Abstract
We investigated the effect of N348I alone and with M184V on nonnucleoside reverse transcriptase inhibitor (NNRTI) drug susceptibility and replicative capacity in B and non-B HIV-1 isolates. N348I reduced the susceptibility to all NNRTI drugs across subtypes. The replication capacity of all viruses in a variety of cell lines was impaired by N348I. Interestingly, the N348I and M184V double mutation compensated for the reduced NNRTI drug susceptibility observed in the N348I single mutant and marginally improved viral replicative capacity.
The nonnucleoside reverse transcriptase (RT) inhibitors (NNRTIs) nevirapine (NVP), efavirenz (EFV), and etravirine (ETV) bind to a hydrophobic cavity located near the RT active site and act as allosteric inhibitors of DNA polymerization. Mutations in the binding pocket are selected during failure of NNRTI therapy, and the long-term virologic response to sequential NNRTI treatment, even with the newer NNRTI ETV, is poor, particularly when multiple mutations are present (8, 9, 10, 20). Recently, mutations in the C-terminal domain of RT, including the “connection subdomain” and the RNase (RNase H) domain, have been identified and implicated in resistance to RT inhibitors (3, 15). In particular, N348I mutation in the connection subdomain confers reduced susceptibility to EFV and NVP, as well as the nucleoside analogues zidovudine (ZDV) and didanosine (DDI), and is highly associated with M184V and several thymidine analogue-associated mutations in HIV-1-infected individuals (6, 15, 25, 28).
However, there remain few data on the impact of N348I on drug susceptibility across non-subtype B viruses, and its impact on susceptibility to the newer NNRTI ETV is unclear. These issues are of particular importance for second-line treatment strategies in resource-limited settings.
To study the role of N348I in drug resistance in B and non-B HIV-1 isolates, HIV-1-positive clinical samples from drug-naïve patients (infected with subtype A [clones A1 and A2], subtype B, or subtype C HIV-1) were obtained by routine drug resistance testing. RT-PCR amplified gag-pol sequences from clinical plasma-derived isolates were amplified, sequenced, and cloned into the NotI and XmaI sites of the replication-defective, envelope-deleted subtype B retroviral vector p8.9NSX as described previously (7, 12, 18), as were subtype-specific gag-pol sequences from molecular clones MJ4 (subtype C) (14) and 94UG114.1.6 (subtype D) (5).
To measure the effect of N348I on drug susceptibility and replicative capacity (RC), virus stocks were generated as described previously (18). HEK 293T cells were infected with viral supernatant in the presence of serial dilutions of RT inhibitors to determine drug susceptibility. Viral replication was determined by measuring luciferase expression using SteadyGlo and a GloMax luminometer (Promega). Fifty percent effective concentrations (EC50s) were determined by linear regression analysis, and the fold change in drug susceptibility was determined by comparing mutant virus and wild-type subtype-specific EC50s.
In order to determine whether there were subtype differences in NNRTI drug susceptibility, we tested 7 wild-type clinical isolates and molecular clones incorporating full-length gag-pol. We observed some differences among subtypes in terms of NNRTI susceptibility, in which the absolute EC50 for subtypes A and D were higher than those for subtypes B and C, reaching around 2-fold higher for EFV and 3-fold higher for NVP in some instances (Table 1). However, because we studied a limited number of viruses of each subtype we cannot imply that this effect applies to these subtypes in general.
TABLE 1.
