Abstract
The generation of non-thienamycin-producing mutants with mutations in the thnL, thnN, thnO, and thnI genes within the thn gene cluster from Streptomyces cattleya and their involvement in thienamycin biosynthesis and regulation were previously reported. Four additional mutations were independently generated in the thnP, thnG, thnR, and thnT genes by insertional inactivation. Only the first two genes were found to play a role in thienamycin biosynthesis, since these mutations negatively or positively affect antibiotic production. A mutation of thnP results in the absence of thienamycin production, whereas a 2- to 3-fold increase in thienamycin production was observed for the thnG mutant. On the other hand, mutations in thnR and thnT showed that although these genes were previously reported to participate in this pathway, they seem to be nonessential for thienamycin biosynthesis, as thienamycin production was not affected in these mutants. High-performance liquid chromatography (HPLC)-mass spectrometry (MS) analysis of all available mutants revealed some putative intermediates in the thienamycin biosynthetic pathway. A compound with a mass corresponding to carbapenam-3-carboxylic acid was detected in some of the mutants, suggesting that the assembly of the bicyclic nucleus of thienamycin might proceed in a way analogous to that of the simplest natural carbapenem, 1-carbapen-2-em-3-carboxylic acid biosynthesis. The accumulation of a compound with a mass corresponding to 2,3-dihydrothienamycin in the thnG mutant suggests that it might be the last intermediate in the biosynthetic pathway. These data, together with the establishment of cross-feeding relationships by the cosynthesis analysis of the non-thienamycin-producing mutants, lead to a proposal for some enzymatic steps during thienamycin assembly.
The β-lactam family is the most important class of antibiotics for clinical use as antimicrobial agents and constitutes a major part of the global antibiotic market (6). This family includes penicillins, cephalosporins, carbapenems, and clavams. The basis of the selective toxicity of β-lactams is their role as inhibitors of bacterial cell wall peptidoglycan biosynthesis. Carbapenem antibiotics constitute a nonconventional class of β-lactams, produced by streptomycetes and Gram-negative bacteria, with important clinical applications, particularly in infections mediated by multidrug-resistant bacteria. They show a broad spectrum of activity and are highly resistant to most of the clinically encountered bacterial β-lactamases. Although the classical β-lactams (penicillins and cephalosporins) and their precursors are industrially produced by fermentation and semisynthesis (9), such a methodology has not been developed for clinically used carbapenems.
Thienamycin (Fig. 1A), the first carbapenem described (14), is the most potent and broadest in spectrum of all natural antibiotics known so far (6). It plays an important clinical role in the treatment of severe nosocomial infectious diseases, as it is active against aerobic and anaerobic bacteria, both Gram positive and Gram negative, including Pseudomonas species. Thienamycin was isolated from a soil species named Streptomyces cattleya NRRL 8057 (14), which interestingly also synthesizes the cephalosporin cephamycin C. However, owing to the low titers of thienamycin production and its chemical instability, commercial production by fermentation is problematic. A more stable derivative produced by chemical synthesis, named imipenem (N-formidoylthienamycin), is the product of choice for clinical use. Imipenem and other clinically useful carbapenems (meropenem, ertapenem, and doripenem, etc.) are currently made by total organic synthesis, and they are among the most expensive antibiotics on the market. Hence, there is interest in an understanding of the genetics and biochemistry of carbapenem biosynthesis to develop fermentation-based semisynthetic methodologies for the production of carbapenem antibiotics or intermediates useful for their preparation (8). Although the biosynthetic pathways leading to the bicyclic nuclei of penicillins, cephalosporins, and the clavam clavulanic acid have been extensively studied (for a review, see reference 19), the pathways for the highly substituted carbapenems, such as thienamycin, are less well understood.
FIG. 1.
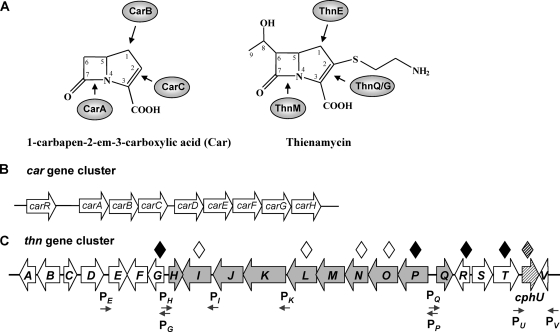
Carbapenems and biosynthetic gene clusters. (A) Chemical structure of 1-carbapenem-2-em-3-carboxylic acid (Car) and thienamycin and enzymes proposed to be involved in carbapenem ring formation. (B) Organization of the car gene cluster for Car biosynthesis in Pectobacterium carotovorum. The arrows indicate the order and direction of the transcription of the genes. (C) Organization and putative physical boundaries of the thn gene cluster for thienamycin biosynthesis in S. cattleya. Arrows indicate the order and direction of the transcription of the previously described thn gene cluster (25). Among them, gray arrows represent the ThnI-dependent genes integrating the ThnI regulon (27, 28). The cross-hatched arrow indicates a gene not required for thienamycin biosynthesis but essential for cephamycin biosynthesis (27), here named cphU (formerly thnU). White diamonds represent previously generated mutants affected in thienamycin production. Black diamonds indicate mutants generated in this work. The shaded diamond indicates a previously generated mutant affected in cephamycin C production. Small arrows indicate promoter regions identified (see the text for details).
