Abstract
Raltegravir is highly efficacious in the treatment of HIV-1 infection. The prevalence and impact on virologic outcome of low-frequency resistant mutations among HIV-1-infected patients not previously treated with raltegravir have not been fully established. Samples from HIV treatment-experienced patients entering a clinical trial of raltegravir treatment were analyzed using a parallel allele-specific sequencing (PASS) assay that assessed six primary and six secondary integrase mutations. Patients who achieved and sustained virologic suppression (success patients, n = 36) and those who experienced virologic rebound (failure patients, n = 35) were compared. Patients who experienced treatment failure had twice as many raltegravir-associated resistance mutations prior to initiating treatment as those who achieved sustained virologic success, but the difference was not statistically significant. The frequency of nearly all detected resistance mutations was less than 1% of viral population, and the frequencies of mutations between the success and failure groups were similar. Expansion of pre-existing mutations (one primary and five secondary) was observed in 16 treatment failure patients in whom minority resistant mutations were detected at baseline, suggesting that they might play a role in the development of drug resistance. Two or more mutations were found in 13 patients (18.3%), but multiple mutations were not present in any single viral genome by linkage analysis. Our study demonstrates that low-frequency primary RAL-resistant mutations were uncommon, while minority secondary RAL-resistant mutations were more frequently detected in patients naïve to raltegravir. Additional studies in larger populations are warranted to fully understand the clinical implications of these mutations.
Raltegravir (RAL) is the first of a new class of antiretroviral drugs that target HIV integrase, demonstrating potent suppression of viral replication in HIV-1-infected individuals. RAL is highly effective for both treatment-naïve and -experienced patients (5, 10, 23). However, the development of drug resistance mutations can render the drug ineffective (5, 8). Primary mutations conferring RAL resistance develop through three independent pathways: Q148R/K/H, N155H and Y143R/C (5, 7, 8, 14, 15). They can affect integrase activity and viral replication. Many secondary mutations specific for each pathway are also identified (15). Those mutations themselves have little effect on RAL susceptibility. They, however, can augment RAL resistance when they are present together with primary mutations and affect viral replication capacity (2, 13, 15).
Studies of other antiretroviral drugs such as protease inhibitors (PIs) and reverse transcriptase inhibitors (RTIs) have shown that the presence of minority drug-resistant viruses before treatment may be associated with poor treatment responses following antiretroviral therapy (11, 16, 21, 22, 24). Recent studies have shown that minority RAL-associated drug-resistant mutations can also be detected in patients before treatment (2, 3, 12, 13, 17, 18). In these studies, however, drug-resistant mutations were mostly identified by population or clonal sequencing methods that are not sensitive for minority mutation populations present at frequencies less than 1% (2, 13, 17). A highly sensitive allele-specific PCR assay was used to detect lower-frequency resistant mutations, but only three sites were analyzed (3). In addition, whether different RAL resistance mutations are present in the same viral genome is unknown. Improved understanding of the prevalence and potential impact on treatment outcome of low-frequency RAL-resistant mutations in HIV-1-infected patients who have not been previously treated with RAL may help determine which patients are the best candidates for RAL.
We recently developed a highly sensitive parallel allele-specific sequencing (PASS) assay that can detect minority drug-resistant populations present in 0.1% to 0.01% of the viral population by simultaneously analyzing thousands of viral genomes in a single assay (1). In addition, this new technology allows for the identification of linkages of multiple drug-resistant mutations on individual viral genomes. In this study, we assayed baseline pre-RAL treatment samples for the presence of 12 primary and secondary RAL-resistant mutations from three pathways and evaluated the impact on RAL-based salvage treatment outcomes.
MATERIALS AND METHODS
Patient plasma samples.