Effect of N348I mutation on NNRTI drug susceptibility of molecular clones and clinical isolates of HIV-1a
| Subtype | NVP |
EFV |
ETV |
|||
|---|---|---|---|---|---|---|
| Mean EC50 ± SEM | Fold change (P value) | Mean EC50 ± SEM | Fold change (P value) | Mean EC50 ± SEM | Fold change (P value) | |
| Molecular clones | ||||||
| p8.9 B (WT)b | 55.64 ± 7.35 | 2.73 ± 1.39 | 1.31 ± 0.16 | |||
| p8.9 B (N348I) | 235.36 ± 19.83 | 4.23 (<0.02) | 7.69 ± 3.75 | 2.81 (NS) | 2.75 ± 0.25 | 2.10 (<0.02) |
| MJ4 C (WT) | 65.85 ± 4.2 | 2.82 ± 1.23 | 1.20 ± 0.18 | |||
| MJ4 C (N348I) | 287.33 ± 13.05 | 4.36 (<0.05) | 7.65 ± 3.02 | 2.71 (NS) | 2.05 ± 0.31 | 1.71 (NS) |
| 94ug114.1.6 D (WT) | 153.8 ± 5.26 | 5.73 ± 2.63 | 2.47 ± 0.23 | |||
| 94 μg 114.1.6 D (N348I) | 584.9 ± 12.29 | 3.81 (<0.02) | 15.88 ± 6.72 | 2.77 (NS) | 7.26 ± 0.93 | 2.94 (<0.02) |
| Clinical isolates | ||||||
| A 1 (WT) | 157.56 ± 17.69 | 5.76 ± 2.29 | 2.39 ± 0.31 | |||
| A 1 (N348I) | 849.24 ± 41.56 | 5.39 (<0.02) | 20.03 ± 7.38 | 3.47 (NS) | 6.52 ± 0.89 | 2.72 (<0.02) |
| A2 (WT) | 146.67 ± 13.48 | 5.19 ± 0.75 | 2.77 ± 0.77 | |||
| A2 (N348I) | 838.95 ± 50.99 | 5.72 (<0.02) | 24.24 ± 2.21 | 4.67 (<0.05) | 8.22 ± 1.43 | 2.97 (NS) |
| B (WT) | 47.73 ± 4.61 | 2.57 ± 0.54 | 1.79 ± 0.42 | |||
| B (N348I) | 265.26 ± 21.56 | 5.56 (<0.02) | 9.99 ± 2.48 | 3.89 (<0.05) | 4.52 ± 0.73 | 2.53 (NS) |
| C (WT) | 69.96 ± 3.25 | 2.63 ± 0.33 | 2.26 ± 0.15 | |||
| C (N348I) | 281.94 ± 10.02 | 4.03 (<0.02) | 12.09 ± 4.91 | 4.60 (NS) | 4.26 ± 0.25 | 1.89 (<0.05) |
Data shown are mean drug EC50s (nM) for both mutant and wild-type viruses and mean fold differences between mutant and wild-type values. Standard errors of the mean from three independent experiments are shown. Statistical analyses using Student's t test compared the N348I mutant virus with the wild-type virus. NS, no statistically significant difference.
WT, wild type.
The N348I mutation was introduced into wild-type RT by site-directed mutagenesis (Stratagene QuikChange), and RT drug susceptibility assays were performed to determine the impact on susceptibility in the different HIV-1 backbones. Apart from ZDV, the N348I mutation had a minimal effect on NRTI susceptibility (tenofovir [TDF], abacavir [ABC], lamivudine [3TC], emtricitabine [FTC], didanosine [DDI], and stavudine [D4T]) across all of the subtypes investigated (data not shown). The EC50s of ZDV increased 1.7- to 3-fold across subtypes when N348I was compared to the wild-type counterpart. Contrary to other findings (6, 28), N348I did not confer reduced DDI susceptibility (EC50 range, 0.9- to 1.0-fold). However, the N348I mutation conferred reduced susceptibility to NVP, EFV, and ETV across all viruses (Table 1), namely, 5-fold for NVP and 2.9-fold for ETV in some instances, falling within the clinical cutoff for ETV as defined by other assay systems (19). For each of the viruses, N348I had a more pronounced statistically significant effect on NVP susceptibility. N348I conferred resistance to EFV and ETV to a lesser extent, and the effect was statistically significant only for virus A2 (subtype A) and B for EFV and virus A1 (subtype A), p8.9 (B), C, and D for ETV. These findings regarding NNRTI susceptibility extend those of others, who limited their analyses to subtype B HIV-1 (6, 22, 28). Although we recognize subtype differences, we cannot conclude that the N348I effect is subtype specific due to the sample number and the extent of variability within subtype B.
Since N348I has been reported to be replaced by wild-type virus after treatment cessation (6), we examined whether this mutation had an effect on viral replication. RC was determined by titration of virus stocks on SupT1, Jurkat, and HEK 293T cells in the absence of drug as previously described (18).