Several carbapenem gene clusters have been identified from producer organisms, contributing to the knowledge of biosynthesis and regulation of these antibiotics (for a review, see reference 4). Carbapenems are biosynthesized via a different metabolic pathway than that of the classical β-lactams. Instead of the condensation of precursors by an ACV [δ-(l-α-aminoadipyl)-l-cysteinyl-d-valine] synthetase followed by cyclization by an isopenicillin N synthetase (IPNS), β-lactam ring formation in carbapenems occurs through an alternative mechanism involving a β-lactam synthetase. The gene cluster (car) for the simplest natural carbapenem, 1-carbapen-2-em-3-carboxylic acid (Fig. 1A and B), was first discovered in the plant pathogen Pectobacterium carotovorum (formerly Erwinia carotovora) (22). Similar clusters have since been found in both the opportunistic human pathogen Serratia marcescens (5) and the entomopathogenic bacterium Photorhabdus luminescens (7). Nine genes comprise the cluster; five of them are structural genes (carABCDE), although only three genes (carABC) are absolutely required for carbapenem-3-carboxylic acid (Car) biosynthesis (18, 23): CarB catalyzes the first step in the pathway, generating the pyrrolidine ring after precursor condensation; CarA catalyzes the formation of the β-lactam ring; and, finally, CarC converts the carbapenam to carbapenem (for details, see reference 4). CarD and CarE were proposed previously to catalyze the oxidation of l-proline to provide l-glutamate semialdehyde (l-GSA) (31) as a precursor for the condensation reaction catalyzed by CarB. The CarF and CarG proteins are involved in β-lactam resistance, and the role of CarH is still unknown. CarR is a transcriptional activator involved in the regulation of the gene cluster.
The gene cluster (thn) for the biosynthesis of the complex carbapenem thienamycin (Fig. 1A and C) was cloned from S. cattleya (25), opening up the possibility to study its biosynthesis and regulation. After in silico sequence analysis, putative roles were assigned to the gene products. The hypothetical physical boundaries of the thn cluster were initially established as lying beyond the region spanning from thnA to thnV, as judged by the generation of thienamycin-producing mutants in the flanking regions. The involvement in thienamycin biosynthesis was demonstrated after the independent mutagenesis of the structural thnL, thnN, and thnO genes (25) and the regulatory gene thnI (27). A previously reported bioinformatic analysis revealed ThnE and ThnM as likely homologues of CarB and CarA, respectively (25). Recent advances in the study of the biochemistry of some thienamycin enzymatic steps, mainly by in vitro studies with recombinant thn gene products, have contributed to a better understanding of the biosynthetic pathway. ThnE, which encodes a carboxymethylproline synthase, was functionally reported to be responsible for pyrrolidine ring formation in the first step of thienamycin biosynthesis (13). ThnG and ThnQ are members of the same family as CarC, and they have been postulated to catalyze reactions analogous to those of CarC (13, 30). A tentative biosynthetic pathway for the cysteaminyl side chain of thienamycin, in which coenzyme A (CoA) (after the incremental cleavage by ThnR, ThnH, and ThnT) appears to be the source of cysteamine, was proposed (12). In addition, ThnF was found to be most likely responsible for the formation of the S. cattleya cometabolite N-acetylthienamycin in the presence of acetyl-CoA (12). Recombinant versions of ThnG and ThnQ were recently shown to generate in vitro structural diversity in carbapenem antibiotics (2).
Several recent findings in this area concern the regulation of thienamycin biosynthesis. ThnI is a LysR-type transcriptional regulator which constitutes a key factor in controlling thienamycin biosynthesis. It is involved in the regulation of a subgroup of 10 genes in the thn cluster, which constitute the ThnI regulon (27, 28). A putative model for the mechanism of ThnI regulation has been proposed, in which ThnI appears to in part positively autoregulate its own expression (28). However, the regulation of the remaining genes of the cluster is still unknown. Surprisingly, a regulatory gene linked to the thn gene cluster, thnU, turned out to be involved in cephamycin C biosynthesis and not in thienamycin biosynthesis. This gene has been renamed as a cephamycin biosynthesis gene and will here be designated cphU (Fig. 1C). CphU is a Streptomyces antibiotic regulatory protein (SARP) pathway-specific transcriptional activator that regulates the expression of the nonlinked cephamycin structural genes pcbAB and cmcI (27).
