failed PI therapy, as their viruses had known major protease resistance mutations. Thus, Gag alone from treated patients can confer reduced PI susceptibility, as well as contribute to replication capacity of viruses with PI-resistant protease. Gag also contributes significantly to PI resistance by enhancing the effect of mutations in protease (6, 28). There is increasing evidence that differences in PI susceptibility can be influenced by natural variation within HIV, such as differences in gag. The PI susceptibility of full-length gag and protease from wild-type (treatment-naïve) HIV-1 strains of different subtypes varies from that of standard subtype B. Gag was again shown to be the main contributor to this phenotype (12, 15).
Our previous study on the relationship between Gag and protease from a highly drug-resistant clinical sample (termed “mutant”) showed that the coevolved mutant Gag was able to restore the replication capacity of the multi-PI-resistant protease mutant virus. Mapping the regions of Gag that contributed to this recovery, we identified that the amino-terminal half of mutant Gag, matrix (MA), and part of the capsid protein (CA) restored the replication capacity of the protease mutant. The same region when expressed with a WT protease also had reduced susceptibility to several PIs (28). We therefore studied the changes found in mutant matrix and partial CA in order to determine which caused the improvement to the replication capacity of the protease mutant and reduced PI susceptibility.
MATERIALS AND METHODS
Resistance vectors.
Resistance vectors based on an HIV-1 retroviral vector system (1, 25, 34) were used to study replication capacity and drug susceptibility, as previously described (28). Briefly, resistance vectors were produced by transfection of confluent HEK293T cells with three plasmids: p8.9NSX, a derived gag-pol expression vector; pMDG, encoding vesicular stomatitis virus G protein; and pCSFLW, encoding firefly luciferase. Pseudovirus-containing supernatants were collected at 48 and 72 h posttransfection.
Site-directed mutagenesis.
Site-directed mutagenesis was carried out by standard molecular biology techniques, whereby the desired change was introduced by PCR using appropriate primers and Pfu Turbo enzyme (Stratagene), following the manufacturer's instructions. Amplified DNA was enriched by DpnI digest of template DNA, and plasmids were screened for the presence of the required sequence by standard DNA sequencing following transformation into Escherichia coli and plasmid miniprep (Qiagen).
Replication capacity.
The replication capacity of resistance test vectors was determined by titration of serial dilutions on HEK293T cells, adjusted for p24 levels by enzyme-linked immunosorbent assay (ELISA) to control for transfection efficiency, and quantification of luciferase 48 h after infection. Luciferase activity was measured with SteadyGlo (Promega) and a GloMaxMulti luminometer (Promega) following the manufacturer's instructions and expressed as a percentage of that of the WT. The viruses compared were produced and titrated in parallel for each experiment. Comparison of wild-type viruses produced and titrated in parallel results in variation of less than 5%. Statistical significance was determined by a Student's t test with a two-tailed distribution.
p24 ELISA.
A p24 enzyme linked immunosorbent assay (ELISA) was carried out as previously described (28), using reagents supplied by Aalto Bioreagents.
Drug susceptibility.
PI susceptibility was determined as previously described (28). Briefly, HEK293T cells were transfected as described above, and 16 h posttransfection, cells were harvested and seeded in the presence of different concentrations of PIs. Resistance vectors were harvested 24 h later and used to infect fresh target HEK293T cells. Virus replication was determined by measuring the luciferase expression in target cells 48 h postinfection and expressed relative to that of no-drug controls. Fifty percent effective concentrations (EC50s) were determined by linear regression analysis. Results are expressed as n-fold change in the EC50 compared to that of WT subtype B (p8.9NSX) and are the means of at least two separate experiments. Statistical significance was determined as described above for replication capacity. The protease inhibitor drugs used in this study, amprenavir (APV), atazanavir (ATV), darunavir (DRV), indinavir (IDV), lopinavir (LPV), nelfinavir (NFV), saquinavir (SQV), and tipranavir (TPV), were obtained from the NIH AIDS Research and Reference Reagent Program.
Molecular modeling.
Modeling studies were performed with the PyMol molecular graphics software (7), with a previously determined structure of subtype B HIV-1 matrix as the starting structure (Protein Data Bank identification [ID] 1HIW) (13). The R76K, Y79F, and T81A mutations were modeled with the mutagenesis tool. The best possible rotamers were selected so as to avoid van der Waals clashes. Hydrogen bond donor and acceptor atoms that were within h-bonding distance were analyzed to determine h-bond interactions.
Sequence covariation.
HIV-1 subtype B protease and matrix sequences were retrieved from the Los Alamos HIV sequence database (19). Protease and matrix sequences were translated to amino acids, aligned independently with ClustalW (33), and then concatenated where sequences shared a common identifier. The resulting alignment contained 670 sequences. Covariation analysis was performed by calculating the Jaccard index (JI) and deriving a Z score by comparing observed JI scores with JI scores derived from a random model (5,000 simulated sequences) (30). Z scores were used to estimate the probability of each result and then corrected for multiple testing by using a false discovery rate (FDR) (14) set to P = 0.001.
