Abstract
For the first time, the mechanism of action of microcin L (MccL) was investigated in live bacteria. MccL is a gene-encoded peptide produced by Escherichia coli LR05 that exhibits a strong antibacterial activity against related Enterobacteriaceae, including Salmonella enterica serovars Typhimurium and Enteritidis. We first subcloned the MccL genetic system to remove the sequences not involved in MccL production. We then optimized the MccL purification procedure to obtain large amounts of purified microcin to investigate its antimicrobial and membrane properties. We showed that MccL did not induce outer membrane permeabilization, which indicated that MccL did not use this way to kill the sensitive cell or to enter into it. Using a set of E. coli and Salmonella enterica mutants lacking iron-siderophore receptors, we demonstrated that the MccL uptake required the outer membrane receptor Cir. Moreover, the MccL bactericidal activity was shown to depend on the TonB protein that transduces the proton-motive force of the cytoplasmic membrane to transport iron-siderophore complexes across the outer membrane. Using carbonyl cyanide 3-chlorophenylhydrazone, which is known to fully dissipate the proton-motive force, we proved that the proton-motive force was required for the bactericidal activity of MccL on E. coli. In addition, we showed that a primary target of MccL could be the cytoplasmic membrane: a high level of MccL disrupted the inner membrane potential of E. coli cells. However, no permeabilization of the membrane was detected.
Microcins (Mcc) are bacteriocins secreted by members of the Enterobacteriaceae family, in particular Escherichia coli strains. They constitute a class of low-molecular-mass antimicrobial peptides (<10 kDa) that exhibit a narrow antimicrobial spectrum of activity directed against bacterial species phylogenetically related to the producing strains. They are, therefore, believed to be involved in microbial competition within the intestinal tract. We classified these peptides in two classes (20, 39). Class I, which includes (to date) MccB17, MccC7/C51, MccJ25, and presumably MccD93, encompasses very-low-molecular-mass peptides (<5 kDa) that are highly posttranslationally modified. Class II includes higher-molecular-mass peptides (ranging from 5 to 10 kDa) and now is subdivided into two subclasses (16): class IIa microcins contain disulfide bonds but no further posttranslational modification (MccL, MccV, and Mcc24), and class IIb microcins are linear peptides that may carry a C-terminal posttranslational modification (MccE492, MccM, MccH47, and MccI47) and correspond to the family of siderophore-microcins (57).
The broad variety of microcin structures results in a diversity of mechanisms of action, such as the inhibition of vital enzymatic functions and damage to the inner membrane. However, the initial recognition pathways are common to several microcins. Except for MccB17, which uses an outer membrane porin, OmpF, to be transferred into the periplasm (31), the uptake of all studied microcins is dependent on outer membrane receptors of iron-siderophores and their associated energy transduction system, the TonB-ExbB-ExbD inner membrane complex (11). MccJ25 was shown to require the ferrichrome receptor FhuA to interact with the outer membrane (51). While MccE492, MccH47, and MccM would bind the three receptors of catecholate siderophore, FepA, Fiu, and Cir in Escherichia coli and FepA, IroN, and Cir in Salmonella Typhimurium (11, 38, 57), MccV recognition would need the Cir outer membrane protein only (12). These microcins then would be actively transferred into the periplasm via the Ton system, which uses the proton-motive force energy from the inner membrane (8, 41). After initial recognition on sensitive bacteria, microcins act on their cellular target(s) to exert their antibacterial activity. Class I microcins were shown to target intracellular enzymes responsible for DNA/RNA structure or synthesis. Therefore, these peptides need to cross the inner membrane. The small-size microcins MccB17 and MccJ25 require an inner membrane protein, SbmA, to be internalized into the bacterial cytoplasm (31, 52), where they target the DNA gyrase (27, 58) and the RNA polymerase (14, 61), respectively. MccJ25 also was reported to disrupt inner membrane integrity in Salmonella enterica serovar Newport (46), in liposomes (45), and in uncharged phospholipids monolayers (3). MccC7 targets translation by blocking the function of the aspartyl-tRNA synthetase (34), but its cytoplasmic membrane receptor still is unknown (23). Among class II microcins, only MccE492, MccH47, and MccV were studied and were shown to target the cytoplasmic membrane. MccE492 inserts into the inner membrane and destabilizes the membrane potential by pore formation (13, 15, 30). MccV was reported to abolish the E. coli membrane potential, but pore formation was not demonstrated (60). Laviňa and collaborators proposed a cytoplasmic membrane target for MccH47: the Fo proton channel of the ATP synthase was necessary for the MccH47 antibacterial activity (47, 56).
We isolated microcin L (MccL) from a microcinogenic strain, E. coli LR05, that produced three other microcins, MccJ25, MccB17, and MccD93, and also had immunity to MccV (20, 49). MccL exhibits a strong antibacterial activity against related Enterobacteriaceae, including Salmonella enterica serovars Typhimurium and Enteritidis (20, 40). The MccL cluster consists of four genes: one structural gene, mclC, one immunity gene, mclI, and two export genes, mclA and mclB, with a strong relatedness to the ABC transporter proteins and accessory factors involved in the secretion of MccE492, H47, 24, and V. The mclC structural gene encodes a 105-amino-acid precursor with a 15-amino-acid N-terminal extension ending with a Gly-Ala motif upstream of the cleavage site (40). MccL is a 90-amino-acid peptide of 8,884 Da. It is an anionic and hydrophobic peptide with more than 45% nonpolar amino acids and without any posttranslational modification. It contains a high content of glycine (15.6%) and four cysteines engaged in two intramolecular disulfide bridges, conferring to the mature microcin a high stability. The N-terminal region has significant homologies with several Gram-positive bacteriocins, and the C-terminal 32-amino-acid sequence is 87.5% identical to that of MccV (18, 40). At this stage, the MccL mechanism of action was not elucidated.
In this work, we reduced the 13.5-kb DNA fragment bearing the MccL genetic system in an attempt to achieve a strain with a plasmid containing only the 4-kb MccL genetic system. We then used the obtained strain to optimize the purification procedure to obtain large amounts of MccL purified to homogeneity to investigate its antimicrobial and membrane properties. For the first time, the MccL mechanism of action was investigated. We determined the proteins involved in MccL uptake in sensitive E. coli and S. enterica. Moreover, we proved that the microcin induced the disruption of the proton-motive force at the cytoplasmic membrane. However, no membrane permeabilization was detected.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Escherichia coli K-12 and Salmonella enterica strains used in this study are listed in Table 1. Unless otherwise stated, all strains were grown in brain heart infusion medium at 37°C. Luria-Bertani (LB) rich medium and M63 minimal medium supplemented with glucose (2 g/liter), MgSO4 (1 mM), and thiamine (1 mg/liter) were prepared as described by Miller (35). Nutrient medium was used for the activity assays. Soft and normal solid media were prepared by adding 0.6 or 1.2% (wt/vol) agar, respectively. Antibiotics were added to media at the following final concentrations: ampicillin (AMP), 100 μg/ml; tetracycline (TET), 20 μg/ml; kanamycin (KAN), 30 μg/ml; streptomycin (STR), 100 μg/ml; and nalidixic acid (NAL), 40 μg/ml.
TABLE 1.