The aim of this work was to gain further insight into the characterization of the thienamycin biosynthetic pathway in S. cattleya and determine the genes essential for thienamycin biosynthesis. High-performance liquid chromatography (HPLC)-mass spectrometry (MS) analysis and cross-feeding relationships of several mutants of thn genes revealed some putative intermediates and the order of some enzymatic steps in the biosynthetic pathway. An increase in thienamycin production was found for one of the structural gene mutants. In addition, we provide evidence supporting that some genes previously reported to be part of the thienamycin gene cluster are not essential for thienamycin biosynthesis.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and cloning vectors.
Streptomyces cattleya NRRL 8057, the thienamycin and cephamycin C producer, was used in this study. The S. cattleya thnL, thnN, thnO, and thnI non-thienamycin-producing mutants (25, 27) and the ΔthnD-cphU non-thienamycin- and non-cephamycin-producing mutant (L. E. Núñez, unpublished data) were also included in this work (Fig. 1C). Escherichia coli DH10B (Invitrogen) was used as a cloning host. E. coli ET12567 (dam dcm hsdS), as a source of nonmethylated DNA (20) to prevent plasmid degradation by the methylation restriction system of S. cattleya, was used, harboring pUB307 (11) as the donor for intergeneric conjugation. Staphylococcus aureus ATCC 6538P, a thienamycin-sensitive and cephamycin C-resistant strain, and E. coli ESS, a strain supersensitive to all β-lactams, were used for bioassays. For sporulation, S. cattleya bacteria were grown in Bennet medium, and for antibiotic production, S. cattleya bacteria were grown in R5A-minus sucrose medium using an inoculum previously grown in tryptic soy broth (TSB) medium (Merck), as previously described (27). Cosmid cosCAT25 (25) was used as a source of S. cattleya thn gene cluster DNA. The apramycin resistance-encoding gene aac(3)IV was obtained from pEFBA (10) and pUO9090 (M. C. Martín, unpublished results). pUC18 (Pharmacia), pBSK (Stratagene), and pUK21 (34) were used as E. coli cloning vectors, and cosmid pHZ1358 (32) was used for E. coli-S. cattleya intergeneric conjugation. When needed, antibiotics were added to a final concentration of 25 μg/ml for thiostrepton, apramycin, kanamycin, chloramphenicol, or nalidixic acid; 100 μg/ml for ampicillin; and 20 μg/ml for tobramycin.
DNA manipulations, sequencing, and analysis.
Total DNA isolation, plasmid DNA preparations, restriction endonuclease digestions, ligations, and other DNA manipulations were performed according to standard procedures for E. coli (29) and for Streptomyces (16). Intergeneric conjugation from E. coli ET12567/pUB307 into S. cattleya was done according to procedures previously described (21). Southern analysis, hybridization, and detection were performed according to procedures reported previously (29), and the labeling of DNA probes was performed with digoxigenin (DIG) using the DIG DNA labeling and detection kit (Roche). DNA sequencing was performed on double-stranded DNA templates by using the dideoxynucleotide chain termination method and the Thermo Sequenase labeled primer cycle sequencing kit with 7-deaza-dGTP (Amersham Biosciences) and an ALF-Express automatic DNA sequencer (Pharmacia). Computer-assisted database searching and sequence analyses were made by using the GCG sequence analysis software package from the University of Wisconsin Genetics Computer Group and the BLAST program (1).
Gene replacement experiments.
As a general rule for insertional inactivation, gene replacement experiments were carried out by the insertion of the aac(3)IV gene encoding the apramycin resistance cassette. In all cases, the aac(3)IV gene was inserted within the coding region of the gene to be mutagenized and in the same direction of transcription. The introduction of DNA into S. cattleya was achieved by using the bifunctional cosmid pHZ1358 (32) through intergeneric conjugation from E. coli ET12567/pUB307 as described previously (21). After the introduction of the construct into S. cattleya by intergeneric conjugation, transconjugants in which a double crossover occurred were identified by their resistance to apramycin and susceptibility to thiostrepton.
(i) Mutagenesis of thnP.