RESULTS
We have previously reported that matrix and part of capsid (amino acids [aa] 1 to 240, numbered according to HXB2 Gag) from a treatment-experienced patient infected with subtype B HIV-1 can restore the replication capacity of the highly PI-resistant protease from the same patient; sequences from this patient sample are termed mutant (GenBank accession no. FJ224363). The same region of Gag also confers reduced susceptibility to PIs (28). The changes in this sequence (up to an SpeI restriction enzyme site in capsid at codon 240) compared to HXB2 are as follows: K30R, R76K, Y79F, T81A, T84V, E93D, I94V, D102E, H124N, and N126S and an insertion (Q116TQ) in matrix as well as I138M and S173M in capsid (numbered according to HXB2 Gag). After studying an amino acid alignment (between the mutant and HXB2) and the crystal structure of HIV-1 matrix/capsid (21, 32), two regions were selected for further study. (i) The first region contained insertion of 2 amino acids at amino acid 116 (Q116TQ) that extended a disordered loop containing the protease cleavage site between matrix and capsid. (ii) The second consisted of four changes (R76K, Y79F, T81A, and T84V) clustered together within a short region of nine amino acids that are within α-helix-4 of matrix (21, 32); we therefore speculated that this group of changes could be acting together. Position 84 is polymorphic in the two HIV-1 strains commonly used in HIV drug resistance assays—T in HXB2 and V in NL4-3. We therefore decided that this position would be unlikely to contribute to the phenotype, and it was not studied further, leaving R76K, Y79F, and T81A.
Replication capacity.
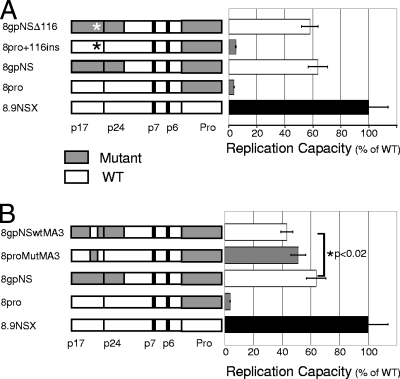
Descriptions of the constructs used in this study are provided in Table 1. The 116TQ insertion from mutant matrix was introduced into the WT gag of construct p8pro (expressing mutant protease in a WT background), creating construct p8pro+116. The insertion was also removed from the construct containing mutant matrix/part capsid and protease (p8gpNS) to create p8gpNSΔ116. Resistance vectors based on the WT (p8.9NSX), the protease mutant (p8pro), mutant matrix/partial capsid and protease (p8gpNS), and the two new constructs created by site-directed mutagenesis as described above were produced by transfection. These single-cycle resistance vectors express luciferase once the vector-packaged genome has been reverse transcribed and integrated into the chromosomal DNA of target cells. As previously described, the protease mutant alone had a replication capacity of around 4% of wild type, a 25-fold reduction in replication capacity; this deficiency could be substantially rescued by mutant matrix and part of capsid to give a replication capacity of 63% of wild type (Fig. 1A). The addition of the 116 insertion into a wild-type gag (construct p8pro + 116) did not have a significant effect on the impaired replication capacity caused by the protease mutant (Fig. 1A, compare gray bars in bar chart), nor did removal of the aa 116 insertion from mutant matrix (construct p8gpNSΔ116) decrease the rescue caused by the remaining changes in mutant matrix and part of capsid (Fig. 1A, compare white bars in bar chart).
TABLE 1.