Baseline plasma samples from 84 treatment-experienced patients in a phase III trial (MK-0518-019 study) were studied. Before patients started the RAL-containing salvage regimen, all had failed first-line treatment, and multiple drug-resistant mutations were present in reverse transcriptase and protease genes. Forty-four patients were responders, and 40 patients experienced virologic failure in accordance with the study protocol (23). Written informed consent was obtained from the individuals whose blood samples were collected. The study was approved by the Duke University Institutional Review Board.
Viral RNA extraction and cDNA synthesis.
One milliliter of each plasma sample was concentrated by ultracentrifugation at 32,000 rpm for 3 h at 4°C. The virus pellet was then resuspended in 200 μl of supernatant, and the viral RNA (vRNA) was extracted using a PureLink viral RNA/DNA minikit (Invitrogen, Carlsbad, CA). The vRNA was eluted into 17 μl of the elution buffer and used for cDNA synthesis using Superscript III (Invitrogen, Carlsbad, CA) and the primer IN-REVII 5′-CCTAGTGGGATGTGTACTTCTGA-3′ in a 40-μl reaction volume.
Detection of drug-resistant mutations by PASS.
The PASS assay was performed as previously described (1). Briefly, 20 μl of 6% acrylamide gel mix, containing 1 μM acrydite-modified reverse primer (5′Acr-ACACAATCATCACCTGCCATCTGTTT-3′), cDNA template, 0.3% diallyltartramide, 5% rhinohide polyacrylamide gel strengthener (Molecular Probes, Eugene, OR), 0.1% ammonium persulfate (APS), 0.1% TEMED (N,N,N′,N′-tetramethylethylenediamine), and 0.2% bovine serum albumin (BSA), was used to cast a gel on a bind-saline (Amersham Biosciences, Piscataway, NJ)-treated glass slide. Various amounts of cDNA (5 μl to 18.5 μl) were used for the PASS assay to obtain an optimal number of viral genomes (between 1,000 and 2,000, or as many as possible with low-viral-load samples) in each assay. The in-gel PCR amplification was then performed in a PTC-200 thermal cycler with a mix of 1 μM forward primer (5′-GAATTGGAGGAAATGAACAAGTAGATAAATTAG-3′), 0.1% Tween 20, 0.2% BSA, 1× PCR buffer, 100 μM deoxynucleoside triphosphate (dNTP) mix, 3.3 U of Jumpstart Taq DNA polymerase (Sigma, St. Louis, MO), and H2O (up to 300 μl) under a sealed SecurSeal chamber (Grace Bio-Labs, Inc., Bend, OR). The PCR conditions were as follows: 94°C for 3 min; 65 cycle of 94°C for 30 s, 56°C for 45 s, and 72°C for 3 min; 72°C for 6 min.
After PCR amplification, single-base extension (SBE) was performed with mutant and wild-type (WT) bases distinctively labeled with Cy3 and Cy5, respectively, using the primers that annealed just upstream of the targeted mutation sites. Six primary mutations (Q148R, Q148K, Q148H, N155H, Y143R, and Y143C) and six secondary mutations (G140S, G140A, L74M, E92Q, T97A, and Y143H) were selected for analysis based on our sequence database and previously published results (12, 20). The amplified viral genomes in each gel were then sequentially interrogated by 12 SBE reactions for targeted drug-resistant mutations. After each SBE, the gel was scanned to acquire images with a GenePix 4000B microarray scanner (Molecular Devices, Sunnyvale, CA).
PASS data analysis.
The two channel images (Cy5 for the WT base and Cy3 for the mutant base) acquired from each PASS assay were first cropped with Picture Window Pro3.5 to remove the edge area containing no signals. The cropped images were then analyzed with the Progenesis PG200 (Nonlinear Dynamics, Durham, NC) software. After background subtraction, normalization, and spot filter setting, only unambiguous spots at either channel were included for further analysis. The normalized pixel count data at multiple mutation sites at each spot were exported into an Excel file with a unique identifier. By comparing each spot's normalized values at both channels, the base was classified as either WT or mutant. Finally, the linkage pattern of all mutations on each viral genome was determined by compiling mutation information at all analyzed sites with the Linksys program developed in-house using macros in Excel.