Acquisition of N348I caused impaired RC in B and non-B viruses compared to that of the wild-type virus in SupT1, Jurkat, and HEK 293T cells whether the virus was derived from clinical isolates or characterized molecular clones (Table 2). The effect of N348I on RC was similar across all subtypes and cell lines and found to be statistically significant. Replication was reduced by as much as 58% and 65% in HEK 293T cells for subtype A (A1) and C (MJ4) mutant viruses, respectively. Strong impairment of viral replication conferred by N348I has also been observed in wild-type subtype B in MT-2 and SupT1 cells but not in PM1 cells and peripheral blood mononuclear cells (6).
TABLE 2.
Effect of N348I mutation on RC of molecular clones and clinical isolates in different cell linesa
| Subtype | Mean % of wild-type RC ± SEM (fold decrease), P value |
||
|---|---|---|---|
| HEK 293T cells | Jurkat cells | SupT1 cells | |
| Molecular clones | |||
| p8.9 B | 55.57 ± 4.07 (1.80), <0.02 | 54.16 ± 5.5 (1.85), <0.02 | 62.91 ± 4.49 (1.59), <0.02 |
| MJ4 C | 34.65 ± 2.28 (2.89), <0.02 | 23.26 ± 1.57 (4.30), <0.02 | 26.35 ± 0.44 (3.80), <0.02 |
| 94ug114.1.6 D | 68.33 ± 0.94 (1.46), <0.02 | 67.55 ± 3.03 (1.48), <0.02 | 73.38 ± 2.66 (1.36), <0.02 |
| Clinical isolates | |||
| A1 | 42.28 ± 4.39 (2.37), <0.02 | 31.28 ± 4.28 (3.20), <0.02 | 37.80 ± 3.49 (2.65), <0.02 |
| A2 | 46.86 ± 1.31 (2.13), <0.02 | 40.25 ± 0.07 (2.48), <0.02 | 47.46 ± 0.84 (2.11), <0.02 |
| B | 73.20 ± 3.00 (1.37), <0.02 | 67.12 ± 4.32 (1.49), <0.02 | 81.92 ± 2.25 (1.22), <0.02 |
| C | 50.39 ± 4.16 (1.98), <0.02 | 41.87 ± 1.19 (2.39), <0.02 | 61.17 ± 3.91 (1.63), <0.02 |
Data are from mean RC values for mutant viruses which were calculated from the number of relative light units per nanogram of input p24 and expressed as a percentage of the wild-type value. Fold decreases in RC are shown in parentheses for comparisons of mutant and wild-type values. The standard error of the mean from two independent experiments is shown. Statistical analyses using Student's t test compared the N348I mutant virus with the wild-type virus.
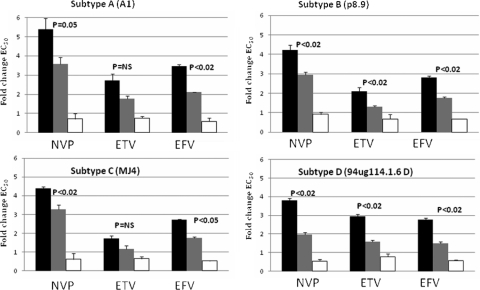
Since N348I commonly exists with M184V in clinical isolates (11) and the impact of both mutations on NNRTI susceptibility has not been reported, we introduced both mutations into RT by site-directed mutagenesis to determine phenotypic effects. Surprisingly, the reduced NNRTI drug susceptibility conferred by N348I alone was reversed across all subtypes when it was coexpressed with M184V (Fig. 1). M184V alone had a minimal effect on NNRTI susceptibility. The fold increase in NNRTI susceptibility conferred by M184V on N348I mutant virus was similar across subtypes and was statistically significant for NVP and EFV compared with N348I alone and only in subtype B and D viruses for ETV. Interestingly, M184V has previously been shown to hypersensitize a virus to NNRTIs (13, 24, 27).
FIG. 1.
NNRTI drug susceptibility conferred by N348I plus or minus M184V in B and non-B HIV-1 clinical isolates or molecular clones. Data are mean n-fold changes in drug EC50s for mutant viruses compared to those for the wild-type virus across subtypes ± the standard error of the mean from two independent experiments. Statistical analyses using Student's t test compared subtype-specific N348I plus M184V mutant virus with N348I mutant virus. NS, no statistically significant difference. Black bars, N348I; grey bars, N348I plus M184V; white bars, M184V.