Names and descriptions of the constructs used in this study
| Construct name | Descriptiona | Figure(s) |
|---|---|---|
| 8pro | Mutant protease with WT Gag | 1 and 2 |
| 8gpNS | Mutant protease with MA, partial CA to aa 240, and the remainder of Gag WT | 1 and 2 |
| 8pro+116 | Mutant protease WT Gag with mutant aa 116 insertion | 1A |
| 8gpNSΔ116 | 8gpNS with aa 116 insertion removed | 1A |
| 8proMutM3 | 8pro with amino acid changes R76K, Y79F, and T81A (as in mutant) | 1B |
| 8gpNSwtMA3 | 8gpNS with amino acid changes K76R, F79Y, and A81T (as in HXB2) | 1B |
| 8proMA76K | 8pro with R76K | 2A |
| 8proMA79F | 8pro with Y79F | 2A |
| 8proMA81A | 8pro with T81A | 2A |
| 8gpNS76R | 8gpNS with K76R | 2A |
| 8gpNS79F | 8gpNS with Y79F | 2A |
| 8gpNS81T | 8gpNS with A81T | 2A |
| 8pMA76K79F | 8pro with R76K and Y79F | 2B |
| 8pMA79F81A | 8pro with Y79F and T81A | 2B |
| 8pMA76K81A | 8pro with R76K and T81A | 2B |
| 8gpNS76R79Y | 8gpNS with K76R and F79Y | 2B |
| 8gpNS79Y81T | 8gpNS with F79Y and A81T | 2B |
| 8gpNS76R81T | 8gpNS with K76R and A81T | 2B |
| 8gNS | 8.9NSX (WT) with mutant MA and CA to aa 240 (WT protease) | 3A |
| 8gNSΔ116 | 8gNS with mutant aa 116 insertion removed | 3A |
| 8.9+116 | 8.9NSX (WT) with mutant aa 116 insertion (WT protease) | 3B |
| 8gNSwtMA3 | 8gNS with K76R, F79Y, and A81T | 3C |
| 8.9MutMA3 | 8.9NSX (WT) with mutant R76K, Y79F, and T81A (WT protease) | 3D |
| 8MA76K | 8.9NSX (WT) with mutant R76K (WT protease) | 4A |
| 8MA79F | 8.9NSX (WT) with mutant Y79F (WT protease) | 4A |
| 8MA81A | 8.9NSX (WT) with mutant T81A (WT protease) | 4A |
| 8MA76K79F | 8.9NSX (WT) with mutant R76K and Y79F(WT protease) | 4B |
| 8MA76K81A | 8.9NSX (WT) with mutant Y76K and T81A (WT protease) | 4B |
| 8MA79F81A | 8.9NSX (WT) with mutant Y79F and T81A (WT protease) | 4B |
All constructs are in 8.9 NSX backbone (wild type). 8pro and 8p constructs have mutant protease with wild-type gag and specific changes to matrix, as listed. 8gpNS constructs have mutant gag (matrix and partial capsid) from the NotI to the SpeI sites and mutant protease, as well as specific changes within matrix, as listed. 8gNS constructs have mutant gag from the NotI to SpeI site and wild-type protease; specific changes within matrix are listed.
FIG. 1.
Replication capacity of mutant matrix and partial capsid constructs with mutant protease. Recombinant resistance vectors containing wild-type (white) and mutant (shaded) regions were titrated by serial dilution. The luciferase signal is shown relative to wild type after normalization for p24 protein levels. The names and schematic representations of gag-protease constructs are shown. Protease cleavage sites are shown: cleavage sites and p1 and p2 spacer peptides are depicted by heavy lines. The functional Gag proteins matrix (p17), capsid (p24), nucleocapsid (p7), and p6 (p1 and p2 not shown) are listed, as is protease (Pro). Error bars show the standard error of the mean. The mean was derived from 11 data points for all constructs, except 8pro, which was derived from 9 data points. (A) Effect of mutant insertion at amino acid 116 of matrix, shown by an asterisk in the schematic (8pro+116ins and 8gpNSΔ116). (B) The effect of changing three amino acids together at positions 76, 79, and 81 of matrix is shown as a gray box (mutant) in an otherwise white wild-type gag (8proMutMA3) and as a white box (WT) in an otherwise gray mutant matrix (8gpNSwtMA3).
The three selected amino acid changes in mutant matrix, namely, R76K, Y79F, and T81A, were introduced together into construct p8pro, producing p8proMutMA3. A reciprocal construct with amino acids 76, 79, and 81 of mutant matrix back-mutated to the HXB2 wild type (K76R, F79Y, and A81T) was also made by site-directed mutagenesis of p8gpNS, producing p8gpNSwtMA3. The three changes in an otherwise wild-type gag background with the protease mutant (p8proMutMA3) gave a replication capacity of 51% of wild type: over a 10-fold increase compared to the protease mutant alone (p8pro) and similar to the replication capacity of p8gpNS (Fig. 1B). Changing the same three amino acids in mutant matrix to those of HXB2 (p8gpNSwtMA3) caused a significant reduction in replication capacity from 63% to 43% of wild type (P < 0.02) (Fig. 1B, compare white bars). However, this change was not as great as the increase caused by introducing the three mutant positions into wild-type Gag (p8pro replication capacity of 4% of wild type and p8proMutMA3 replication capacity of 51%) (Fig. 1B).
Given the impact of the three mutations on recovery of replication capacity, we explored the role of each position. Individual changes were made to introduce the mutant amino acid within a wild-type gag gene linked to the protease mutant (p8proMA76K, p8proMA79F, and p8proMA81A) as well as by introducing the HXB2 amino acid to mutant matrix in the context of mutant matrix/partial capsid linked to the protease mutant (p8gpNS76R, p8gpNS79Y, and p8gpNS81T). Introduction of the mutant amino acids to wild-type matrix had very minor effects (Fig. 2A, compare gray bars). Changing these single amino acids in mutant matrix to those found in HXB2 in the context of mutant matrix/partial capsid and protease did not have any significant effect (Fig. 2A, compare white bars).