Statistical analysis.
Poisson regression analysis was performed when total mutation sites was the dependent variable using PROC GENMOD in SAS v9.1 (SAS, Inc. Cary, NC). Logistic regression analysis was performed when any mutation was the dependent variable using PROC LOGISTIC in SAS v9.1. In these analyses, the base 10 log of viral load and the natural log of the number of genomes were used instead of the raw values, as they are presumed not to be on a linear scale.
RESULTS
Minority drug-resistant mutations were present in RAL treatment-naïve patients.
The RAL-associated drug-resistant mutations in 84 baseline samples were determined by PASS. Among them, 44 were from the treatment success patients and 40 were from the treatment failure patients. Samples with fewer than 10 detected viral genomes were excluded for analysis (8 treatment success patients and 5 treatment failure patients), since minority drug-resistant populations (<10%) could not be determined. Data obtained from 35 treatment failure patients and 36 treatment success patients were analyzed (Table 1). The average baseline viral loads were 4.73 log10 copies/ml and 4.99 log10 copies/ml in treatment success and failure groups, respectively (P > 0.05).
TABLE 1.
Number of patients with detected low-frequency RAL-resistant mutationsa
| Treatment group | No. (%) of patients |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Q148R/K/H pathway |
N155H pathway |
Y143R/C pathway |
||||||||||
| Q148R | Q148K | Q148H | G140S | G140A | N155H | L74 M | E92Q | T97A | Y143R | Y143C | Y143H | |
| Treatment success (n = 36) | 0 | 0 | 2 | 5 | 0 | 0 | 2 | 0 | 4 | 0 | 1 | 2 |
| Treatment failure (n = 35) | 2 | 1 | 1 | 9 | 0 | 1 | 3 | 1 | 5 | 2 | 3 | 4 |
| Total (n = 71) | 2 (2.8) | 1 (1.4) | 3 (4.2) | 14 (19.7) | 0 | 1 (1.4) | 5 (7.0) | 1 (1.4) | 9 (12.7) | 2 (2.8) | 4 (5.6) | 6 (8.4) |
Boldface type indicates primary mutations.
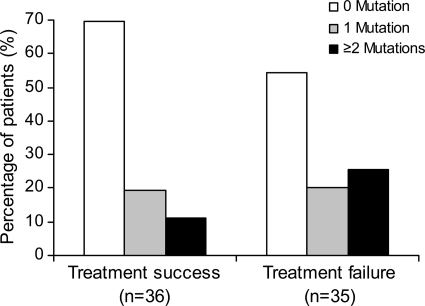
An average of 630 (14 to 3,636) and 1,131 (14 to 3,903) viral genomes were analyzed in the treatment success and failure groups, respectively. All of the mutations assessed were identified in at least one sample except G140A. Overall, more patients in the treatment failure group had baseline resistance mutations (46%) than in the treatment success group (31%), although this difference was not statistically significant. Single RAL resistance mutations were present in equal numbers of treatment success and failure patients, but the presence of two or more mutations was found in more than twice as many treatment failure patients (26%) as treatment success patients (11%) (Fig. 1). One treatment success patient was found to carry three mutations (G140S, T97A, and Y143H), while five treatment failure patients had three mutations (G140S, E92Q, and T97A; G140S, L74M, and T97A; or Q148R, T97A, and Y143R) or four mutations (Q148K, T97A, Y143C, and Y143H or G140S, T97A, Y143C, and Y143H). Among all patients with three or more mutations, only the T97A mutation detected at baseline was also detected by population sequencing at virologic failure in one patient. Linkage analysis showed that none of the multiple mutations were present in the same virus genome.
FIG. 1.
Percentage of patients with different numbers of RAL-resistant mutations. RAL-resistant mutations were determined by PASS in all patients. The percentages of patients with 0 (white), 1 (gray), or more (black) mutations were compared between treatment success (n = 36) and treatment failure (n = 35) groups.