The viral RC deficit of N348I alone in SupT1, Jurkat, and HEK 293T cells was reduced slightly for a few of the N348I and M184V double mutants, but not to wild-type levels (Table 3), and was statistically significant only for subtype C virus (MJ4) in Jurkat (P = 0.01) and Sup T1 cells (P = 0.02) compared with N348I alone. Interestingly, M184V alone reduced the RC of subtype A, B, C, and D viruses in Jurkat and SupT1 cells compared with that in HEK 293T cells. Similarly, M184V variants in vivo have been reported to be 4 to 8% less fit than the wild-type virus (17), due partly to a decreased ability to use natural nucleotide substrates (2). However, estimates of HIV-1 fitness vary considerably, depending on the methodology and genetic backbone used (4, 26).
TABLE 3.
Effect of N348I plus or minus M184V on RC of B and non-B clinical isolates or molecular clones in HEK 293T, Jurkat, and SupT1 cellsa
| Subtype and mutation(s) | Mean % of wild-type RC ± SEM (fold decrease) |
||
|---|---|---|---|
| HEK 293T cells | Jurkat cells | SupT1 cells | |
| A1 | |||
| N348I | 42.28 ± 4.39 (2.37) | 31.28 ± 4.28 (3.20) | 37.80 ± 3.49 (2.65) |
| N348I + M184V | 48.47 ± 5.51 (2.06) | 44.97 ± 5.34 (2.22) | 58.93 ± 4.44 (1.70) |
| M184V | 75.91 ± 3.59 (1.32) | 97.28 ± 3.24 (1.03) | 92.67 ± 4.32 (1.08) |
| p8.9 B | |||
| N348I | 55.57 ± 4.07 (1.80) | 54.16 ± 5.5 (1.85) | 62.91 ± 4.49 (1.59) |
| N348I + M184V | 65.19 ± 2.73 (1.53) | 72.15 ± 3.48 (1.39) | 80.78 ± 1.21 (1.24) |
| M184V | 83.49 ± 3.46 (1.20) | 98.93 ± 2.43 (1.01) | 98.94 ± 2.13 (1.01) |
| MJ4 C | |||
| N348I | 34.65 ± 2.28 (2.89) | 23.26 ± 1.57 (4.30) | 26.35 ± 0.44 (3.80) |
| N348I + M184V | 51.81 ± 5.97 (1.93) | 60.83 ± 4.27 (1.64) | 67.29 ± 5.14 (1.49) |
| M184V | 81.99 ± 3.02 (1.22) | 96.52 ± 3.58 (1.04) | 98.43 ± 1.13 (1.02) |
| 94ug114.1.6 D | |||
| N348I | 68.33 ± 0.94 (1.46) | 67.55 ± 3.03 (1.48) | 73.38 ± 2.66 (1.36) |
| N348I + M184V | 77.07 ± 4.49 (1.30) | 81.35 ± 3.65 (1.23) | 80.54 ± 3.46 (1.24) |
| M184V | 88.30 ± 0.02 (1.13) | 97.37 ± 1.26 (1.03) | 99.67 ± 0.32 (1.00) |
Data are mean RC values of mutant viruses which were calculated from the number of relative light units per nanogram of input p24 and expressed as a percentage of the wild-type value. Fold decreases in RC are shown in parentheses for comparisons of mutant and wild-type values. The standard error of the mean from two independent experiments is shown.
In this study, albeit having only looked at a few isolates, we demonstrate an impact of N348I on NNRTI susceptibility across subtypes. The mechanism of N348I resistance has been experimentally proven (21), in that N348I decreases catalytic efficiency and causes in vitro resistance to NVP by decreasing inhibitor binding. Differences among RTs from different clades can be due to small differences in the binding capacity of N348I mutants. Less clear is the role of RNase H in NNRTI resistance. It has been proposed that a decrease in RNase H activity due to connection subdomain mutations preserves template RNA and provides more time for NNRTI dissociation from RT, resulting in resumption of DNA synthesis and enhanced NNRTI resistance (1, 4, 16). However, NNRTI resistance cannot always be linked to a defect in RNase H (21, 23).