FIG. 2.
Contribution to replication capacity of single amino acid changes and pairs of amino acid changes in matrix with mutant protease. Recombinant resistance vectors containing wild-type (white) and mutant (gray) regions were titrated by serial dilution as described in the legend to Fig. 1. The names and schematic representations of gag-protease constructs are shown as in Fig. 1. Error bars show the standard error of the mean. The mean was derived from 7 or more data points for all constructs, except 8proMA81A, which was derived from 5 data points. (A) Effect on the replication capacity of the protease mutant by individual changes at positions 76, 79, and 81 to mutant amino acids (shown with a gray asterisk) to wild-type matrix (white). Shown is the effect on replication capacity of mutant matrix and partial capsid with the protease mutant by individual changes (shown with a white asterisk) within mutant matrix (gray). (B) The effect on replication capacity of the protease mutant of changing pairs of amino acids together (76 and 79, 79 and 81, and 76 and 81) within matrix is shown as a gray oval (mutant) in an otherwise white wild-type gag gene or as a white oval (WT) in an otherwise gray mutant matrix.
Since individual changes to matrix did not have as great an effect as changes R76K, Y79F, and T81A together, the contribution of pairs of these changes was determined. As before, reciprocal pairs of changes were made to wild-type gag and mutant matrix, both linked to the protease mutant, and replication capacity was measured. Introduction of both R76K and Y79F into wild-type gag increased the replication capacity of the protease mutant from 4% to 6.8% (Fig. 2B), while the Y79F and T81A change to wild-type Gag increased the replication capacity of the protease mutant to 20.4%. The largest increase in replication capacity was produced by changing R76K and T81A, increasing the replication capacity to 33% (compare gray bars in Fig. 2B; P < 0.02 for all comparisons). When reversing the mutants at these positions within the clinically derived mutant matrix, the K76R and F79Y changes (8gpNS76R79Y) had the greatest impact, reducing the replication capacity of mutant matrix/partial capsid linked to the protease mutant from 59% of wild type to 30% (P < 0.02). The other pairs of changes had less effect: F79Y and A81T (8gpNS79Y81T) and K76R and A81T (8gpNS76R81T) had similar replication capacity values of 46.5% and 47.3% of wild type, respectively (P < 0.05 for both; see the white bars in Fig. 2B). Since the effects on replication capacity by double-point mutations in a wild-type gag gene differ from the reciprocal mutations in mutant matrix (Fig. 2B), other changes within the mutant must also contribute to the rescue of replication capacity observed.
Protease inhibitor susceptibility.
The PI susceptibility of the retroviral vectors containing mutant matrix, mutant matrix with specific changes to wild type, or wild type with specific mutant changes was determined (Fig. 3). In contrast to the replication capacity experiments, all of the PI susceptibility constructs had wild-type protease; therefore, any differences in PI susceptibility were only due to the changes made within the amino half of Gag. Removal of the 116 insertion within mutant matrix (construct 8gNSΔ116) did not have a notable effect on the PI susceptibility of mutant matrix/partial capsid (8gNS shown in Fig. 3A), nor did introduction of the 116 insertion into wild-type gag-pol (8.9+116ins) (Fig. 3B).
FIG. 3.
Protease inhibitor susceptibility of mutant matrix and partial capsid constructs, all with wild-type protease. Susceptibility is shown as the change in EC50 compared to wild type (p8.9NSX); a value of 1 indicates the same susceptibility as wild type. The names and schematics of constructs are shown above each bar chart: the wild type is shown as white and the mutant as gray. Error bars represent the standard error of the mean from at least two separate experiments. The mean for constructs 8gNS is derived from five replicates, 8gNSD116 and 8.9+116ins from two replicates, and 8NSwtMA3 and 8.9MutMA3 from three replicates. (A) PI susceptibility of mutant matrix and partial capsid (8gpNS) and with the insertion at aa 116 removed, shown with an asterisk in the schematic (8gpNSΔ116). (B) PI susceptibility of the wild type with the mutant 116 insertion, shown with an asterisk in the schematic (8.9+116ins). (C) PI susceptibility of mutant matrix and partial capsid (8gpNS) and with 3 aa changes (76R, 79Y, and 81T), shown as a white (WT) box in the gray mutant background. (D) PI susceptibility of the wild type with 3 aa changes (76K, 79F, and 81A), shown as a gray box in an otherwise white background.