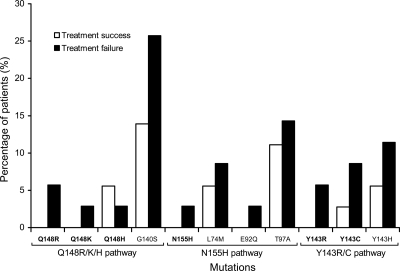
Detection of primary RAL resistance mutations at baseline was uncommon, being identified in fewer than 10% of patients in the treatment failure group (2.9% to 8.6%) and even less frequently among patients in the treatment success group. For the latter group, only two primary mutations (Q148H and Y143C) were detected in 5.6% and 2.8% of patients, respectively (Fig. 2). N155H is considered a major primary resistance mutation that confers resistance to RAL (5, 15). This mutation was detected in only one treatment failure patient at baseline (Table 1). Greater numbers of primary resistance mutations from the other two pathways were identified in the treatment failure group than in the treatment success group: 4 versus 2 for the Q148R/K/H pathway and 5 versus 1 for Y143R/C pathway. More than one primary mutation was detected in three treatment failure patients (patient A with Q148K and Y143C, patient B with Q148R and Y143R, and patient C with Q148H and N155H) but in only one treatment success patient (patient D with Q148H and Y143C). None of these differences were statistically significant.
FIG. 2.
Percentage of patients with RAL-resistant mutations in treatment success and treatment failure patients. The frequency of each RAL-resistant mutation from three pathways was compared between treatment success (white) and treatment failure (black) patients.
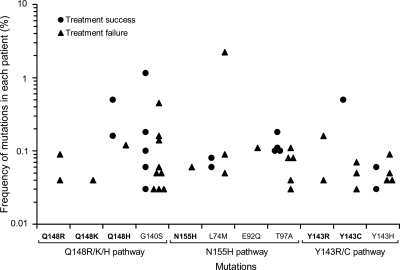
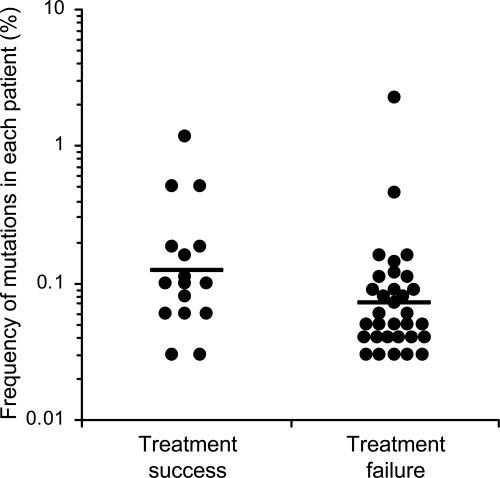
While primary mutations were rare, baseline secondary mutations were more frequently detected in both groups of patients, being seen in almost three times as many patients as was noted with primary mutations (35 versus 13). As with primary mutations, more secondary mutations were found in the treatment failure group than the treatment success group (22 versus 13), although this difference was not statistically significant (Table 1 and Fig. 2). Two secondary mutations were found to be present in more than 10% of the patients: G140S (19.7%) and T97A (12.7%). All detected mutations were present at very low frequencies (<1%), except two secondary mutations in two patients (2.2% for L74M and 1.1% for G140S) (Fig. 3). There were no significant differences in frequencies of minority mutations between the treatment success and the treatment failure groups (Fig. 4).
FIG. 3.
Frequency of minority RAL-resistant mutations in each patient. The percentage of each RAL-resistant mutation was determined by PASS in each patient. Each symbol represents the percentage of one mutation in either treatment success (circles) or treatment failure (triangles) patients.
FIG. 4.