Wild-type and mutant bacterial strains used in this study
| Strain | Genotype/phenotype | Source or reference |
|---|---|---|
| Escherichia coli K-12 | ||
| MC4100 | F−araD Δ(argF lac)U169 rpsL relA flb deoC1 ptsF25 rbsR (Strr) | 26 |
| MC4100 pL102 | MccL (Strr Ampr) | 40 |
| MC4100 pL104 | MccL (pL102 derivative) (Strr Ampr) | This work |
| TG1 | supE thi-1Δ(lac-proAB)Δ(mcrB-hsdSM)5(rK− mK−)[F′ traD36 proAB lacIqZΔM15] | Stratagene |
| ML35p | lacI lacY lacZ+ pBR322 (Ampr) | 33 |
| H1443 | MC4100 aroB(Strr) | 26 |
| MS172 | H1443 fhuE::λplacMu53 (Strr Kanr) | 54 |
| ZI311 | H1443 fecA zag::Tn10 (Strr Tetr) | K. Hantke |
| H873 | H1443 fepA::Tn10 (Strr Tetr) | K. Hantke |
| H1594 | H1443 fiu::Mud1X (Strr Ampr) | 25 |
| VR42 | H1443 cir (Strr) | 26 |
| H1728 | H1443 fiu::Mud1X cir (Strr Ampr) | 26 |
| H1729 | H1443 cir fhuA fiu::Mud1X (Strr Ampr) | 54 |
| H1875 | H1443 fepA::Tn10 cir::Mud1X (Strr Tetr Ampr) | 26 |
| H1876 | H1443 fepA::Tn10 fiu::Mud1X cir (Strr Tetr Ampr) | 26 |
| H1877 | H1443 fepA::Tn10 fiu::Mud1X (Strr Tetr Ampr) | 26 |
| MnTM | MC4100 sdaC::Tn10 (Strr Tetr) | 21 |
| AB2847 | aroB tsx thi lamB (Ampr) | K. Hantke |
| AB2847pT04 | ColM | K. Hantke |
| P8 | AB2847 fhuA | F. Moreno |
| W3110 | F− IN(rrnD-rrnE)1 | 1 |
| KP1032 | W3110 tonB::kan (Kanr) | 1 |
| KP1036 | W3110 exbB::Tn10 (Tetr) | 1 |
| KP1118 | W3110 exbB::Tn10 exbD::Tn phoA (Tetr Kanr) | 1 |
| KP1143 | W3110 exbD::Tn phoA (Kanr) | 1 |
| GM1 | ara Δ(pro-lac) thi F′ pro lac | 1 |
| KP1082 | GM1 Δ(ana-trp-tonB) | 1 |
| KP1037 | GM1 exbB::Tn10 (Tetr) | K. Postle |
| TPS13 | GM1 tolQ | K. Postle |
| KP1038 | GM1 tolQ exbB::Tn10 (Tetr) | K. Postle |
| DB503 | MC4100 malE16-1 Δara714 | 4 |
| AB4 | DB503 manY::Tn10 | 4 |
| AB3 | DB503 ΔmanXYZ::cat | 4 |
| Salmonella enterica serovar Typhimurium | ||
| ATCC14028 | Wild-type strain | 44 |
| AJB64 | ATCC14028 Nalr iroN::pGP704 aroA::Tn10 (Ampr Tetr) | 44 |
| AR8412 | ATCC14028 cir::MudJ (Kanr) | 44 |
| WR1726 | ATCC14028 fepA::Tn10dTc (Ampr) | 44 |
| WR1727 | ATCC14028 fepA::Tn10dTc iroN::pGP704 (Ampr Tetr) | 44 |
| WR1728 | ATCC14028 fepA::Tn10dTc iroN::pGP704 cir::MudJ (Ampr Tetr Kanr) | 44 |
| A36 | Wild-type strain, phage type DT36 | 36 |
| WR1174 | A36 tonB::MudJ (Kanr) | 36 |
| Salmonella enterica serovar Enteritidis | ||
| SE147 Nalr | Wild-type strain, phage type 4 (Nalr) | 44 |
| WR1425 | SE147 Nalr fepA::Tn10dTc (Tetr) | 44 |
| WR1434 | SE147 Nalr fepA::Tn10dTc iroN::pGP704 (Ampr Tetr) | 44 |
| WR1530 | SE147 Nalr cir::MudJ (Kanr) | 44 |
| WR1458 | SE147 Nalr iroN::pGP704 cir::MudJ (Ampr Kanr) | 44 |
| WR1529 | SE147 Nalr tonB::MudJ (Kanr) | 44 |
DNA extraction.
All plasmids were extracted from E. coli using the Qiagen plasmid midi kit or QIAprep spin miniprep kit, depending on the quantity required, or following the rapid protocol of Birnboim and Doly (5).
DNA sequencing.
Each sequencing reaction was carried out in a total volume of 10 μl containing 500 ng DNA plasmid, 0.4 μM primer, and 4 μl BigDye terminator ready reaction mix (ABI Prism BigDye terminator v3.1 ready reaction cycle sequencing kit from Applied Biosystems). Twenty-five sequencing cycles (96°C for 10 s, 50°C for 5 s, and 60°C for 4 min) in a Perkin-Elmer thermal cycler were followed by purification on a DyeEx column (Qiagen). Ten μl of template suppression reagent then was added to each sample, and a denaturing cycle (94°C, 5 min) was performed before electrophoresis in an ABI Prism 310 DNA sequencer (Applied Biosystems).
pL102 shortening.
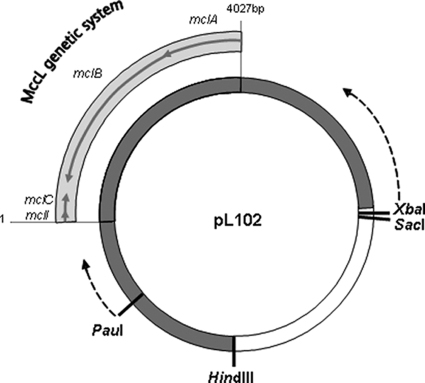
Plasmid pL102 (Fig. 1), carrying the MccL genetic system, was constructed in a previous work (49) by inserting, in the pUC19 vector, a SalI-HindIII DNA fragment of about 13.5 kb from the wild-type strain E. coli LR05. pL104, bearing the MccL genetic system, was constructed in this work by reducing the 13.5-kb fragment of pL102 to 5.5 kb. To shorten the 5′ end of the pL102 insert, 10 μg pL102 was digested with 40 U SacI and XbaI in SacI buffer (Eurogentec) at 37°C for 4 h. After the heat inactivation of the restriction enzymes, the linearized plasmid (4.75 μg) was subjected to the ExoIII nuclease according to the ExoIII/S1 kit (Fermentas) at 30°C for 0, 5, 10, 12, 14, 16, and 18 min and then in 1-min increments from 20 to 36 min. After S1 nuclease digestion for 30 min at room temperature, the reaction was stopped and a ligation with T4 DNA ligase was performed according to the ExoIII/S1 kit (Fermentas) protocol. Competent E. coli TG1 cells were prepared with CaCl2, and transformations were performed by following the molecular cloning manual protocols (53). Cells were grown on LB agar with ampicillin. The plasmid that has been subjected to 12 min of ExoIII action, called p12′, contained the shortest insert with a complete MccL genetic system. This plasmid (5.7 μg) then was digested with 30 U HindIII (Eurogentec) in Fermentas buffer R (required for the second restriction digest) for 3 h at 37°C. The single-strand ends generated were filled with α-phosphorothioate deoxynucleotides and the DNA polymerase I Klenow fragment according to the ExoIII/S1 kit protocol. The DNA then was digested with 15 U PauI (Fermentas) for 2 h at 37°C. We then used the same protocol as that described before, except for the ExoIII incubation times (from 0 to 22 min, with 2-min timed aliquots). A plasmid containing a 5.5-kb insert bearing the MccL genetic system was selected for the following works and called pL104.
FIG. 1.
pL102 presentation and shortening description. pL102 is composed of a 13.5-kb DNA fragment (▪) inserted in the pUC19 vector (□).The mclC structural gene, the mclI immunity gene, and the mclA and mclB export genes constitute the MccL genetic system. The restriction sites used for the insert shortening were XbaI/SacI on one side and HindIII/PauI on the other side. The HindIII-PauI fragment was deleted. The dotted arrows indicate the start points and the direction of the shortening.
Purification and production of microcin L.
Flasks (2,000 ml) containing 400 ml of M63 broth were seeded with a 2% (vol/vol) inoculum from an overnight culture of E. coli MC4100 pL104 and incubated at 37°C for 6 h with shaking (150 rpm). The culture was centrifuged (19,000 × g, 15 min, 4°C), and the supernatant was directly loaded onto Sep-Pak plus environmental C18 cartridges (Waters). First, the elution was performed with methanol and the collected fraction was diluted 1:3 (vol/vol) in water before being subjected to a second solid-phase extraction onto the same cartridges. After a washing step, first with water and then with 50% methanol in water, MccL was eluted with 45% acetonitrile in water (partly purified MccL). This 45% acetonitrile Sep-Pak fraction then was concentrated under vacuum in a Maxy Dry plus concentrator (Heto), and filtered samples (2 ml) were further subjected to reverse-phase high-performance liquid chromatography (RP-HPLC) on a C8 Silice Uptisphere column (5 μm, 300 Å; Interchim). Separation was performed with the following acetonitrile gradient in acidified water (0.1% trifluoroacetic acid [TFA]): 30% for 10 min, 30 to 50% for 20 min, 50% for 5 min, 50 to 100% for 10 min, and a step at 100% for 5 min, all at a flow rate of 0.7 ml/min. Absorbance was monitored at 214 nm. The peak fraction of MccL, eluted during the step at 50% acetonitrile, was diluted 1:1 (vol/vol) in water and subjected to an additional and identical RP-HPLC analysis. All of the collected fractions containing pure MccL were pooled, concentrated under vacuum, and stored at −20°C. Concentrations of peptide solutions were determined by the bicinchoninic acid method (BC assay; Uptima-Interchim) using bovine serum albumin as the standard and by absorbance determination at 280 nm (MccL ɛ280, 19,730 M−1·cm−1) (37). Both methods gave equivalent results. To control the peptide purity, purified MccL samples were analyzed with a Micromass QTOF II mass spectrometer as previously described (40).