We propose that the C-terminal domains of RT, which include RNase H and the connection subdomain, should be considered in genotypic and phenotypic analyses to evaluate new determinants of resistance to existing and new RT inhibitors, particularly regarding the full spectrum of subtypes.
Nucleotide sequence accession numbers.
The GenBank accession number for the clone A1 clinical isolate is EU927369. The nucleotide sequences for subtype A (clone A2), subtype B, and subtype C HIV-1 clinical isolates have been submitted to GenBank (1421568).
Acknowledgments
We thank Nigel Temperton, University of Kent, for pCSFLW and Didier Trono, EPFL, Switzerland, for pCMV-Δ8.91 and pMDG. The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program: ZDV, TDF, ABC, 3TC, FTC, DDI, D4T, NVP, EFV, and ETV. pMJ4 was from Thumbi Ndung'u, Boris Renjifo, and Max Essex, and p94UG114.1.6 was from Beatrice Hahn and Feng Gao and the UNAIDS Network for HIV Isolation and Characterization.
This work was funded by the United Kingdom Medical Research Council. Funding from the European Community Seventh Framework Program (FP7/2007-2013) under the project Collaborative HIV and Anti-HIV Drug Resistance Network, grant agreement 223131, is also acknowledged.
Footnotes
Published ahead of print on 31 January 2011.
REFERENCES
- 1.Biondi, M. J., G. L. Beilhartz, S. McCormick, and M. Götte. 2010. N348I in HIV-1 reverse transcriptase can counteract the nevirapine-mediated bias toward RNase-H cleavage during plus-strand initiation. J. Biol. Chem. 285:26966-26975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deval, J., et al. 2004. Mechanistic basis for reduced viral and enzymatic fitness of HIV-1 reverse transcriptase containing both K65R and M184V mutations. J. Biol. Chem. 279:509-516. [DOI] [PubMed] [Google Scholar]
- 3.Ehteshami, M., and M. Götte. 2008. Effects of mutations in the connection domain and RNase-H domains of HIV-1 reverse transcriptase on drug susceptibility. AIDS Rev. 10:224-235. [PubMed] [Google Scholar]
- 4.Frost, S. D., M. Nijhuis, R. Schuurman, C. A. Boucher, and A. J. Brown. 2000. Evolution of lamivudine resistance in human immunodeficiency virus type 1-infected individuals: the relative roles of drift and selection. J. Virol. 74:6262-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao, F., et al. 1998. A comprehensive panel of near-full-length clones and reference sequences for non-subtype B isolates of human immunodeficiency virus type 1. J. Virol. 72:5680-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hachiya, A., et al. 2008. Amino acid mutation N348I in the connection subdomain of human immunodeficiency virus type 1 reverse transcriptase confers multiclass resistance to nucleoside and nonnucleoside reverse transcriptase inhibitors. J. Virol. 82:3261-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ikeda, Y., L. M. Ylinen, M. Kahar-Bador, and G. J. Towers. 2004. Influence of gag on human immunodeficiency virus type 1 species-specific tropism. J. Virol. 78:11816-11822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kagan, R. M., P. Sista, T. Pattery, L. Bacheler, and D. A. Schwab. 2009. Additional HIV-1 mutation patterns associated with reduced phenotypic susceptibility to etravirine in clinical samples. AIDS 23:1602-1605. [DOI] [PubMed] [Google Scholar]
- 9.Maïga, A. I., et al. 2010. Resistance-associated mutations to etravirine (TMC-125) in antiretroviral-naïve patients infected with non-B HIV-1 subtypes. Antimicrob. Agents Chemother. 54:728-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marcelin, A. G., et al. 2010. Factors associated with virological response to etravirine in nonnucleoside reverse transcriptase inhibitor-experienced HIV-1-infected patients. Antimicrob. Agents Chemother. 54:72-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCormick, A., et al. 2009. Prevalence and in vitro characteristics of reverse transcriptase (RT) N348I mutation in non-B subtypes in the absence of viral load monitoring, in the DART study (NORA substudy), poster 643. 16th Conference on Retroviruses and Opportunistic Infections, Montréal, Canada. http://www.ctu.mrc.ac.uk/dart/files/DART%20N348I%20poster%20CROI%202009.pdf.