Comparison of frequencies of minority RAL-resistant mutations between treatment success and treatment failure groups. Geometric means of minority drug resistance mutation percentages were compared between treatment success and treatment failure groups using a two-tailed t test (P = 0.06).
Pre-existing RAL-associated drug-resistant mutations were selected in some treatment failure patients.
Expansion of pre-existing minority drug-resistant mutations was detected by a population-based genotypic assay in six patients after viral rebound occurred (Table 2). The N155H mutation was present in only one patient at baseline, and it expanded following treatment failure and became detectable by population sequencing, which requires a mutation present at >20% of the population (25). Interestingly, the primary RAL-associated Q148H mutation was also present at baseline in the same patient, but it was not detected after treatment failure. All other expanded pre-existing mutations were secondary mutations. G140S was found in nine patients at baseline. However, the expansion of this mutation was only observed in three patients by population genotypic sequencing after viral rebound. These results demonstrate that pre-existing minority RAL-associated resistance mutations can expand to become a major viral population under drug selection pressure, but in general, this occurred in only a small subset of treatment failure patients.
TABLE 2.
RAL resistant mutations detected at baseline and after treatment failure
| Mutation | No. of patients with the mutation |
|
|---|---|---|
| Baselinea | Treatment failureb | |
| Q148R | 2 | 0 |
| Q148K | 1 | 0 |
| Q148H | 1 | 0 |
| G140S | 9 | 3 |
| G140A | 0 | 0 |
| N155H | 1 | 1 |
| L74M | 3 | 1 |
| E92Q | 1 | 0 |
| T97A | 5 | 1 |
| Y143R | 2 | 0 |
| Y143C | 3 | 0 |
| Y143H | 4 | 0 |
Determined by PASS.
Determined by populaiotn sequencing.
Low-frequency mutations detected by PASS were higher than the assay error rate.
A total of 48 drug-resistant mutation events were identified in 27 patients prior to treatment. In most cases, one or two mutations were detected in a patient. Six patients were found to have three or more mutations. The maximum number of detected drug-resistant mutations in any single patient was four. Since the frequencies of those minority mutations were very low, it was important to determine if they represented real mutations present in the patients or mutations generated by the assay itself.
To address this issue, we determined mutation rates for all possible misincorporations at each base using both DNA and RNA templates in the PASS assay. When a plasmid DNA containing the partial HIV-1 pol gene was used as the template, misincorporations were only detected at A (to G), T (to C) and C (to A) bases after nearly two million bases were analyzed (Table 3). The overall mutation rate for the DNA template was 0.6 × 10−5 in the PASS assay. Since we detected drug-resistant mutations from viral RNA, we next determined the assay error rate with the RNA template that was generated by an in vitro T7 transcription system. Mutations were detected at all possible misincorporation scenarios except one (T to G) (Table 4). The error rates were highest at three misincorporation pairs (T:C, A:C, and G:A). The overall mutation rate for the RNA template was 5.5 × 10−5, which was about 10-fold higher than that for the DNA template. Of note, the error rate with the RNA template also included the mutations that were contributed by T7 RNA polymerase. Thus, the actual mutation rate with the RNA templates should be lower in the PASS assay. Compared to the assay background for the specific misincorporation at each base, nearly all frequencies of detected resistant mutations in this study were at least threefold higher than the background. In only one case, the frequency of resistant mutations was less than 3 fold (Y143C; 2.7-fold). The data showed that the mutations detected by the PASS assay most likely represent the real mutations present in patients.
TABLE 3.