Microcin activity.
MccL activity was assayed by adapting the agar well diffusion assay previously described by Sablé et al. (48). Sterile glass rings (4-mm inside diameter) were placed on solid nutrient plates (15 ml) and filled with 20 μl of samples to be tested. After the total diffusion of the samples, rings were removed and plates were overlaid with 5 ml of soft nutrient agar seeded with 107 CFU/ml of the target strain. The clear zones of growth inhibition were measured after overnight incubation at 37°C. Unless otherwise stated, E. coli MC4100 was used as the standard indicator strain. Inhibition assays were done in duplicate.
Determination of MIC and MBC.
The minimum inhibitory concentration (MIC) was determined by the standard macrodilution broth method as described by Sahm and Washington (50). We checked that the highest concentration of acetonitrile in broth did not affect the growth of tested bacteria. The minimum bactericidal concentration (MBC) was determined from tubes showing complete inhibition. A nutrient agar plate was seeded on the surface with 0.1 ml from each clear tube and incubated for 24 h at 37°C. The MBC was defined as the lowest concentration in the tubes giving no growth on a nutrient plate afterwards.
Protoplast preparation and assay with microcin L.
Protoplasts of wild-type and mutant E. coli cells were prepared according to the protocol described by Weiss (59). E. coli strains were grown aerobically overnight at 37°C in nutrient broth. The cells were harvested by centrifugation (5,000 × g, 10 min), washed once with 10 mM sodium phosphate buffer (pH 8.0), and resuspended in the same buffer supplemented by 0.5 M sucrose to obtain an optical density at 600 nm (OD600) of 0.4. A solution of 20 mg/ml lysozyme was added very slowly to reach a final concentration of 20 μg/ml, and cells were incubated at room temperature for 10 min (spheroplast formation). The mixture then was diluted 1:1 (vol/vol) with 10 mM sodium phosphate buffer (pH 8.0), and EDTA was added to a final concentration of 1 mM. After incubation at room temperature for 15 min, the protoplasts were ready for the survival assays with MccL. The OD600 was adjusted to 0.2 by diluting the cells in 10 mM sodium phosphate buffer supplemented with 250 mM sucrose (pH 8.0). MccL was added to 1 ml of protoplasts at a final concentration of 0.6 μM. After incubation for 1 h at 37°C, the cells were plated on LB agar and CFU were counted after overnight incubation at 37°C. Data, averages from three independent experiments, are expressed as the percentage of surviving cells calculated in relation to control cells not treated with MccL (100% surviving cells).
Influence of proton-motive force in microcin activity.
The influence of proton-motive force in microcin activity was determined according to the liquid growth inhibition assay in the presence of carbonyl cyanide 3-chlorophenylhydrazone (CCCP) as described by Destoumieux-Garzon et al. (15). Briefly, microcin-susceptible E. coli MC4100 cells in stationary growth phase were harvested by centrifugation and resuspended in M63 medium to an OD600 of 0.035 (4 × 107 CFU/ml). Two μl CCCP was added to the cell suspension (2 ml) to a final concentration of 20 μM. After 5 min of incubation at 37°C, MccL was added at a final concentration of 0.6 μM. CCCP alone and MccL alone were used as negative and positive controls, respectively. Aliquots were removed regularly and plated on LB agar, and CFU were counted after overnight incubation at 37°C. Data are the averages from three independent experiments.
Outer membrane permeabilization assay.
Outer membrane permeability was determined as described by Eriksson et al. (17) by measuring the access of extracellular nitrocefin to the periplasm of E. coli ML-35p cells. This strain, which carries a plasmid-borne gene for periplasmic β-lactamase, was inoculated into 20 ml of fresh nutrient medium in 100 ml flasks (2%, vol/vol). After growth for 2 h at 37°C with shaking (150 rpm), cells were centrifuged (5,000 × g, 5 min), washed twice, and suspended in 10 mM sodium phosphate (pH 7.4) to an OD600 of 0.35 (4 × 108 CFU/ml). The permeabilization assays were carried out using 96-well microtiter plates with a final volume of 100 μl per well (10 μl of washed bacteria diluted in 90 μl of buffer containing 50 μg/ml nitrocefin and MccL). Following the addition of MccL, nitrocefin cleavage was monitored by light absorption measurements at 495 nm for 20 min at 37°C. For a positive control, we substituted melittin (1 μM) for MccL. A basal β-lactamase activity was detected with the MccL solvent and deduced from assays. Data are the averages from three independent experiments.
Inner membrane permeabilization assay.
The permeabilization of the inner membrane was determined by the hydrolysis of extracellular o-nitrophenyl β-d-galactopyranoside (ONPG) by cytoplasmic β-galactosidase produced constitutively by E. coli ML-35p (32, 33). This strain lacks the membrane permease that transports the substrate across the inner membrane. Bacteria were prepared and assays were carried out as previously described for outer membrane permeabilization assays, replacing nitrocefin with ONPG to a final concentration of 2 mM. ONPG hydrolysis was monitored by measuring the o-nitrophenol (ONP) release at 420 nm in the presence of peptide.
Membrane depolarization.
The cytoplasmic membrane depolarization activity of MccL was performed according to the Zhang et al. (62) protocol using the membrane potential-sensitive cyanine dye DiSC3(5). This dye fits into the membrane with high membrane potential gradient and then forms aggregates that involve self quenching, resulting in decreased fluorescence. Membrane disruptions generate membrane potential dissipation and then DiSC3(5) releasing into the medium, causing an increased fluorescence. E. coli MC4100 cells in the mid-logarithmic phase were collected by centrifugation, washed twice in 10 mM sodium phosphate (pH 7.4), and resuspended in the same buffer to an OD600 of 0.35. Cells were diluted 1:10 (vol/vol) with the same buffer with 1 μM DiSC3(5) and 0.2 mM EDTA (pH 8.0) and then incubated for 20 to 30 min in the dark at room temperature to allow dye uptake through the outer membrane and quenching. KCl (100 mM) then was added to equilibrate the cytoplasmic and external K+ concentrations. MccL was added to yield a final concentration of 6 μM. For the positive control, melittin (3.5 μM) replaced MccL. The fluorescence reading was monitored by using a Hitachi F-2500 fluorescence spectrophotometer at an excitation wavelength of 643 ± 2.5 nm and an emission wavelength of 666 ± 2.5 nm.
RESULTS
Genetic environment of microcin L genes and pL104 construction.
The MccL genetic system belongs to a 13.5-kb DNA fragment inserted in pUC19. The resulting recombinant plasmid was called pL102 (49). About 5 kb was sequenced previously in the middle of the insert, and four genes implied in the MccL production were identified: a structural gene, mclC, an immunity gene, mclI, and two export genes, mclA and mclB (Fig. 1) (40). To construct a strain with a plasmid bearing the complete MccL genetic system and only this genetic system, we first sequenced the 13.5-kb insert (GenBank accession number AY237108.2). This sequence was compared to DNA data banks using the BLAST software to identify genes or sequences potentially implied in the regulation of the MccL production. No such sequences were detected. However, interestingly, about 1,400 bp upstream from mclI, a sequence showing 99% identity to the colicin M immunity gene (cmi) was found. This 354-bp gene exhibited three nucleotides different from the data bank cmi gene sequences (G127→T, G150→A, and G249→A [GenBank accession no. CP001232] or T357→A [GenBank accession no. FJ664733] or C49→T [GenBank accession no. FJ664773]). To check that this gene was functional, colicin M resistance was assayed by a cross-streaking test, as previously described (49), using the E. coli pT04 strain, which produces colicin M. This strain inhibited the growth of E. coli MC4100 but not the growth of MC4100pL102, showing that pL102 possesses a gene conferring colicin M resistance.
As no potentially regulating sequence was found in the MccL genetic system vicinity, we shortened the 13.5-kb DNA fragment from both ends with the Fermentas ExoIII/S1 deletion kit. We selected the plasmid with the shortest insert containing the complete MccL genetic system. It remained only 218 bp upstream from the mclI gene but about 1,200 bp downstream from the mclA gene. We did not succeed in obtaining shorter plasmids without a deletion in the MccL genetic system. This plasmid was called pL104 and was used to transform E. coli MC4100, giving the MC4100pL104 strain. As we determined, this strain was able to produce MccL.