Changing the three amino acids at positions 76, 79, and 81 of mutant matrix to those found in HXB2 (8gNSwtMA3) led to a reduction in LPV susceptibility from 7.2 for mutant matrix and partial capsid (8gNS) to 3.3-fold for the same construct with positions 76, 79, and 81 as in HXB2 (8gNSwtMA3), both relative to wild type. Conversely, inserting these three changes into a wild-type background (8.9MutMA3) resulted in reduced PI susceptibility for APV, ATV, IDV, LPV, and NFV (Fig. 3D).
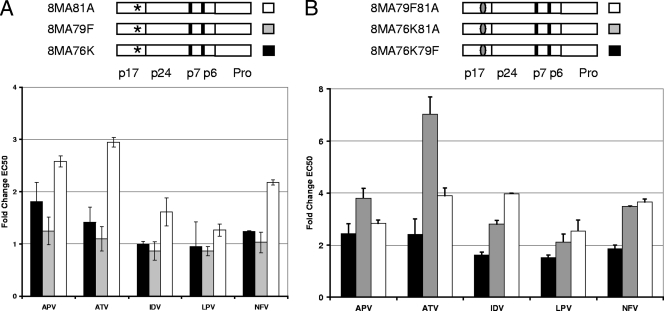
Since changing three amino acids altered PI susceptibility, the contribution of single changes was determined. When studying each position alone, only the T81A-containing virus (8MA81A) had slightly reduced susceptibility to APV, ATV, and NFV, the three drugs that showed the most change when all three amino acids were changed (Fig. 4A). As with the replication capacity experiments, these results suggested that no single change was responsible for the phenotype observed when all three amino acid changes are present in matrix; therefore, the effects of changes to pairs of amino acids in a WT viral vector were determined. Three constructs (8MA76K79F, 8MA76K81A, and 8MA79F81A) were tested for their susceptibility to PIs (Fig. 4B). The largest effect seen was with the R76K T81A dual mutant (8MA76K81A), for which the EC50 for ATV was increased by 7-fold. More modest changes of between 3- and 4-fold were seen for the other drugs with the R79F T81A dual mutant (Fig. 4B). The results from individual changes and pairs of changes suggest that the effect of the three amino acid positions occurs as a result of them functioning together.
FIG. 4.
Protease inhibitor susceptibility of matrix constructs with single mutations or pairs of mutations, all with wild-type protease. Susceptibility is shown as described in the legend to Fig. 3. Error bars represent the standard error of the mean from two separate experiments. (A) PI susceptibility of single mutations to aa 76, 79, and 81 of matrix (8MA76K, 8MA79F, and 8MA81A, respectively), shown by an asterisk in the schematics. (B) PI susceptibility of pairs of mutations (8MA76K79F, 8MA76K81A, and 8MA79F81A), shown by gray ovals in an otherwise white (wild-type) background.
Molecular modeling.
Amino acids 76, 79, and 81 of matrix identified above as contributing to replication capacity of the protease mutant and protease inhibitor susceptibility (with a wild-type protease) all lie within α-helix-4 of matrix of previously determined structures (13, 21). Molecular modeling indicates that each of the residues (aa 76, 79, and 81) participates in a hydrogen-bonding network within matrix. Residue R76 forms a water-mediated hydrogen bond to G80 within helix 4, a hydrogen bond to G71, and a potential salt bridge to E73 in the loop preceding helix 4. Y79 forms a hydrogen bond across to Q65 in helix 3, while T81 forms a hydrogen bond to S77 within helix 4 (Fig. 5A). The changes found in mutant matrix are predicted to result in the loss of most of these hydrogen-bonding networks. Change R76K could maintain one of the hydrogen bonds but not coordinate all three as in the wild type (Fig. 5B). Y79F can no longer form a hydrogen bond to helix 3, and T81A can no longer form a hydrogen bond within helix 4 (Fig. 5B).
FIG. 5.
Molecular modeling showing the changes studied in HIV-1 matrix (MA). (A) Crystal structure of HIV-1 matrix. Helix 4 is shown in yellow stick form with amino acids 76, 79, and 81 in green showing van der Waals surfaces. Hydrogen bonds are shown with dashed lines. (B) Molecular model of MA showing the mutant changes at 76, 79, and 81. Lys76 can no longer coordinate three independent hydrogen bonds; the single maintained bond is shown. Phe79 can no longer form a hydrogen bond to helix 3. Ala81 can no longer form a hydrogen bond within helix 4. The position of the insertion at 116 is also shown.
Sequence covariation.
Amino acids 76, 79, and 81 of matrix showed significant covariation between the three possible pairwise combinations of mutations (Table 2). The matrix mutations also covaried with polymorphisms in protease, most noticeably with protease inhibitor resistance positions 54 and 90 (Table 2). There was also evidence that at least two (usually all three) of the matrix amino acids covaried with polymorphisms in positions within protease thought to compensate for selection of drug resistance mutations; these included amino acids 10, 20, 33, 46, and 72. The common protease inhibitor resistance mutation at position 82 showed no significant covariation with any of the three matrix mutations (R. E. Myers, data not shown).