Error rates at each nucleoside with DNA templates in the PASS assay
| Expected base | Misinorporated base | No. of misincorporations | No. of analyzed genomes | Error rate |
|---|---|---|---|---|
| A | T | 0 | 177,000 | <0.6 × 10−5 |
| C | 0 | 177,000 | <0.6 × 10−5 | |
| G | 2 | 177,000 | 1.1 × 10−5 | |
| T | A | 0 | 177,000 | <0.6 × 10−5 |
| C | 2 | 177,000 | 1.1 × 10−5 | |
| G | 0 | 177,000 | <0.6 × 10−5 | |
| C | A | 7 | 102,000 | 6.9 × 10−5 |
| T | 0 | 102,000 | <1.0 × 10−5 | |
| G | 0 | 102,000 | <1.0 × 10−5 | |
| G | A | 0 | 177,000 | <0.6 × 10−5 |
| T | 0 | 177,000 | <0.6 × 10−5 | |
| C | 0 | 177,000 | <0.6 × 10−5 | |
| Overall | 11 | 1,899,000 | 0.6 × 10−5 |
TABLE 4.
Error rates at each nucleoside with RNA templates in the PASS assay
| Expected base | Misinorporated base | No. of misincorporations | No. of analyzed genomes | Error rate |
|---|---|---|---|---|
| A | T | 2 | 96,000 | 2.1 × 10−5 |
| C | 4 | 96,000 | 4.2 × 10−5 | |
| G | 9 | 96,000 | 9.5 × 10−5 | |
| T | A | 1 | 75,000 | 1.3 × 10−5 |
| C | 1 | 75,000 | 1.3 × 10−5 | |
| G | 0 | 75,000 | <1.3 × 10−5 | |
| C | A | 10 | 102,000 | 9.8 × 10−5 |
| T | 20 | 102,000 | 19.6 × 10−5 | |
| G | 3 | 102,000 | 2.9 × 10−5 | |
| G | A | 6 | 96,000 | 6.3 × 10−5 |
| T | 4 | 96,000 | 4.2 × 10−5 | |
| C | 1 | 96,000 | 1.0 × 10−5 | |
| Overall | 61 | 1,107,000 | 5.5 × 10−5 |
DISCUSSION
Low-frequency mutations in HIV integrase that are associated with resistance to raltegravir were detected in 38% of pretreatment baseline samples from 71 patients enrolled in the BENCHMRK study based on the analysis of a large number of viral genomes from each patient using the highly sensitive PASS assay. Primary mutations were uncommon: a total of 13 mutations were identified in 9 (12.7%) patients, while secondary mutations were more often detected: 35 mutations in 24 (33.8%) patients. Patients experiencing RAL treatment failure had twice as many RAL-associated mutations as the treatment success patients. More patients in the treatment failure group were also found to carry two or more resistance mutations than in the treatment success group. However, the differences observed between the treatment success and failure groups were not statistically significant, probably because the number of patients studied had insufficient power to detect a significant difference. It is also possible that these low-frequency resistance mutations do not contribute significantly to virologic failure.
Two or more mutations were detected in about 20% of the patients. However, none of them were found in single viral genomes by linkage analysis. Our data confirmed that it was extremely rare for two or more mutations to be present in the same virus genome, as proposed by Colgrove and Japour (4).
Previous studies have shown that primary RAL-associated mutations were not detectable at baseline using less-sensitive population or clonal-based assays (2, 13, 17). This is not unexpected, since our data showed that primary mutations are rare and only present at frequencies less than 1% of the viral population, which are below the detection limits for those assays. However, rare primary mutations were found when sequences from a large number of infected individuals were analyzed (12, 18). Secondary mutations were more commonly detected in the baseline samples, but they were found to have little impact on susceptibility to integrase inhibitors in the absence of specific primary mutations (2, 13). Using a more sensitive allele-specific real-time PCR method, Charpentier et al. studied low-frequency variants with specific primers for the primary mutations Q148H, Q148R, and N155H (detection limit, 0.1%, 0.1%, and 0.05%, respectively) in 91 patients (3). At baseline, the Q148R variant was detected at a low level (0.4% of the viral population) in most patients (81% in treatment-experienced and 86% in naïve patients), but the Q148H and N155H mutations were not detected. The rate of the Q148R mutation was significantly higher than that observed in our study. The clinical significance of pre-existing drug-resistant mutations is uncertain, as the relationship between treatment failure and baseline low-frequency resistance mutations has not been conclusively established (2, 3, 13, 17).