Microcin L production and purification.
A suitable method of production and purification, reported in Materials and Methods, was established by improving our previously described protocol (40) to produce sufficient amounts of pure MccL for detailed studies of its biological activity. MccL was purified to homogeneity from the early-stationary-phase culture supernatant of the E. coli MC4100pL104 producer. Partly purified MccL, obtained by solid-phase extraction on Sep-Pak plus environmental C18 cartridges, was concentrated under vacuum and then subjected to an accurate separation by two successive C8 RP-HPLC assays. After each separation, only one fraction, corresponding to one OD peak, presented antimicrobial activity, showing that no other antimicrobial peptides were copurified with MccL. From the final C8 RP column, MccL eluted in a single peak at 50% (vol/vol) acetonitrile-0.1% (vol/vol) TFA. The peptide purity was checked by mass spectrometry, and the primary structure as well as the disulfide bridge assignments were controlled as described by Pons et al. (40), showing the presence of one single molecule identical to the pure MccL produced by native E. coli strain LR05. Finally, a concentration under vacuum of the pure MccL led to a higher final yield of the microcin. As there were no other antibacterial peptides in the partly purified MccL, we used it to determine its antibacterial activity against the outer and inner membrane receptor mutants and the Ton system mutants. Throughout the remainder of this study, pure and quantified MccL samples were used for the characterization of MccL activity and mechanism of action.
No permeabilization of the bacterial membranes by microcin L. (i) Outer membrane.
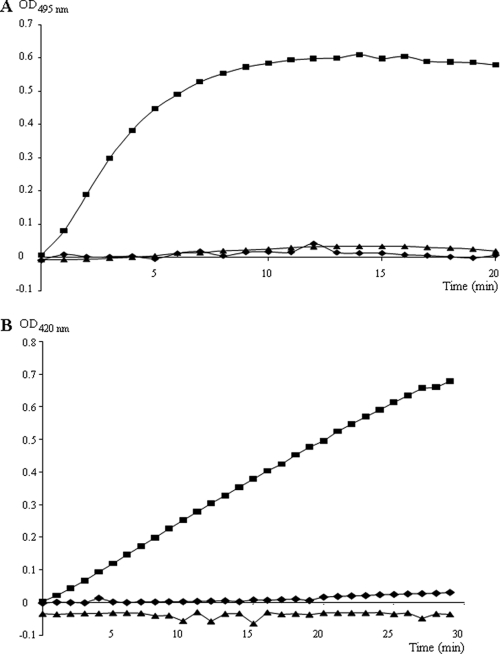
To localize the MccL activity on sensitive cells, we had to know if this microcin acted directly on the outer membrane or if the peptide needed to penetrate in the periplasm and/or further in the cytoplasm to be able to kill cells. The activity of pure MccL on the outer membrane was investigated using MccL-sensitive E. coli strain ML-35p. E. coli ML-35p produces a periplasmic β-lactamase. If the outer membrane of these bacteria is damaged, the β-lactamase activity can be detected extracellularly. Beforehand, we confirmed the bactericidal activity of MccL on E. coli ML-35p by the determination of the MIC (160 nM) and the minimum bactericidal concentration (MBC; 640 nM). We then tested the effect of MccL on the outer membrane of ML-35p. In the assay, the hydrolysis of the β-lactamase substrate, nitrocefin, which was added to the culture medium, was monitored at 495 nm in a spectrophotometer (Fig. 2A). Two concentrations of MccL (1 and 10 μM) were tested, and no activity of β-lactamase was observed for 20 min of incubation, whereas in the presence of 1 μM melittin, a strong membrane-disrupting agent used as a positive control, an immediate and significant β-lactamase activity was detected. This result indicated that MccL did not act by permeabilizing the outer membrane of susceptible cells.
FIG. 2.
In vivo bacterial membrane permeabilization assay. Two concentrations of MccL were tested against E. coli ML-35p: 1 μM (⧫) and 10 μM (▴). Melittin (1 μM; ▪) was used as a positive control. (A) Assay on the outer membrane with 50 μg/ml nitrocefin. (B) Assay on the inner membrane with 2 mM ONPG.
(ii) Cytoplasmic membrane.
An assay similar to the outer membrane permeability assay was used to study MccL activity on the inner membrane permeability. The activity of the cytoplasmic β-galactosidase, constitutively expressed in lactose permease-deficient E. coli ML-35p, was detected extracellularly when the inner membrane of the target bacteria was damaged. The hydrolysis of ONPG, the β-galactosidase substrate, into ONP was monitored spectrophotometrically (Fig. 2B). Melittin, used as a positive control, induced a significant β-galactosidase activity within the first minutes of incubation, showing a permeabilization of the inner membrane, whereas no β-galactosidase activity was detected during the 30 min of incubation of E. coli ML35p with 1 or 10 μM MccL. These results indicated that MccL bactericidal activity was not linked to the bacterial membrane permeability.
Specific outer membrane receptor protein in microcin L-sensitive cells.
Several siderophores, used by enteric bacteria to provide intracellular iron, have specific outer membrane receptor proteins also used by colicins and microcins (11, 38, 51). To identify the outer membrane receptor(s) necessary for the uptake of MccL, the activity of microcin was tested against a set of E. coli and Salmonella enterica mutants lacking siderophore receptors (Table 2). E. coli mutants P8, MS172, and ZI311, deleted for the receptor of ferrichrome (FhuA), Fe3+-coprogen (FhuE), and Fe3+-dicitrate (FecA), respectively, were as sensitive to MccL as the control strains AB2847, MC4100, and H1443 (Table 2). In addition, strains mutated in the gene fepA (mutant H873) or fiu (mutant H1594), coding for a catecholate siderophore receptor, showed an equivalent MccL sensitivity. On the other hand, only the mutants lacking the catecholate siderophore receptor Cir were fully resistant to MccL. Salmonella enterica strains possess their own Fe3+-catecholate receptor, named IroN, in addition to FepA and Cir, which also are present in E. coli but do not have Fiu receptors. S. Typhimurium and S. Enteritidis fepA and/or iroN mutants were sensitive to MccL, while mutants deleted for the Cir outer membrane receptor were fully resistant (Table 2). In conclusion, only the catecholate siderophore receptor Cir, from E. coli and Salmonella enterica, was identified as a receptor for MccL.
TABLE 2.
Antibacterial activity of MccL against wild-type and receptor mutant strains
| Parent strain | Derived mutant | Relevant receptor gene(s) | Inhibition zone (mm) |
|---|---|---|---|
| Escherichia coli | |||
| AB2847 | 23 | ||
| P8a | fhuA | 23 | |
| MC4100 | 23 | ||
| H1443 | 24 | ||
| MS172a | fhuE | 22 | |
| ZI311a | fecA | 25 | |
| H1876a | fepA, fiu, cir | 0 | |
| H873a | fepA | 24 | |
| H1594a | fiu | 23 | |
| H1877a | fepA, fiu | 25 | |
| VR42a | cir | 0 | |
| H1875a | fepA, cir | 0 | |
| H1728a | fiu, cir | 0 | |
| H1729a | fhuA, fiu, cir | 0 | |
| DB503 | 16 | ||
| AB4b | manY | 15 | |
| AB3b | manXYZ | 16 | |
| MC4100 | 16 | ||
| MnTMb | sdaC | 18 | |
| Salmonella Typhimurium ATCC 14028 | |||
| 8 | |||
| WR 1726a | fepA | 6 | |
| AJB 64a | iroN | 15 | |
| AR 8412a | cir | 0 | |
| WR 1727a | fepA, iroN | 10 | |
| WR 1728a | fepA, iroN, cir | 0 | |
| Salmonella Enteritidis SE 147 Nalr | |||
| 8 | |||
| WR 1425a | fepA | 11 | |
| WR 1434a | fepA, iroN | 11 | |
| WR 1530a | cir | 0 | |
| WR 1458a | iroN, cir | 0 |
Strains with a mutated siderophore receptor of the outer membrane.
Strains with a mutated cytoplasmic membrane receptor.
Ton system implication in microcin L activity.