TABLE 2.
Sequence covariation analysis
| Covariation and aa positiona | MA 2 aa positionb | Z scorec | JI score derived fromd: |
SE | Mutation occurrence as determined frome: |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sequence data | Random model | Sequence data |
Random model |
||||||||
| XY | XO | YO | XY | XO | YO | ||||||
| Matrix-matrix (MA 1) | |||||||||||
| 76 | 79 | 39.05 | 0.300 | 0.269 | 0.001 | 134 | 218 | 94 | 899 | 1,533 | 916 |
| 76 | 81 | 61.20 | 0.081 | 0.048 | 0.001 | 29 | 323 | 6 | 124 | 2,308 | 165 |
| 79 | 81 | 47.84 | 0.087 | 0.054 | 0.001 | 21 | 207 | 14 | 108 | 1,707 | 181 |
| Protease-matrix (PR 1) | |||||||||||
| 54 | 79 | 12.71 | 0.046 | 0.040 | 0.001 | 11 | 10 | 217 | 77 | 125 | 1,738 |
| 54 | 81 | 57.49 | 0.120 | 0.019 | 0.002 | 6 | 15 | 29 | 9 | 193 | 280 |
| 90 | 76 | 5.05 | 0.039 | 0.037 | 0.000 | 14 | 11 | 338 | 93 | 106 | 2,339 |
| 90 | 79 | 22.03 | 0.050 | 0.038 | 0.001 | 12 | 13 | 216 | 74 | 125 | 1,741 |
| 90 | 81 | 52.46 | 0.111 | 0.025 | 0.002 | 6 | 19 | 29 | 12 | 187 | 277 |
MA 1, matrix mutation position 1; PR 1, protease mutation position 1.
MA 2, matrix mutation position 2.
Z score: (sequence data JI score − random model JI score)/sequence data JI score SE. The greater the value of the Z score, the greater the evidence that two amino acids covary. However, the results shown are all statistically significant based on the magnitude of the Z score compared with a standard normal distribution corrected for multiple testing.
Jaccard index (JI): JI = XY/XY + XO + YO. Shown are the JI scores calculated from both sequence data and the random data model.
For mutation occurrence, XY represents the occurrence of mutations 1 and 2 in the same sequence, XO represents the number of sequences with mutation 1 alone, and YO represents the number of sequences with mutation 2 alone.
DISCUSSION
In a previous study, we showed that the amino-terminal regions of Gag from a subtype B HIV-1 isolate derived from a highly drug-experienced patient can play a role in the replication capacity of the cognate protease as well as contribute to reduced PI susceptibility (28). We have now identified a trio of changes in matrix, namely, R76K, Y79F, and T81A, which, when introduced into WT Gag are sufficient to recover most of the fitness deficit caused by the drug-resistant protease. In contrast, reversal of these mutations to the HXB2 consensus amino acids within the cognate patient-derived Gag did not completely reverse this recovery, signifying additional sites in matrix/capsid responsible for fitness compensation. Of note, these three mutations also contributed to the reduced susceptibility to LPV of the patient-derived matrix/partial capsid. Removal of these three mutations within the patient-derived sequence did not significantly alter the susceptibility to other PIs. Thus, other than for LPV, the remaining changes within mutant matrix and part of capsid are playing as important a role in PI susceptibility as the three amino acid positions studied here. At the time that the plasma sample was taken, the patient was on a drug combination that included LPV; it is therefore possible that the specific contribution of the three amino acids to LPV susceptibility is the result of selection, providing some advantage to the virus in the presence of this drug.
The impact of the triad of mutations was greater than those of mutant pairs, suggesting a functional interaction between them. Nevertheless, position 81, either alone or in combination, seems to be a major contributor to the phenotype observed. The smallest effect on replication capacity and drug susceptibility was observed when amino acid positions 76 and 79 were altered.
Molecular modeling suggests a structural hypothesis to account for our findings. The three wild-type HXB2 residues are involved in a total of five potential intermolecular bonds, both within helix 4 and in positioning helix 4 relative to the remainder of matrix. Molecular modeling indicates that the majority of these bonds would not maintained in the mutant matrix that would be predicted to result in more flexibility around helix 4 and possibly therefore an altered structure. We hypothesize that the loss of these hydrogen bonds and potential flexibility increases either the affinity or the availability of the matrix-capsid cleavage site with respect to the protease. This would allow the less-efficient drug-resistant protease mutant to cleave a greater proportion of matrix-capsid cleavage sites, giving the rescue phenotype. Such an increased affinity would also reduce the PI susceptibility with a wild-type protease as it would require lower levels of active protease to cleave between matrix and capsid, thus tolerating higher levels of inhibitor, resulting in the reduced PI susceptibility seen here. This is clearly distinct from the mechanism described for CSM A431V, which has a better fit within the enzyme by having increased contacts between the substrate and mutant active site (29). Therefore, factors outside of protease and CSM contribute to PI susceptibility and may play a role in the successful outcome of therapy that includes PI.