In this study, minority drug-resistant mutations were identified by analyzing a large number of viral genomes in each patient. By the nature of the study, the frequencies of such mutations were low. Thus, in many cases, only one or a small number of mutations were detected even when thousands of viral genomes were analyzed. However, since all but one of the frequencies of the detected drug-resistant mutations were at least 3-fold higher than the background of the PASS assay, they likely represent real mutations present in the patients. The result also demonstrates that the value of the PASS assay in studying minority drug-resistant mutations in HIV-1-infected patients. It will be of interest to compare the results generated by PASS and other ultra-deep sequencing methods such as 454 Sequencing Systems in future studies.
Expansion of pretreatment low-level RAL-associated mutations was observed in the rebounding virus following treatment failure. This was identified in 6 of 16 patients in whom baseline minority drug-resistant mutations were detected. The result suggests that these mutations may play a role in development of resistance to raltegravir. However, most expanded mutations were secondary mutations. Since secondary mutations alone generally do not confer drug resistance but instead increase levels of resistance in conjunction with primary resistance mutations to RAL (6, 8, 13, 19), the role of this expansion in RAL resistance is uncertain. Determination of the frequencies of each mutation and linkage relationships with primary mutations from longitudinal samples during the development of drug resistance in patients will help answer this question.
Pre-existing minority drug-resistant mutations have been shown to play an important role in the development of drug resistance in PI- and RTI-treated patients (11, 16, 21, 22, 24). To date, no clear association has been confirmed in integrase inhibitor-treated patients (2, 3, 13, 17). Our study demonstrates that pre-existing low-frequency drug-resistant mutations were present in both the treatment success and failure patients, that the prevalence of resistant mutations varied among patients, and that all mutations identified were present at low frequencies (<1%). It may be important to determine the frequency threshold at which the pre-existing mutations will likely lead to drug resistance (9, 16). It is possible that mutations present at very low frequencies (less than 0.1%) may not be clinically relevant, although this may differ depending on the specific mutation in question. Understanding the clinical implications of pre-existing minority resistant mutations in patients will require longitudinal follow-up on a much larger population of raltegravir-treated patients.
Acknowledgments
We thank Jing Li for software development and Brooke Walker for preparation of the manuscript.
This study was supported by a grant from the Merck Investigator-Initiated Studies Program (IISP 33026) and grants from the National Institutes of Health (GM065057 and AI64518).
Footnotes
Published ahead of print on 20 December 2010.
REFERENCES
- 1.Cai, F., et al. 2007. Detection of minor drug-resistant populations by parallel allele-specific sequencing. Nat. Methods 4:123-125. [DOI] [PubMed] [Google Scholar]
- 2.Ceccherini-Silberstein, F., et al. 2010. Secondary HIV-1 integrase resistance mutations, found as minority quasispecies in integrase therapy naive patients, have little or no effect on susceptibility to integrase inhibitors. Antimicrob. Agents Chemother. 54:3938-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charpentier, C., et al. 2010. High frequency of integrase Q148R minority variants in HIV-infected patients naive of integrase inhibitors. AIDS 24:867-873. [DOI] [PubMed] [Google Scholar]
- 4.Colgrove, R., and A. Japour. 1999. A combinatorial ledge: reverse transcriptase fidelity, total body viral burden, and the implications of multiple-drug HIV therapy for the evolution of antiviral resistance. Antiviral Res. 41:45-56. [DOI] [PubMed] [Google Scholar]
- 5.Cooper, D. A., et al. 2008. Subgroup and resistance analyses of raltegravir for resistant HIV-1 infection. N. Engl. J. Med. 359:355-365. [DOI] [PubMed] [Google Scholar]