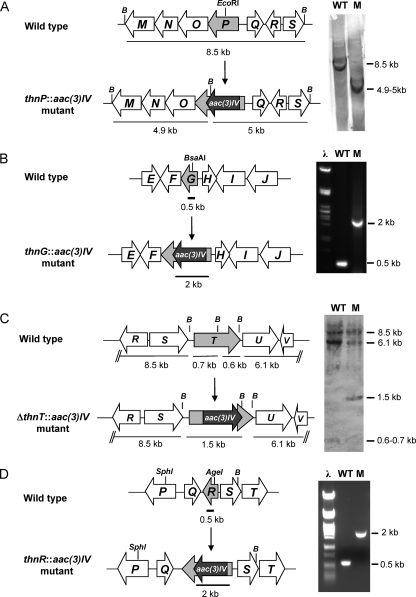
To generate a mutant containing a thnP deletion mutation, a 6.5-kb BglII-BamHI DNA fragment carrying thnP from cosCAT25 was cloned into pUK21 previously digested with the same restriction enzymes. The resulting plasmid was digested with EcoRI, and the generated sticky ends were filled in with Klenow polymerase to obtain blunt ends. The aac(3)IV gene (apramycin resistance cassette) obtained from pEFBA, after EcoRV-SmaI digestion (blunt ends), was then inserted into the blunt-ended EcoRI located within the thnP coding region. An 8-kb insert from this construct was isolated after SpeI digestion and cloned into the XbaI-digested conjugative vector pHZ1358 to generate pHZLE7F, which was introduced into S. cattleya to generate the thnP disruption mutant by gene replacement via homologous recombination (Fig. 2A).
FIG. 2.
Generation of mutants in thn genes. Shown are the organizations of chromosomal regions of the wild type and the mutant strains and genetic analysis by Southern analysis (A and C) and by PCR (B and D). Only relevant restriction sites are indicated, and B indicates BamHI. The gray arrow represents the mutated gene, and the black arrow indicates the aac(3)IV gene, encoding the apramycin resistance marker (apr cassette). (A) Insertion of the apr cassette into the thnP coding region and Southern hybridization of BamHI-digested genomic DNA from the wild type (WT) and the thnP::aac(3)IV mutant (M) using the 8.5-kb BamHI fragment as a probe. (B) Insertion of the apr cassette into the thnG coding region and PCR analysis of genomic DNA from the wild type and the thnP::aac(3)IV mutant using oligonucleotides ThnG-R and ThnG-F. (C) Deletion of the internal fragment of thnT and replacement with the apr cassette (see Materials and Methods) and Southern hybridization of BamHI-digested genomic DNA from the wild type and the ΔthnT::aac(3)IV mutant using pHZMRT as a probe. (D) Insertion of the apr cassette into the thnR coding region and PCR analysis of genomic DNA from the wild type and the thnR::aac(3)IV mutant using oligonucleotides ThnR-R and ThnR-F.
(ii) Mutagenesis of thnG.
To construct a mutant containing a thnG disruption mutation, a 5.8-kb fragment carrying thnG was cloned from cosCAT25 into pUC18 previously digested with the same restriction enzymes. The resulting plasmid was digested with BsaAI located within the thnG coding region. The apramycin resistance cassette, obtained from pEFBA as a BamHI (blunt ended)-SmaI fragment, was then inserted into the BsaAI-digested plasmid. A 7.3-kb EcoRI-HindIII fragment was subcloned into pBSK previously digested with the same enzymes. The fragment was then obtained after BamHI digestion of this construct and cloned into BamHI-digested pHZ1358 to generate pHZLE1cF, which was then introduced into S. cattleya to generate the thnG disruption mutant (Fig. 2B).
(iii) Mutagenesis of thnT.
To generate a mutant containing a deletion in the thnT coding region, two fragments of approximately 2 kb containing part of thnT and the flanking genes MR1 and MR2 were amplified from cosCAT25 by PCR with high-fidelity Pfx DNA polymerase (Invitrogen) using primer pairs MR1-F/MR1-R and MR2-F/MR2-R, respectively. MR1 and MR2 carry synthetic restriction sites (BglII-SphI and NotI-XbaI, respectively) to facilitate cloning into pUO9090 at both sites of the apramycin resistance cassette (Apr)-encoding gene, aac(3)IV. The amplified MR1 fragment was digested with BglII-SphI and cloned into pUO9090, previously digested with the same restriction enzymes, upstream of aac(3)IV, resulting in pUOMR1apr. In the next step, the amplified MR2 fragment was digested with NotI-XbaI and cloned into pUOMR1apr, previously digested with the same restriction enzymes, downstream of aac(3)IV. Finally, the resulting construct, pUOMR1aprMR2, was digested with SpeI and the MR1aprMR2 fragment (5.5 kb) recovered after agarose gel electrophoresis, blunt ended with Klenow polymerase (Roche), and cloned into the BamHI-digested (blunt-ended) conjugative vector pHZ1358 to generate pHZMRT. This plasmid was introduced into S. cattleya to generate a thnT deletion mutant (Fig. 2C).
(iv) Mutagenesis of thnR.
To construct a mutant containing a thnR disruption mutation, a 3.9-kb BamHI-SphI fragment comprising thnR and the flanking genes was cloned from cosCAT25 into pUC18 previously digested with the same restriction enzymes. The resulting plasmid was digested with AgeI and blunt ended with Klenow polymerase. Afterwards, the apramycin resistance cassette (1.5 kb), isolated from pEFBA as an EcoRV-SmaI blunt-end fragment, was inserted into the AgeI site (blunt ended) located within the thnR coding region. A 5.4-kb fragment was isolated after PvuI digestion, blunt ended, and cloned into BamHI-digested (blunt-ended) vector pHZ1358 to generate pHZMR2R. This plasmid was introduced into S. cattleya to generate a thnR disruption mutant (Fig. 2D).