In Gram-negative bacteria, the TonB protein, associated with cytoplasmic membrane proteins ExbB and ExbD, transduces the proton-motive force of the cytoplasmic membrane to transport iron-siderophore complexes across the outer membrane. The Cir protein belongs to the large family of TonB-dependent outer membrane receptors (9). Therefore, the activity of MccL was tested against different mutants of the Ton system (Table 3). In E. coli and Salmonella strains, all tested tonB mutants were fully resistant to MccL. These results showed that MccL antibacterial activity was TonB dependent. Surprisingly, the exbB exbD double mutant still was sensitive to MccL, whereas the ExbB and ExbD cytoplasmic proteins are necessary to produce a functional Ton complex. As it was previously demonstrated that in the TolA transduction system, TolQ and TolR were analogues of ExbB and ExbD, respectively (6, 10, 11), the antibacterial activity of MccL was tested against isogenic strains mutated in the tolQ gene. When only the exbB or tolQ gene was mutated, MccL bactericidal activity was maintained, whereas the exbB tolQ double mutation conferred to the target bacteria a high resistance to MccL. This result suggested that the function of the ExbB protein could be replaced by its homologue TolQ in TonB-dependent MccL activity.
TABLE 3.
Antibacterial activity of MccL against wild type and Ton system mutant for E. coli and Salmonella enterica strains
| Parent strain | Derived mutant | Genotype | Inhibition zone (mm) |
|---|---|---|---|
| Escherichia coli | |||
| W3110 | 20 | ||
| KP1032 | tonB | 0 | |
| KP1036 | exbB | 20 | |
| KP1143 | exbD | 19 | |
| KP1118 | exbB, exbD | 20 | |
| GM 1 | 22 | ||
| KP1082 | tonB | 0 | |
| KP1037 | exbB | 19 | |
| TPS13 | tolQ | 20 | |
| KP1038 | exbB, tolQ | 9 | |
| Salmonella Typhimurium A36 | |||
| 9 | |||
| WR 1174 | tonB | 0 | |
| Salmonella Enteritidis SE 147 Nalr | |||
| 8 | |||
| WR1529 | tonB | 0 |
Membrane potential-dependent activity of microcin L.
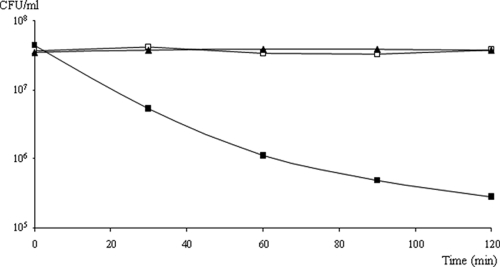
To further study the effect of the membrane energetic state on MccL activity, we monitored MccL activity on sensitive E. coli MC4100 cells (4 × 107 CFU/ml) in the presence or in the absence (control) of the protonophore carbonyl cyanide m-chlorophenyl hydrazone (CCCP) (Fig. 3). The concentration of CCCP (20 μM) used in this assay was known to fully dissipate the proton-motive force. MccL was used in this bactericidal assay at a final concentration of 0.6 μM (15× MIC) (40) to prevent dose-dependent effects. The control with only CCCP showed that the dissipation of the proton-motive force alone did not lead to bacterial death. While MccL induced a rapid loss of the viability of E. coli MC4100 of about 2 logs within 2 h in the absence of CCCP, the presence of 20 μM CCCP prevented the MccL activity. This clearly indicated that the proton-motive force was required for the bactericidal activity of MccL on E. coli.
FIG. 3.
Influence of the protonophore CCCP on the bactericidal activity of MccL. E. coli MC4100 was incubated with only 20 μM CCCP (□), 0.6 μM MccL and CCCP solvent (▪), or both CCCP and MccL (▴).
Microcin L activity on protoplast cells.
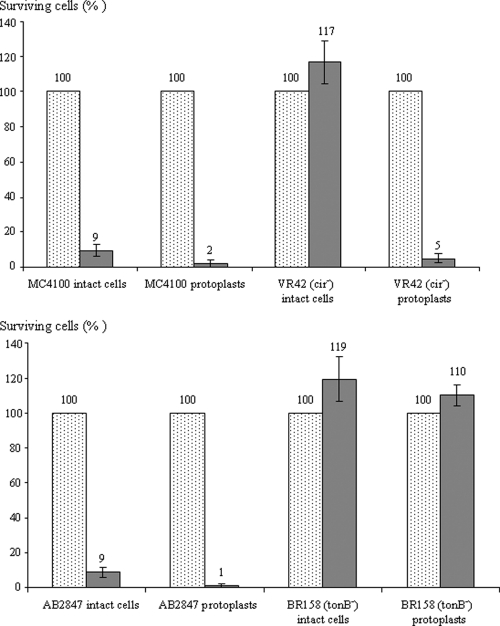
To know if the outer membrane was necessary for MccL activity, microcin was tested against protoplast cells (Fig. 4). Protoplasts were prepared by removing the outer membrane and the peptidoglycan layer. The cytoplasmic membrane of protoplasts was not damaged, and the TonB protein, anchored in the cytoplasmic membrane, was still present. First, the wild-type strains MC4100 and AB2847, transformed into protoplasts, were as sensitive to MccL as the control intact cells, indicating that MccL killed the sensitive cells in the absence of the outer membrane. Moreover, cir mutant VR42 became sensitive to MccL when cells were transformed into protoplasts. All of these results confirmed that the outer membrane was not the target of MccL and that Cir protein was necessary only for the microcin to reach the periplasm of sensitive cells (Table 2). To further study the role of TonB in the MccL activity, protoplasts of tonB mutants were used as target bacteria of MccL. Surprisingly, these cells remained as fully resistant to microcin as the control intact cells. This indicated that the presence of the TonB protein was essential to the MccL bactericidal activity, even in the absence of the outer membrane.
FIG. 4.
Bactericidal activity of MccL against protoplasts of E. coli wild-type and mutant strains. Cells were counted before (dotted bars) and 1 h (solid bars) after the addition of 0.6 μM MccL. MC4100 and AB2847 are positive controls for MccL activity, whereas VR42 and BR158 are negative controls. Data are the averages from three independent experiments.
Has microcin L a cytoplasmic membrane receptor?
To determine potential MccL receptor(s) on the cytoplasmic membrane, two types of membrane proteins were studied: the mannose permease, a complex of the three proteins ManX, ManY, and ManZ, and the serine permease SdaC. These permeases were described as MccE492 and MccV receptors, respectively (4, 21). The agar diffusion assay of MccL on E. coli mutants showed that the strains deleted for mannose or serine permease still were sensitive to microcin (Table 2). We could conclude that neither the mannose permease nor the serine permease were necessary for the MccL antibacterial activity.
Depolarization of the cytoplasmic membrane.
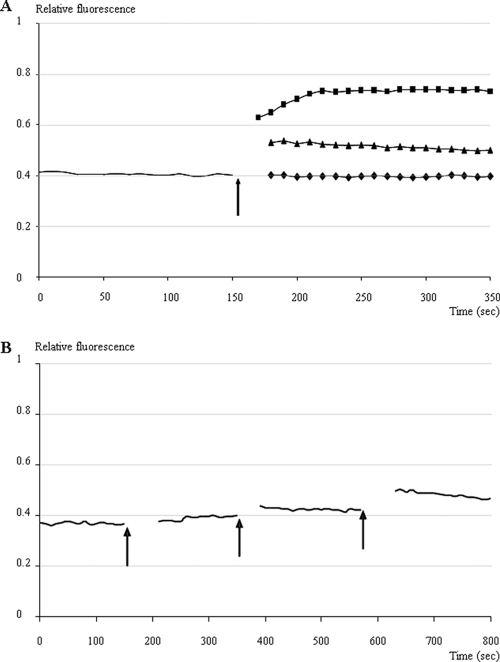
An assay involving the membrane potential-sensitive dye DiSC3(5) was performed to measure the disruption of electrical potential gradient in intact bacteria. E. coli MC4100 was used as a susceptible strain. The fluorescent probe DiSC3(5), which is a caged cation, distributes among cells and the culture medium, depending on the cytoplasmic membrane potential. Once it is inside the cells, it becomes concentrated and quenches its own fluorescence. If peptides form channels or otherwise disrupt the membrane, the membrane potential is dissipated and the DiSC3(5) is released into the medium, which increases the fluorescence, as it can be detected by fluorescent spectrophotometry. In these assays, 0.1 M KCl was added to the buffer to balance the chemical potential of K+ inside and outside the cells. MccL was added during the fluorescence measurements, and the membrane-disrupting agent melittin was used as a control. The addition of MccL caused a rapid increase in fluorescence, although the positive control caused a higher increase in fluorescence (Fig. 5A). Moreover, a dose-dependent effect was observed with successive additions of 2 μM MccL; at each addition of MccL, the fluorescence increased (Fig. 5B). These results demonstrated that MccL dissipated the membrane potential gradient.
FIG. 5.