- 12.Naldini, L., et al. 1996. In vivo gene delivery and stable transduction of non-dividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 13.Napolitano, L., K. Limoli, A. Paquet, M. Haddad, and E. Coakley. 2010. K65R, L74V/I, and M184V mutations are associated with hypersusceptibility to first and next generation NNRTI, poster 549. 17th Conference on Retroviruses and Opportunistic Infections, San Francisco, CA. http://www.retroconference.org/2010/Abstracts/38171.htm.
- 14.Ndung'u, T., B. Renjifo, and M. Essex. 2001. Construction and analysis of an infectious human immunodeficiency virus type 1 subtype C molecular clone. J. Virol. 75:4964-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nikolenko, G. N., et al. 2007. Mutations in the connection domain of HIV-1 reverse transcriptase increase 3′-azido-3′-deoxythymidine resistance. Proc. Natl. Acad. Sci. U. S. A. 104:317-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nikolenko, G. N., K. A. Delviks-Frankenberry, and V. K. Pathak. 2010. A novel molecular mechanism of dual resistance to nucleoside and nonnucleoside reverse transcriptase inhibitors. J. Virol. 84:5238-5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paredes, R., et al. 2009. In vivo fitness cost of the M184V mutation in multidrug-resistant human immunodeficiency virus type 1 in the absence of lamivudine. J. Virol. 83:2038-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parry, C. M., et al. 2009. Gag determinants of fitness and drug susceptibility in protease inhibitor-resistant human immunodeficiency virus type 1. J. Virol. 83:9094-9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peeters, M., et al. 2008. Determination of phenotypic clinical cut-offs for etravirine (ETR): pooled week 24 results of the DUET-1 and DUET-2 trials, abstr. 121. XVIIth International Drug Resistance Workshop, Sitges, Spain. http://www.aegis.com/conferences/hivdrw/2008/121.html.
- 20.Poveda, E., et al. 2007. Prevalence of etravirine (TMC-125) resistance mutations in HIV-infected patients with prior experience of non-nucleoside reverse transcriptase inhibitors. J. Antimicrob. Chemother. 60:1409-1410. [DOI] [PubMed] [Google Scholar]
- 21.Schuckmann, M. M., et al. 2010. The N348I mutation at the connection subdomain of HIV-1 reverse transcriptase decreases binding to nevirapine. J. Biol. Chem. 285:38700-38709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sluis-Cremer, N., K. Moore, J. Radzio, S. Sonza, and G. Tachedjian. 2010. N348I in HIV-1 reverse transcriptase decreases susceptibility to tenofovir and etravirine in combination with other resistance mutations. AIDS 24:317-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spence, R. A., W. M. Kati, K. S. Anderson, and K. A. Johnson. 1995. Mechanism of inhibition of HIV-1 reverse transcriptase by nonnucleoside inhibitors. Science 267:988-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tozzi, V., et al. 2004. Mutations in HIV-1 reverse transcriptase potentially associated with hypersusceptibility to nonnucleoside reverse-transcriptase inhibitors: effect on response to efavirenz-based therapy in an urban observational cohort. J. Infect. Dis. 189:1688-1695. [DOI] [PubMed] [Google Scholar]
- 25.von Wyl, V., et al. 2010. Epidemiological and biological evidence for a compensatory effect of connection domain mutation N348I on M184V in HIV-1 reverse transcriptase. J. Infect. Dis. 201:1054-1062. [DOI] [PubMed] [Google Scholar]
- 26.Wainberg, M. A. 2004. The impact of the M184V substitution on drug resistance and viral fitness. Expert Rev. Anti Infect. Ther. 2:147-151. [DOI] [PubMed] [Google Scholar]
- 27.Whitcomb, J. M., et al. 2002. Hypersusceptibility to non-nucleoside reverse transcriptase inhibitors in HIV-1: clinical, phenotypic and genotypic correlates. AIDS 16:F41-F47. [DOI] [PubMed] [Google Scholar]
- 28.Yap, S. H., et al. 2007. N348I in the connection domain of HIV-1 reverse transcriptase confers zidovudine and nevirapine resistance. PLoS Med. 4(12):e335. [DOI] [PMC free article] [PubMed] [Google Scholar]