Cross-feeding bioassays.
Cross-feeding occurs when a mutant blocked at an earlier step in the pathway converts an intermediate produced by a mutant blocked at a later step in the pathway, resulting in thienamycin production. The cross-feeding experiments were performed mainly as described previously (3, 26). Spore suspensions (20 μl) of non-thienamycin-producing mutants were analyzed by pairs and inoculated close to each other (20 mm) onto R5A-minus sucrose agar plates to establish cross-feeding relationships. After 6 to 10 days of incubation at 28°C, agar strips containing both mutants were cut out and analyzed by a bioassay. The strips were placed onto TSA1/2 (half of the concentration of all components of tryptic soy agar) containing S. aureus ATCC 6538P as the indicator organism to determine whether the diffusional exchange of accumulated intermediates restored the thienamycin production ability of the tested mutants. Inhibition zones or halos corresponding to the restoration of thienamycin production were detected after 1 day of incubation at 37°C.
Analysis of thienamycin and cephamycin C production.
Thienamycin production was determined by a bioassay using thienamycin-sensitive S. aureus strain ATCC 6538P (cephamycin C resistant) as an indicator. The thienamycin concentration was determined with imipenem, the N-formidoyl derivative of thienamycin, as a standard. Cephamycin C production was assayed by using the β-lactam-supersensitive E. coli strain ESS. The identification and quantitative analysis of thienamycin and cephamycin C production were performed by HPLC-MS using chromatographic equipment formed by an Alliance separation module, photodiode array detector 9965, and a ZQ 4000 mass spectrometer (Micromass; Waters). Acetonitrile and 0.1% trifluoroacetic acid (TFA) in water were used as the mobile phase, and a reversed-phase column (Atlantis dC18, 5 μm, 2.1 by 150 mm; Waters) was used for separations. The detection and spectral characterization of peaks were made with Empower software (Waters).
Samples of culture supernatants (5 μl) were eluted with a linear gradient from 0 to 15% acetonitrile over 15 min, followed by an additional 5 min of elution with 100% acetonitrile and a further 10 min of equilibration under initial conditions at a flow rate of 0.25 ml/min. For some analyses performed with a different column (Atlantis T3, 3 μm, 2.1 by 150 mm; Waters), the initial gradient was prolonged to reach 20% acetonitrile in 20 min. Mass detection was done by electrospray ionization in the positive mode, with an ionization voltage of 3 kV, a cone voltage of 15 V, a cone temperature of 100°C, and a desolvation temperature of 250°C.
Detection was performed spectrophotometrically at 309 nm for thienamycin and at 264 nm for cephamycin C and also by selected ion recording (SIR) of the protonated ions (m/z 273 for thienamycin and m/z 447 for cephamycin C). Other molecular ions corresponding to putative intermediates were also detected in some analyses.
RESULTS
An approximately 25.4-kb region of the S. cattleya chromosome (EMBL accession number AJ421798) encoding a series of 22 genes, thnA to thnV, was first proposed (25) to participate in thienamycin biosynthesis and regulation (Fig. 1C). We have used a mutational approach for some thn genes as a way to determine their involvement in the biosynthesis of the complex carbapenem thienamycin. Together with previously generated mutants of the structural genes thnL, thnN, and thnO (25), additional ones were obtained here. All mutants were further analyzed by searching for an accumulation of putative intermediates to establish some steps of the biosynthetic pathway leading to thienamycin molecule assembly.
Concerning transcriptional organization, the ThnI-regulated genes appear to be organized into four operons driven by four promoter regions whose in vivo functionality has been determined (28). PQ and PH (Fig. 1C) drive the expression of the downstream genes as two monocistronic transcripts, termed thnQ and thnH, respectively. PP drives the transcription of thnP, thnO, thnN, thnM, and thnL as the pentacistronic operon thnPONML, whereas PK drives the transcription of thnK, thnJ, and thnI as the tricistronic operon thnKJI. Other putative promoter sequences have also been identified for the ThnI-independent genes (Fig. 1C) by the use of the BPROM bacterial promoter prediction server (Softberry Inc., Mount Kisco, NY). Upstream of the thnG coding region, and divergently oriented to PH, a putative promoter sequence (PG) consisting of a −35 box at TTGCCT (score of 56) and a −10 box at CCGGATAGT (score of 16) was found. Another putative promoter sequence (PE) is located upstream of the thnE initiation codon. It consists of a −35 box at TTGACT (score of 26) and a −10 box at ACCTAGGCT (score of 61). An additional putative promoter sequence has been identified in the thnT-cphU intergenic region (PU), consisting of a −35 box at TTGACC (score of 44) and a −10 box at GCCTACACT (score of 45). Upstream of the thnV initiation codon, a putative promoter sequence (PV) consisting of a −35 box at TTGTCC (score of 31) and a −10 box at CGTTATGAT (score of 73) was also identified. The above-mentioned ThnI-dependent promoters were not localized with this bioinformatic program, with the unique exception of PH, consisting of a putative −35 box at CTGCCG (score of 10) and a −10 box at ACTTACGCT (score of 24).