These data support recent work of others, showing Gag processing intermediates having a strongly transdominant effect on the production of infectious virus (18, 23). Indeed, the presence of as little as 5% of Gag with an uncleavable matrix-capsid junction reduces HIV infectivity by 50% (18). Thus, even a relatively modest increase in cleavage between the matrix and capsid domains of Gag would have a large affect on the amount of infectious virus produced; whether in the context of the replication capacity of the protease mutant or in the presence of protease inhibitor.
In viruses with resistance mutations in protease, it is clearly these mutations that are the main cause of PI resistance, yet Gag also contributes. The changes in PI susceptibility shown here are modest but are the result of alteration of only three amino acids in matrix. It is unlikely that such modest changes alone would cause treatment failure, especially when PIs are pharmacologically boosted with ritonavir. These results support and add to previous published work suggesting that changes within Gag have a role in both the replication capacity of drug-resistant protease and, perhaps more importantly, in the drug susceptibility of viruses with no mutations in protease (6, 12, 15, 28). While we have identified three specific changes (R76K, Y79F, and T81A) that have a role in replication capacity and PI susceptibility, it is important to note that none of the other viruses studied elsewhere shares all three of these changes (12, 15). It is, however, interesting to note that virus AE-Gag62 shares the same K30R (not studied here), R76K, and Y79F changes as our mutant, and these authors report that the amino-terminal region of Gag from this virus caused reduced PI susceptibility (15). Some of the reduced PI susceptibility has been attributed to K165 (within capsid) of the CRF01_AE virus studied (not present in our mutant). Introduction of K165 into CRF01_AE had little effect on replication capacity, while the same change in NL4-3 significantly reduced replication capacity (16). It therefore seems that there is a role for the amino-terminal half of Gag in replication capacity and PI susceptibility, but that the interplay of changes within this region of Gag will be complex and dependent on the other amino acid residues with which they may share intermolecular bonds, as suggested by our molecular modeling and covariation data. The variation between subtypes will also be an important factor to take into account. Further phenotypic studies of the amino-terminal half of Gag along with molecular modeling, or even the determination of more diverse matrix proteins, will be required to fully understand the role of this variable protein in PI susceptibility and replication capacity.
Acknowledgments
We thank Nigel Temperton, University of Kent, for pCSFLW and Didier Trono, EPFL, Switzerland, for pCMV-Δ8.91 (from which is derived p8.9NSX) and pMDG. The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program: APV, ATV, DRV, IDV, LPV, NFV, SQV, and TPV.
This work was supported in part by the Health Protection Agency UK through a Strategic Research and Development grant (project no. 105039). D.P. is partly funded by the NIHR UCLH/UCL Comprehensive Biomedical Research Centre.
Footnotes
Published ahead of print on 13 December 2010.
REFERENCES
- 1.Bainbridge, J. W., et al. 2001. In vivo gene transfer to the mouse eye using an HIV-based lentiviral vector; efficient long-term transduction of corneal endothelium and retinal pigment epithelium. Gene Ther. 8:1665-1668. [DOI] [PubMed] [Google Scholar]
- 2.Borman, A. M., S. Paulous, and F. Clavel. 1996. Resistance of human immunodeficiency virus type 1 to protease inhibitors: selection of resistance mutations in the presence and absence of the drug. J. Gen. Virol. 77:419-426. [DOI] [PubMed] [Google Scholar]
- 3.Condra, J. H., et al. 1996. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J. Virol. 70:8270-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Condra, J. H., et al. 1995. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 374:569-571. [DOI] [PubMed] [Google Scholar]
- 5.Croteau, G., et al. 1997. Impaired fitness of human immunodeficiency virus type 1 variants with high-level resistance to protease inhibitors. J. Virol. 71:1089-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dam, E., et al. 2009. Gag mutations strongly contribute to HIV-1 resistance to protease inhibitors in highly drug-experienced patients besides compensating for fitness loss. PLoS Pathog. 5:e1000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLano, W. L. 2002. The PyMol molecular graphics system.