Depolarization of the bacterial cytoplasmic membrane by MccL. The fluorescence of the dye DiSC3(5) within E. coli MC4100 bacterial suspension was monitored at 37°C with a spectrophotometer (excitation wavelength, 643 ± 2.5 nm; emission wavelength, 666 ± 2.5 nm). (A) The arrow indicates the addition of 6 μM MccL (▴), 3.5 μM melittin (▪) (positive control), or MccL solvent (⧫) (negative control). (B) Each arrow indicates the addition of 2 μM MccL.
DISCUSSION
We report here the mode of import and action of MccL, one of the four microcins produced by wild-type E. coli LR05 isolated from a poultry intestinal tract.
In previous genetic studies, a 13.5-kb HindIII-SalI DNA fragment issued from LR05 DNA plasmid was cloned into pUC19, resulting in pL102. This fragment, containing the MccL gene cluster, directs the production of MccL and immunity to MccL and MccV (49). In this study, the analysis of DNA sequence around MccL genes allowed us to underscore a functional gene conferring colicin M resistance. The nucleotide sequence of this 354-bp gene was almost identical (99%) to the colicin M immunity gene sequences (cmi). Slightly different cmi sequences are described in GenBank, and our cmi sequence differed from the GenBank ones by three nucleotides. Only the G→T mutation at position 127 resulted in a change in the peptide (Val43→Phe). This change does not appear to affect the function of the immunity protein, as pL102 conferred a colicin M resistance to E. coli MC4100pL102. This phenomenon was already observed in wild-type E. coli LR05 for the immunity MccV gene product that differed from sequences in databases by a single amino acid at position 34, without the modification of the immunity protein activity (40), and for the gene encoding the MccB17 precursor with two transitions and an insertion that did not affect microcin maturation and activity (49). Therefore, the wild-type strain E. coli LR05 carries the colicin M immunity gene besides the MccV immunity gene and the genetic determinants of microcins L, B17, J25, and D93 (40, 49). This LR05 wild strain should result from a large genetic rearrangement corresponding to multiple exchanges and the recombination of genetic material between bacteria. The possibility of a horizontal transfer of plasmid genes responsible for microcin biosynthesis was evoked for the microcins MccC51 and MccC7 (19). Similarly, among class IIb microcins, several genes from the MccE492, MccM, MccH47, and MccI47 genetic systems were shown to display similar functions and to be interchangeable (57). In microbial ecosystems, selective pressure might lead to the dissemination of the bacteriocin gene clusters among bacterial populations and to its transfer to different species and genera of bacteria. Consequently, this abundance of microcin and colicin genes in E. coli LR05 may provide the microorganism an important ecological advantage in a competitive bacterial environment such as the intestinal tract, widening the target cell spectrum and providing protection from the antagonistic activity of colicin- and/or microcin-producing strains. These data support prior claims that bacteriocin production plays a significant role in the colonization of E. coli in the gastrointestinal tract (22, 28). However, the persistence of numerous genes encoding microcins, which appear to share the same functions, could be surprising. It is generally accepted that useless genes are eliminated from dynamic genomes, but E. coli does not appear to eliminate several homologous genes present in different microcin gene clusters (57). Each microcin possibly has specific functions, but these remain to be elucidated.
When the DNA sequence of the surrounding regions of the MccL genetic system was compared to sequences in DNA data banks, none of the sequenced open reading frames had a known transcription regulation role. The lack of potentially regulating sequence in the MccL genetic system vicinity allowed us to shorten the 13.5-kb DNA fragment bearing the MccL genes and to construct pL104 containing a 5.5-kb insert directing the production of an active MccL. However, we did not succeed in obtaining a complete MccL genetic system with less than 1,200 bp downstream from the mclA gene. The shorter plasmids always lacked part of the end of the MccL genetic system. Previous evidence suggested that mclA and mclB are detrimental to the cell, since multiple attempts to clone mclA and mclB have failed (40). Furthermore, the CvaB or SecY transmembrane secretory proteins were shown to be deleterious to E. coli cells when overproduced and/or unassociated with their complex partners (7, 29). Thus, as suggested for cvaB or secY, the sequence downstream from the mclA gene may have transcriptional downregulating properties. Whatever the mechanism involved, this sequence seems to be essential for cell survival. Further studies clearly must be developed to investigate the possible regulation of the MccL genetic system.
For the first time, we investigated the MccL mechanism of action in vivo. First, we showed that pure MccL did not damage the outer membrane of susceptible cells. Moreover, the same strains transformed into protoplasts, i.e., without the outer membrane or the peptidoglycan layer, always were sensitive to MccL. These results prove that the outer membrane is not the target of MccL. Since MccL did not act by permeabilizing the outer membrane, we wanted to understand the MccL uptake mechanism across the outer membrane of sensitive cells. In recent years, it has been shown that the iron uptake machineries and their associated energy transduction system, the TonB/ExbB/ExbD inner membrane complex, are required for the recognition and import of various microcins (16). Our study of the mechanism of action of MccL showed that this peptide is recognized by the catecholate siderophore receptor Cir only. The mutants in the cir gene were fully resistant to MccL, while they became sensitive to MccL when cells were transformed into protoplasts, which bypass the recognition step. This result confirms the active role of Cir as the MccL receptor, involved both in peptide recognition and in its translocation across the outer membrane. E. coli as well as Salmonella enterica serovars Typhimurium and Enteritidis, containing the catecholate receptor Cir, are susceptible to MccL. As for MccL, only the Cir outer membrane protein acts as a receptor for MccV (12), while the three receptors FepA, Cir, and Fiu are necessary for the antibacterial activity of class IIb microcins MccE492, MccE492m, MccM, and MccH47 (38, 55, 57). It was determined that a C-terminal domain of microcins E492, H47, and M was essential for the specific binding to FepA, Fiu, and Cir and possibly also for TonB-mediated import in the periplasm (4). Moreover, Azpiroz and Laviña (2) constructed chimeric peptides with exchanged C-terminal sequences of MccV and MccH47 and clearly showed that the C-terminal sequence conferred the uptake properties on the molecules, and that the remaining N-terminal portion was responsible for the specific antibiotic toxicity. We previously showed (40) that MccL presented a high degree of sequence similarity to MccV within the C-terminal regions (87.5%). Therefore, we propose that the C-terminal region of MccL is responsible for the receptor recognition and/or translocation across the outer membrane.
Furthermore, we demonstrated that the tonB mutation in sensitive cells induced a high resistance to MccL, indicating that the TonB protein associated with the Cir receptor is required for the antibacterial activity of MccL. TonB, which spans the periplasmic space, generally is associated with two other partners, ExbB and ExbD, located in the inner membrane, thus forming the TonB-dependent energy transduction system (8, 42). We observed that a double mutation in exbB and exbD genes diminished the sensitivity to MccL but did not confer total resistance to MccL to the target bacteria. This strongly suggests that ExbB and ExbD are not essential for MccL activity. However, we showed that the TolQ protein of the TolA system can replace its homologue ExbB in TonB-dependent MccL activity. Similarly, MccL also could use TolR for its translocation when exbD is inactivated. This complementation between TonB-ExbB-ExbD and TolA-TolQ-TolR transduction systems was demonstrated previously for E. coli susceptibility to group B colicins (10) and was suggested for MccE492 (15), MccM, and MccH47 (57). However, we also observed that the exbB tolQ double mutant was not fully resistant to MccL, which suggests that another, unknown interaction and/or complementation exists.
The TonB-ExbB-ExbD complex is well known to transduce the proton-motive force energy from the inner membrane, where it is generated, to the outer membrane (8, 11, 43). The requirement of TonB for MccL antibacterial activity strongly suggests that energy is required for the microcin mechanism of action. That was clearly confirmed by our bactericidal assays in the presence of CCCP, a protonophore that collapses the proton-motive force, in which MccL activity was prevented by the dissipation of the proton-motive force at the cytoplasmic membrane. As it is well known for the iron-siderophore transport complexes across the outer membrane, the TonB-ExbB-ExbD complex may be necessary for MccL activity to transduce the proton-motive force of the cytoplasmic membrane to the Cir receptor. This energy would be essential for peptide transport across the outer membrane. However, TonB protein has another role, because protoplast cells of tonB mutants remained resistant to the antibacterial peptide. The MccL bactericidal activity is dependent on the TonB protein, even in the absence of the outer membrane. At this stage, the other role of TonB remains to be established.