Generation of mutants affected in thienamycin biosynthesis. (i) Inactivation of thnP.
The predicted thnP product consists of 484 amino acid residues with a molecular mass of 53,734 Da (EMBL accession number CAD18984) and displays sequence similarity with methyltransferases belonging to the radical S-adenosylmethionine (SAM) superfamily (cl10028) and associated with the B12 binding domain (cd02068). Radical SAM enzymes catalyze steps in the biosynthesis of many antibiotics. ThnP also shows similarity to two putative methyltransferases encoded in the thn cluster as well: ThnL (26% identity and 46% similarity) and ThnK (24% identity and 49% similarity).
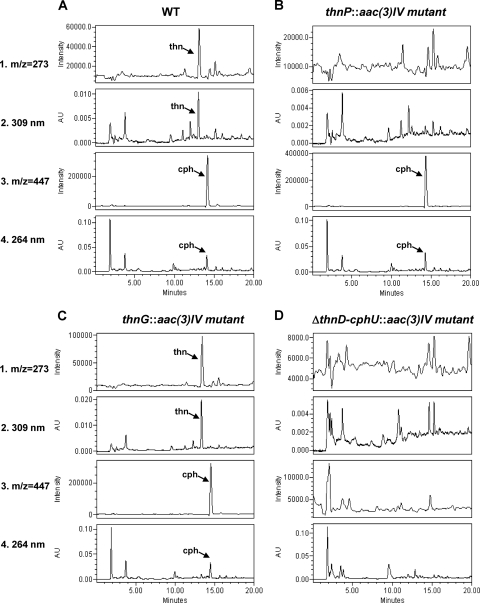
To investigate the role of thnP in thienamycin biosynthesis, a disruption mutant, thnP::aac(3)IV, was generated by inserting the apramycin resistance cassette within the thnP coding region (see Materials and Methods). Chromosomal DNA from this mutant and the wild-type strain was digested and analyzed by Southern analysis using the BamHI 8.5-kb fragment as a probe. An 8.5-kb hybridizing band was detected in the wild-type strain, whereas two positive signals of 4.9 and 5 kb were detected in the mutant (Fig. 2A). Thienamycin production was analyzed in culture supernatants of the thnP disruption mutant after 5 days of growth by bioassays against the thienamycin-sensitive and cephamycin-resistant strain S. aureus ATCC 6539P and also by HPLC-MS (Fig. 3B). An absence of thienamycin production in this mutant demonstrated that thnP is essential for thienamycin biosynthesis. However, the mutant retained the ability to produce cephamycin, thus showing that ThnP is not involved in cephamycin C biosynthesis.
FIG. 3.
MS and HPLC analyses of mutants affected in thienamycin biosynthesis. Shown are data for culture supernatants from the S. cattleya wild-type strain (A), the thnI-disrupted mutant (B), the thnG-disrupted mutant (C), and the ΔthnD-cphU deletion mutant (D). Thienamycin detection was as follows: SIR at m/z 273 (1) and detection at 309 nm (2). Cephamycin C detection was as follows: SIR at m/z 447 (3) and detection at 264 nm (4). The arrows indicate the retention times of the peaks corresponding to thienamycin (thn) and cephamycin C (cph). AU, arbitrary units.
(ii) Inactivation of thnG.
The thnG gene product is a protein of 263 amino acid residues with a molecular mass of 29,188 Da (EMBL accession number CAD18975) and displays sequence similarity to members of the phytanoyl-CoA dioxygenase (PhyH) superfamily (cl02184). ThnG shares 32% identity (45% similarity) with the deduced product of thnQ, another gene belonging to the same family in the thn cluster. ThnG and ThnQ are potential 2-oxoglutarate-dependent dioxygenases proposed to catalyze the desaturation of the carbapenam bicyclic ring to carbapenem, as was shown previously for Car biosynthesis for CarC, which belongs to the same superfamily of enzymes (30).