- 8.Delaugerre, C., et al. 2009. Protease inhibitor resistance analysis in the MONARK trial comparing first-line lopinavir-ritonavir monotherapy to lopinavir-ritonavir plus zidovudine and lamivudine triple therapy. Antimicrob. Agents Chemother. 53:2934-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doyon, L., et al. 1996. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J. Virol. 70:3763-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gatanaga, H., et al. 2002. Amino acid substitutions in Gag protein at non-cleavage sites are indispensable for the development of a high multitude of HIV-1 resistance against protease inhibitors. J. Biol. Chem. 277:5952-5961. [DOI] [PubMed] [Google Scholar]
- 11.Gazzard, B. G., et al. 2008. British HIV Association guidelines for the treatment of HIV-1-infected adults with antiretroviral therapy 2008. HIV Med. 9:563-608. [DOI] [PubMed] [Google Scholar]
- 12.Gupta, R. K., et al. 2010. Full-length HIV-1 Gag determines protease inhibitor susceptibility within in-vitro assays. AIDS 24:1651-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill, C. P., D. Worthylake, D. P. Bancroft, A. M. Christensen, and W. I. Sundquist. 1996. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl. Acad. Sci. U. S. A. 93:3099-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holm, S. 1979. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 6:65-70. [Google Scholar]
- 15.Jinnopat, P., et al. 2009. Impact of amino acid variations in Gag and protease of HIV type 1 CRF01_AE strains on drug susceptibility of virus to protease inhibitors. J. Acquir. Immune. Defic. Syndr. 52:320-328. [DOI] [PubMed] [Google Scholar]
- 16.Kameoka, M., et al. 2010. The role of lysine residue at amino acid position 165 of human immunodeficiency virus type 1 CRF01_AE Gag in reducing viral drug susceptibility to protease inhibitors. Virology 405:129-138. [DOI] [PubMed] [Google Scholar]
- 17.Larrouy, L., et al. 2010. Gag mutations can impact virological response to dual-boosted protease inhibitor combinations in antiretroviral-naive HIV-infected patients. Antimicrob. Agents Chemother. 54:2910-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, S. K., J. Harris, and R. Swanstrom. 2009. A strongly transdominant mutation in the human immunodeficiency virus type 1 gag gene defines an Achilles heel in the virus life cycle. J. Virol. 83:8536-8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leitner, T., et al. 2005. HIV sequence compendium 2005. Los Alamos National Laboratory, Los Alamos, NM.
- 20.Martinez-Picado, J., A. V. Savara, L. Sutton, and R. T. D'Aquila. 1999. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J. Virol. 73:3744-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Massiah, M. A., et al. 1994. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J. Mol. Biol. 244:198-223. [DOI] [PubMed] [Google Scholar]
- 22.Molla, A., et al. 1996. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat. Med. 2:760-766. [DOI] [PubMed] [Google Scholar]
- 23.Muller, B., et al. 2009. HIV-1 Gag processing intermediates trans-dominantly interfere with HIV-1 infectivity. J. Biol. Chem. 284:29692-29703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myint, L., et al. 2004. Gag non-cleavage site mutations contribute to full recovery of viral fitness in protease inhibitor-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 48:444-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naldini, L., et al. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 26.Nijhuis, M., et al. 1999. Increased fitness of drug resistant HIV-1 protease as a result of acquisition of compensatory mutations during suboptimal therapy. AIDS 13:2349-2359. [DOI] [PubMed] [Google Scholar]
- 27.Nijhuis, M., et al. 2007. A novel substrate-based HIV-1 protease inhibitor drug resistance mechanism. PLoS Med. 4:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parry, C. M., et al. 2009. Gag determinants of fitness and drug susceptibility in protease inhibitor-resistant human immunodeficiency virus type 1. J. Virol. 83:9094-9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prabu-Jeyabalan, M., E. A. Nalivaika, N. M. King, and C. A. Schiffer. 2004. Structural basis for coevolution of a human immunodeficiency virus type 1 nucleocapsid-p1 cleavage site with a V82A drug-resistant mutation in viral protease. J. Virol. 78:12446-12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhee, S. Y., T. F. Liu, S. P. Holmes, and R. W. Shafer. 2007. HIV-1 subtype B protease and reverse transcriptase amino acid covariation. PLoS Comput. Biol. 3:e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamiya, S., S. Mardy, M. F. Kavlick, K. Yoshimura, and H. Mistuya. 2004. Amino acid insertions near Gag cleavage sites restore the otherwise compromised replication of human immunodeficiency virus type 1 variants resistant to protease inhibitors. J. Virol. 78:12030-12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang, C., Y. Ndassa, and M. F. Summers. 2002. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat. Struct. Biol. 9:537-543. [DOI] [PubMed] [Google Scholar]
- 33.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wright, E., et al. 2008. Investigating antibody neutralization of lyssaviruses using lentiviral pseudotypes: a cross-species comparison. J. Gen. Virol. 89:2204-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zennou, V., F. Mammano, S. Paulous, D. Mathez, and F. Clavel. 1998. Loss of viral fitness associated with multiple Gag and Gag-Pol processing defects in human immunodeficiency virus type 1 variants selected for resistance to protease inhibitors in vivo. J. Virol. 72:3300-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]