We undertook the identification of an MccL receptor on the cytoplasmic membrane. The inner membrane protein SdaC is involved in serine uptake and also was proposed to serve as a specific inner membrane receptor for MccV, thus helping it locate the inner membrane, a step required for channel formation and the disruption of membrane potential (21). ManYZ is an inner membrane complex that functions together with the cytoplasmic ManX to form the mannose permease involved in the uptake of mannose and related hexoses. ManYZ was identified as necessary for MccE492 antibacterial activity against E. coli, because manYZ mutants were insensitive to the inner membrane depolarization mediated by periplasmic MccE492 (4). In this work, we demonstrated that MccL antibacterial activity did not require the inner membrane protein SdaC or the complex ManXYZ. Despite the high degree of sequence similarity between the MccL and MccV C-terminal regions (40), the two microcins do not share the same specific inner membrane receptor. We hypothesize that the MccL N-terminal sequence, which is quite different from the MccV one, is responsible for its specific antibiotic activity. This is in accordance with Azpiroz and Laviña (2), who showed that the specific toxicity of MccV and MccH47 was conferred by their N-terminal portions.
While most of the gene-encoded antimicrobial peptides, in particular cationic peptides produced by Gram-positive bacteria (24), are believed to inhibit the growth of microorganisms by targeting the cytoplasmic membrane, such a mechanism of action was reported for only few microcins: MccV, MccE492, and MccH47. Cationic peptides generally are able to interact electrostatically with the negatively charged headgroups of bacterial phospholipids and then insert into the cytoplasmic membrane, forming channels or pores that are proposed to lead to the leakage of cell contents and cell death. However, most microcins, like MccL, are anionic peptides. Only McB17, MccV, and putatively Mcc24 are slightly cationic (16). The determination of MccL effects on the permeability of the cytoplasmic membrane to large molecules showed that MccL did not act by permeabilizing the inner membrane, but this microcin was found to dissipate the membrane potential gradient. MccV (60) and MccE492 (15) also were shown to destabilize the membrane potential. However, for MccE492, it is a consequence of the permeabilization of the inner membrane after the insertion of MccE492 into this membrane, as shown in E. coli ML35p (15), or in artificial planar lipid bilayers (30). MccJ25 also was reported to disrupt the inner membrane integrity in S. enterica serovar Newport (46), in liposomes (45), and in uncharged phospholipid monolayers (3). However, these properties were specific to S. enterica serovars and were observed at concentrations much higher than the MIC. MccJ25 was first shown to target intracellular RNA polymerase (14, 61). As with MccJ25, the amount of MccL required for the membrane depolarization on E. coli MC4100 (6 μM) is much higher than the one needed for the minimum bactericidal concentration (90 nM) (40). This suggests that the cytoplasmic membrane destabilization is not fully responsible for MccL bactericidal activity.
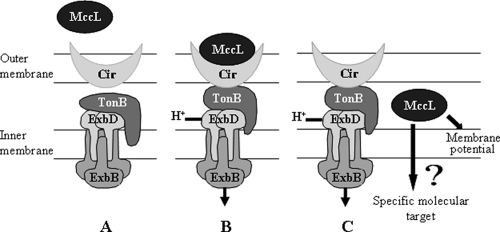
In conclusion, MccL antibacterial activity relies on a complex mechanism of action (Fig. 6). We propose that MccL is recognized at the cell surface by the iron uptake receptor Cir, which promotes its import across the outer membrane following a TonB- and energy-dependent pathway. MccL then could induce a depolarization of the E. coli cytoplasmic membrane, and thereafter MccL would reach a specific molecular target(s). Future studies dedicated to the identification of MccL molecular targets will be of great interest to fully understand the mechanism of the antibacterial activity.
FIG. 6.
Mechanism of action of MccL. (A) Iron uptake receptor Cir and its associated energy transduction system, the TonB-ExbB-ExbD complex. (B) MccL-receptor Cir interaction and transduction of the proton-motive force energy from the inner membrane to the outer membrane. (C) MccL translocation across the outer membrane and action of MccL. A high level of MccL disrupts the inner membrane potential without pore formation. It is possible that MccL has another specific target.
Acknowledgments
We are grateful to D. Belin, K. Hantke, F. Moreno, K. Postle, W. Rabsch, S. Rebuffat, and L. F. Wu for kindly providing bacterial strains.
We dedicate this work to the memory of Anne-Marie Pons.
Footnotes
Published ahead of print on 28 December 2010.
REFERENCES
- 1.Ahmer, B. M., M. G. Thomas, R. A. Larsen, and K. Postle. 1995. Characterization of the exbBD operon of Escherichia coli and the role of ExbB and ExbD in TonB function and stability. J. Bacteriol. 177:4742-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azpiroz, M. F., and M. Laviña. 2007. Modular structure of Microcin H47 and colicin V. Antimicrob. Agents Chemother. 51:2412-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellomio, A., R. G. Oliveira, B. Maggio, and R. D. Morero. 2005. Penetration and interactions of the antimicrobial peptide, microcin J25, into uncharged phospholipid monolayers. J. Colloid. Interface Sci. 285:118-124. [DOI] [PubMed] [Google Scholar]
- 4.Bieler, S., F. Silva, C. Soto, and D. Belin. 2006. Bactericidal activity of both secreted and nonsecreted microcin E492 requires the mannose permease. J. Bacteriol. 188:7049-7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouveret, E., et al. 2002. Analysis of the Escherichia coli Tol-Pal and TonB systems by periplasmic production of Tol, TonB, colicin, or phage capsid soluble domains. Biochimie 84:413-421. [DOI] [PubMed] [Google Scholar]
- 7.Boyer, A. E., and P. C. Tai. 1998. Characterization of the cvaA and cvi promoters of the colicin V export system: iron-dependent transcription of cvaA is modulated by downstream sequences. J. Bacteriol. 180:1662-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun, V. 1995. Energy-coupled transport and signal transduction through the Gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol. Rev. 16:295-307. [DOI] [PubMed] [Google Scholar]
- 9.Braun, V., K. Hantke, and W. Köster. 1998. Bacterial iron transport: mechanisms, genetics, and regulation. Metal Ions Biol. Syst. 35:67-145. [PubMed] [Google Scholar]
- 10.Braun, V., and C. Herrmann. 1993. Evolutionary relationship of uptake systems for biopolymers in Escherichia coli: cross-complementation between the TonB-ExbB-ExbD and the TolA-TolQ-TolR proteins. Mol. Microbiol. 8:261-268. [DOI] [PubMed] [Google Scholar]
- 11.Braun, V., S. I. Patzer, and K. Hantke. 2002. Ton-dependent colicins and microcins: modular design and evolution. Biochimie 84:365-380. [DOI] [PubMed] [Google Scholar]
- 12.Chehade, H., and V. Braun. 1988. Iron-regulated synthesis and uptake of colicin V. FEMS Microbiol. Lett. 52:177-182. [Google Scholar]
- 13.de Lorenzo, V., and A. P. Pugsley. 1985. Microcin E492, a low-molecular-weight peptide antibiotic which causes depolarization of the Escherichia coli cytoplasmic membrane. Antimicrob. Agents Chemother. 27:666-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delgado, M. A., M. R. Rintoul, R. N. Farias, and R. A. Salomon. 2001. Escherichia coli RNA polymerase is the target of the cyclopeptide antibiotic microcin J25. J. Bacteriol. 183:4543-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Destoumieux-Garzón, D., et al. 2003. Microcin E492 antibacterial activity: evidence for a TonB-dependent inner membrane permeabilization on Escherichia coli. Mol. Microbiol. 49:1031-1041. [DOI] [PubMed] [Google Scholar]
- 16.Duquesne, S., D. Destoumieux-Garzon, J. Peduzzi, and S. Rebuffat. 2007. Microcins, gene-encoded antibacterial peptides from enterobacteria. Nat. Prod. Rep. 24:708-734. [DOI] [PubMed] [Google Scholar]
- 17.Eriksson, M., P. E. Nielsen, and L. Good. 2002. Cell permeabilization and uptake of antisense peptide-peptide nucleic acid (PNA) into Escherichia coli. J. Biol. Chem. 277:7144-7147. [DOI] [PubMed] [Google Scholar]
- 18.Fath, M. J., L. H. Zhang, J. Rush, and R. Kolter. 1994. Purification and characterization of colicin V from Escherichia coli culture supernatants. Biochemistry 33:6911-6917. [DOI] [PubMed] [Google Scholar]
- 19.Fomenko, D. E., et al. 2003. Microcin C51 plasmid genes: possible source of horizontal gene transfer. Antimicrob. Agents Chemother. 47:2868-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaillard-Gendron, S., et al. 2000. Isolation, purification and partial amino acid sequence of a highly hydrophobic new microcin named microcin L produced by Escherichia coli. FEMS Microbiol. Lett. 193:95-98. [DOI] [PubMed] [Google Scholar]