In order to understand the role of thnG in thienamycin biosynthesis, a disruption mutant, thnG::aac(3)IV, was generated by inserting the apramycin resistance cassette within the thnG coding region. Chromosomal DNAs from this mutant and the wild-type strain were analyzed by a PCR amplification reaction using the oligonucleotides ThnG-R and ThnG-F (Table 1). As shown in Fig. 2B, a PCR product of about 2.0 kb was obtained with the mutant, while for the wild type, the product was 0.5 kb. The 1.5-kb increase in the size of the mutant was due to the insertion of the apramycin resistance cassette, as was verified by sequencing, thus confirming the replacement of the wild-type gene by the mutated one. Analysis of culture supernatants of the mutant after 5 days of growth by bioassay and HPLC-MS analysis (Fig. 3C) revealed that the mutant still produced thienamycin but at a higher yield than that of the wild-type strain. Additional experiments were performed to quantify thienamycin production at different times during growth, both by bioassay and spectrophotometrically by signal integration at 309 nm. These experiments confirmed a 2.5-fold increase in thienamycin production for the mutant strain in comparison with the wild-type strain (which produces around 1 μg/ml). Furthermore, the accumulation of a compound with a molecular mass corresponding to a putative intermediate in the biosynthetic pathway was also detected in this mutant (see below). However, the mutant retained the ability to produce cephamycin at normal levels, thus showing that ThnG is not involved in cephamycin C biosynthesis.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′-3′)a | Description |
|---|---|---|
| ThnG-F | AGGTGCGCATCATCGGCCG | Forward primer for thnG |
| ThnG-R | TGGCGGGGCAGCGAATAGC | Reverse primer for thnG |
| MR1-F | ACGTAGATCTCGATGACCTCGTTCCCG | Forward primer for thnT |
| MR1-R | ACGTGCATGCGAACTCCCTTCAGCCCG | Reverse primer for thnT |
| MR2-F | ACGTGCGGCCGCTCCTCACCCGCGCCGTG | Forward primer for thnT |
| MR2-R | ACGTTCTAGACGCTCCGGCTGTGGGTG | Reverse primer for thnT |
| ThnR-F | TTCCGACCTGGCTGACCGGC | Forward primer for thnR |
| ThnR-R | AGCTCCCGCACCAGGAACCC | Reverse primer for thnR |
The restrictions sites BglII (AGATCT), SphI (GCATGC), NotI (GCGGCCGC), and XbaI (TCTAGA) used for cloning are in boldface type.
Identification of genes not essential for thienamycin biosynthesis.
The finding that the regulatory gene cphU linked to the thn cluster is involved in cephamycin C biosynthesis and not in thienamycin biosynthesis raised the question of whether other genes in the cluster initially considered to be involved in thienamycin biosynthesis were not part of the thn cluster. In order to redefine more precisely the right physical boundary of the thn gene cluster, two novel mutants were independently generated in the thnT and thnR coding regions within the S. cattleya genome. The thnT gene is adjacent to the cephamycin regulatory gene cphU. thnR is adjacent to thnQ, the distal gene in this side of the cluster involved in thienamycin biosynthesis, since it belongs to the ThnI regulon (27, 28).
(i) Generation of a thnT deletion mutant.
The predicted thnT gene product consists of 399 amino acid residues with a molecular mass of 39,125 Da (EMBL accession number CAD18988) and displays significant similarity to members of the DmpA/ornithine acetyltransferase (OAT) superfamily of aminopeptidases (cl00603). DmpA shows similarity in the catalytic mechanism to N-terminal nucleophile (Ntn) hydrolases, which are enzymes that catalyze the cleavage of amide bonds through the nucleophilic attack of the side chain of an N-terminal serine, threonine, or cysteine. Based on bioinformatic database analysis, it was initially proposed that ThnT might be involved in cysteine transferase during thienamycin biosynthesis (25). Enzymatic assays have shown that recombinant ThnT is able to hydrolyze pantetheine to yield cysteamine, and a putative role in generating the cysteaminyl side chain in thienamycin biosynthesis was proposed (12).
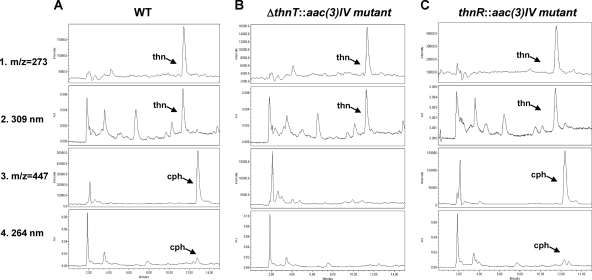
To uncover the role of thnT in S. cattleya, a deletion mutant was generated in the chromosome by gene replacement. The apramycin resistance cassette aac(3)IV was inserted within the thnT coding region, generating a deletion mutant, ΔthnT::aac(3)IV (see Materials and Methods). Chromosomal DNA from this mutant and the wild-type strain was digested with BamHI. Southern blotting experiments were performed by using the pUOMR12 construct as a probe. In addition to 8.5-, 6.1-, and 0.6-kb positive signals found in both strains, a differential hybridizing band of 1.5 kb was detected for the deletion mutant, whereas a 0.7-kb band was found for the wild-type strain (Fig. 2C), confirming the replacement of the wild-type gene by the mutated one. As a consequence of the mutation, a 1.1-kb deletion was produced in the thnT coding region (nucleotides 24,800 to 25,900 in the sequence) (EMBL accession number AJ421798). Analysis of culture supernatants of the mutant, after 5 days of growth, by bioassay against S. aureus ATCC 6538P revealed that the thnT deletion mutant was still able to produce thienamycin (data not shown). Bioassays against Escherichia coli ESS (sensitive to both thienamycin and cephamycin C) showed a clear reduction in the halo diameter in comparison to that of the wild-type strain (data not shown), thus suggesting that the biosynthesis of cephamycin was affected. HPLC-MS analysis (Fig. 4B) confirmed that this mutant retained the ability to produce thienamycin but had lost the ability to produce cephamycin. This result revealed that ThnT is not essential for thienamycin biosynthesis and, as previously shown for cphU, that it is involved in cephamycin C biosynthesis.