- 6.Delelis, O., et al. 2009. The G140S mutation in HIV integrases from raltegravir-resistant patients rescues catalytic defect due to the resistance Q148H mutation. Nucleic Acids Res. 37:1193-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delelis, O., et al. 2010. Impact of Y143 HIV-1 integrase mutations on resistance to raltegravir in vitro and in vivo. Antimicrob. Agents Chemother. 54:491-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fransen, S., et al. 2009. Loss of raltegravir susceptibility by human immunodeficiency virus type 1 is conferred via multiple nonoverlapping genetic pathways. J. Virol. 83:11440-11446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao, F., and D. Wang. 2007. Minor drug-resistant HIV populations and treatment failure. Future Virol. 2:293-302. [Google Scholar]
- 10.Hicks, C., and R. M. Gulick. 2009. Raltegravir: the first HIV type 1 integrase inhibitor. Clin. Infect. Dis. 48:931-939. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, J. A., et al. 2008. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naive populations and associate with reduced treatment efficacy. PLoS Med. 5:e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lataillade, M., J. Chiarella, and M. J. Kozal. 2007. Natural polymorphism of the HIV-1 integrase gene and mutations associated with integrase inhibitor resistance. Antivir. Ther. 12:563-570. [PubMed] [Google Scholar]
- 13.Low, A., et al. 2009. Natural polymorphisms of human immunodeficiency virus type 1 integrase and inherent susceptibilities to a panel of integrase inhibitors. Antimicrob. Agents Chemother. 53:4275-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malet, I., et al. 2008. Mutations associated with failure of raltegravir treatment affect integrase sensitivity to the inhibitor in vitro. Antimicrob. Agents Chemother. 52:1351-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Métifiot, M., C. Marchand, K. Maddali, and Y. Pommier. 2010. Resistance to integrase inhibitors. Viruses 2:1347-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Metzner, K. J., et al. 2009. Minority quasispecies of drug-resistant HIV-1 that lead to early therapy failure in treatment-naive and -adherent patients. Clin. Infect. Dis. 48:239-247. [DOI] [PubMed] [Google Scholar]
- 17.Miller, M. D., R. M. Danovich, and M. Witmer. 2009. Emerging patterns of resistance to integrase inhibitors, abstr. 125. 16th Conf. Retrovir. Opportunistic Infect.
- 18.Myers, R. E., and D. Pillay. 2008. Analysis of natural sequence variation and covariation in human immunodeficiency virus type 1 integrase. J. Virol. 82:9228-9235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quercia, R., E. Dam, D. Perez-Bercoff, and F. Clavel. 2009. Selective-advantage profile of human immunodeficiency virus type 1 integrase mutants explains in vivo evolution of raltegravir resistance genotypes. J. Virol. 83:10245-10249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhee, S. Y., et al. 2008. Natural variation of HIV-1 group M integrase: implications for a new class of antiretroviral inhibitors. Retrovirology 5:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shafer, R. W. 2009. Low-abundance drug-resistant HIV-1 variants: finding significance in an era of abundant diagnostic and therapeutic options. J. Infect. Dis. 199:610-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simen, B. B., et al. 2009. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J. Infect. Dis. 199:693-701. [DOI] [PubMed] [Google Scholar]
- 23.Steigbigel, R. T., et al. 2008. Raltegravir with optimized background therapy for resistant HIV-1 infection. N. Engl. J. Med. 359:339-354. [DOI] [PubMed] [Google Scholar]
- 24.Svarovskaia, E. S., et al. 2007. Low-level K65R mutation in HIV-1 reverse transcriptase of treatment-experienced patients exposed to abacavir or didanosine. J. Acquir. Immune Defic. Syndr. 46:174-180. [DOI] [PubMed] [Google Scholar]
- 25.Van Laethem, K., et al. 1999. Phenotypic assays and sequencing are less sensitive than point mutation assays for detection of resistance in mixed HIV-1 genotypic populations. J. Acquir. Immune Defic. Syndr. 22:107-118. [DOI] [PubMed] [Google Scholar]