- 21.Gérard, F., N. Pradel, and L.-F. Wu. 2005. Bactericidal activity of colicin V is mediated by an inner membrane protein, SdaC, of Escherichia coli. J. Bacteriol. 187:1945-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gillor, O., I. Giladi, and M. A. Riley. 2009. Persistence of colicinogenic Escherichia coli in the mouse gastrointestinal tract. BMC Microbiol. 9:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.González-Pastor, J. E., J. L. San Millan, M. A. Castilla, and F. Moreno. 1995. Structure and organization of plasmid genes required to produce the translation inhibitor microcin C7. J. Bacteriol. 177:7131-7140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hancock, R. E. W. 1997. Peptide antibiotics. Lancet 349:418-422. [DOI] [PubMed] [Google Scholar]
- 25.Hantke, K. 1987. Ferrous iron transport mutants in Escherichia coli K-12. FEMS Microbiol. Lett. 44:53-57. [DOI] [PubMed] [Google Scholar]
- 26.Hantke, K. 1990. Dihydroxybenzoylserine-a siderophore for E. coli. FEMS Microbiol. Lett. 55:5-8. [DOI] [PubMed] [Google Scholar]
- 27.Heddle, J. G., et al. 2001. The antibiotic microcin B17 is a DNA gyrase poison: characterisation of the mode of inhibition. J. Mol. Biol. 307:1223-1234. [DOI] [PubMed] [Google Scholar]
- 28.Kerr, B., M. A. Riley, M. W. Feldman, and B. J. Bohannan. 2002. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature 418:171-174. [DOI] [PubMed] [Google Scholar]
- 29.Kihara, A., Y. Akiyama, and K. Ito. 1995. FtsH is required for proteolytic elimination of uncomplexed forms of SecY, an essential protein translocase subunit. Proc. Natl. Acad. Sci. U. S. A. 92:4532-4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lagos, R., M. Wilkens, C. Vergara, X. Cecchi, and O. Monasterio. 1993. Microcin E492 forms ion channels in phospholipid bilayer membrane. FEBS Lett. 321:145-148. [DOI] [PubMed] [Google Scholar]
- 31.Laviña, M., A. P. Pugsley, and F. Moreno. 1986. Identification, mapping, cloning and characterization of a gene (sbmA) required for microcin B17 action on Escherichia coli K-12. J. Gen. Microbiol. 132:1685-1693. [DOI] [PubMed] [Google Scholar]
- 32.Lehrer, R. I., A. Barton, and T. Ganz. 1988. Concurrent assessment of inner and outer membrane permeabilization and bacteriolysis in E. coli by multiple-wavelength spectrophotometry. J. Immunol. Methods 108:153-158. [DOI] [PubMed] [Google Scholar]
- 33.Lehrer, R. I., et al. 1989. Interaction of human defensins with Escherichia coli. Mechanism of bactericidal activity. J. Clin. Investig. 84:553-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Metlitskaya, A., et al. 2006. Aspartyl-tRNA synthetase is the target of peptide nucleotide antibiotic microcin C. J. Biol. Chem. 281:18033-18042. [DOI] [PubMed] [Google Scholar]
- 35.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria, p. 218-220. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Mirold, S., et al. 1999. Isolation of a temperate bacteriophage encoding the type III effector protein SopE from an epidemic Salmonella typhimurium strain. Proc. Natl. Acad. Sci. U. S. A. 96:9845-9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pace, C. N., F. Vajdos, L. Fee, G. Grimsley, and T. Gray. 1995. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4:2411-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patzer, S. I., M. R. Baquero, D. Bravo, F. Moreno, and K. Hantke. 2003. The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors FepA, Cir, Fiu and IroN. Microbiology 149:2557-2570. [DOI] [PubMed] [Google Scholar]
- 39.Pons, A. M., I. Lanneluc, G. Cottenceau, and S. Sablé. 2002. New developments in non-post translationally modified microcins. Biochimie 84:531-537. [DOI] [PubMed] [Google Scholar]
- 40.Pons, A. M., et al. 2004. Genetic analysis and complete primary structure of microcin L. Antimicrob. Agents Chemother. 48:505-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Postle, K. 1990. TonB and the gram-negative dilemma. Mol. Microbiol. 4:2019-2025. [DOI] [PubMed] [Google Scholar]
- 42.Postle, K. 1993. TonB protein and energy transduction between membranes. J. Bioenerg. Biomembr. 25:591-601. [DOI] [PubMed] [Google Scholar]
- 43.Postle, K., and R. J. Kadner. 2003. Touch and go: tying TonB to transport. Mol. Microbiol. 49:869-882. [DOI] [PubMed] [Google Scholar]
- 44.Rabsch, W., et al. 2003. Role of receptor proteins for enterobactin and 2,3-dihydroxybenzoylserine in virulence of Salmonella enterica. Infect. Immun. 71:6953-6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rintoul, M. R., B. F. de Arcuri, and R. D. Morero. 2000. Effects of the antibiotic peptide microcin J25 on liposomes: role of acyl chain length and negatively charged phospholipids. Biochim. Biophys. Acta 1509:65-72. [DOI] [PubMed] [Google Scholar]
- 46.Rintoul, M. R., B. F. de Arcuri, R. A. Salomon, R. N. Farias, and R. D. Morero. 2001. The antibacterial action of microcin J25: evidence for disruption of cytoplasmic membrane energization in Salmonella newport. FEMS Microbiol. Lett. 204:265-270. [DOI] [PubMed] [Google Scholar]
- 47.Rodríguez, E., and M. Laviña. 2003. The proton channel is the minimal structure of ATP synthase necessary and sufficient for microcin h47 antibiotic action. Antimicrob. Agents Chemother. 47:181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sable, S., A. M. Pons, S. Gendron-Gaillard, and G. Cottenceau. 2000. Antibacterial activity evaluation of microcin J25 against diarrheagenic Escherichia coli. Appl. Environ. Microbiol. 66:4595-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sablé, S., et al. 2003. Wild Escherichia coli isolate producing microcins B, D, J, and L. Cloning of genes for MccL production and immunity. Can. J. Microbiol. 49:357-361. [DOI] [PubMed] [Google Scholar]
- 50.Sahm, D. F., and J. A. Washington. 1991. Antibacterial susceptibility tests: dilution methods, p. 1105-1116. In A. Balows, W. J. Hausler, K. L. Herrmann, Jr., H. D. Isenberg, and H. J. Shadomy (ed.), Manual of clinical microbiology, 5th ed. American Society for Microbiology, Washington, DC.
- 51.Salomón, R. A., and R. N. Farias. 1993. The FhuA protein is involved in microcin 25 uptake. J. Bacteriol. 175:7741-7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salomón, R. A., and R. N. Farias. 1995. The peptide antibiotic microcin 25 is imported through the TonB pathway and the SbmA protein. J. Bacteriol. 177:3323-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning. A Laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 54.Sauer, M., K. Hantke, and V. Braun. 1987. Ferric-coprogen receptor FhuE of Escherichia coli: processing and sequence common to all TonB-dependent outer membrane receptor proteins. J. Bacteriol. 169:2044-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas, X., et al. 2004. Siderophore peptide, a new type of post-translationally modified antibacterial peptide with potent activity. J. Biol. Chem. 279:28233-28242. [DOI] [PubMed] [Google Scholar]
- 56.Trujillo, M., E. Rodriguez, and M. Laviña. 2001. ATP synthase is necessary for microcin H47 antibiotic action. Antimicrob. Agents Chemother. 45:3128-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vassiliadis, G., D. Destoumieux-Garzón, C. Lombard, S. Rebuffat, and J. Peduzzi. 2010. Isolation and characterization of two members of the siderophore-microcin family, microcins M and H47. Antimicrob. Agents Chemother. 54:288-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vizán, J. L., C. Hernandez-Chico, I. del Castillo, and F. Moreno. 1991. The peptide antibiotic microcin B17 induces double-strand cleavage of DNA mediated by E. coli DNA gyrase. EMBO J. 10:467-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weiss, R. L. 1976. Protoplast formation in Escherichia coli. J. Bacteriol. 128:668-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang, C. C., and J. Konisky. 1984. Colicin V-treated Escherichia coli does not generate membrane potential. J. Bacteriol. 158:757-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yuzenkova, J., et al. 2002. Mutations of bacterial RNA polymerase leading to resistance to microcin J25. J. Biol. Chem. 277:50867-50875. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, L., P. Dhillon, H. Yan, S. Farmer, and R. E. Hancock. 2000. Interactions of bacterial cationic peptide antibiotics with outer and cytoplasmic membranes of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:3317-3321. [DOI] [PMC free article] [PubMed] [Google Scholar]