FIG. 4.
HPLC-MS analysis of mutants not affected in thienamycin biosynthesis. Shown are data for culture supernatants from the S. cattleya wild-type strain (A), the ΔthnT deletion mutant (B), and the thnR-disrupted mutant (C). Thienamycin and cephamycin detection (1, 2, 3, and 4) is described in the legend of Fig. 3.
(ii) Generation of a thnR disruption mutant.
ThnR is a protein of 240 amino acid residues and a molecular mass of 26,447 Da (EMBL accession number CAD18986), which displays sequence similarity with members of the Nudix hydrolase superfamily (cd02883). This superfamily includes enzymes found in organisms of all three kingdoms of life, and it catalyzes the hydrolysis of nucleoside diphosphates linked to other moieties, X. ThnR displays a well-conserved domain of cd03426, coenzyme A pyrophosphatase (CoAse), which functions by catalyzing the elimination of oxidized inactive CoA that can inhibit CoA-utilizing enzymes. Although thnR does not resemble any previously known antibiotic biosynthetic gene, it was included in the thn cluster based on its physical situation, surrounded by homologous biosynthetic and regulatory antibiotic genes (25). Enzymatic assays have shown that recombinant ThnR is able to cleave CoA to 4-phosphopantetheine, and a putative role in thienamycin biosynthesis was proposed previously (12).
To more clearly define the thn cluster physical boundary and learn more about the role of thnR in S. cattleya, a disruption mutant was obtained for this gene. The thnR::aac(3)IV mutant was generated by the insertion of the apramycin resistance cassette within the thnR coding region. Chromosomal DNA from the mutant and the wild-type strain was obtained and analyzed by a PCR amplification reaction using the oligonucleotides ThnR-F and ThnR-R as primers (Table 1). As shown in Fig. 2D, a PCR product of about 2.0 kb was obtained for the mutant, while for the wild type, the product was 0.5 kb. The 1.5-kb increase was due to the insertion of the apramycin resistance cassette in the mutant, as was verified by sequencing, thus confirming the replacement of the wild-type gene by the mutated one. Analysis of culture supernatants of the mutant by bioassay and HPLC-MS analysis (Fig. 4C) revealed that the mutation in thnR has no effect on thienamycin or cephamycin production. This indicates that thnR is not essential for thienamycin or cephamycin biosynthesis and might represent the physical boundary of the thienamycin gene cluster.
Cross-feeding experiments.
Since antibiotics are usually the products of stepwise linear biosynthetic pathways, it is often possible to carry out cosynthesis tests between mutants blocked at different steps in the pathway. This is based on the assumption that intermediates accumulated by the blocked mutants are freely diffusible and capable of being taken up by other mutants grown in close proximity. Thus, a mutant blocked later in the pathway (secretor) might accumulate an intermediate that may be able to enter a mutant blocked earlier (convertor), providing a missing intermediate for continuing synthesis in the latter strain.
To establish cross-feeding relationships, cosynthesis analysis of the nonproducing mutants in the thnL, thnN, thnO, and thnP genes blocked in the thienamycin biosynthetic pathway was carried out. Pairs of mutants were plated close to each other on agar plates, and after incubation, strips were analyzed by using S. aureus as an indicator strain to detect a restoration of thienamycin production (see Materials and Methods). Wild-type S. cattleya was used as a positive control, and the different mutants were used as negative ones. Initially, several pairs of mutants were found to show cross-complementation that restored thienamycin production, and halos were found spanning both mutants at the same time. This indicated that some intermediates were stable and were able to cross the cell membrane and diffuse between cells and through the agar. After establishing the conditions for each pair of mutants, distance and time of incubation, one-way complementation was obtained. As shown in Fig. 5, successful cross-feedings were observed by the appearance of an inhibition zone in the thnL, thnO, and thnN mutants. Five pairs gave a positive result: thnL versus thnP, thnO versus thnL, thnN versus thnL, thnN versus thnP, and thnN versus thnO. However, no cross-feeding was detectable for the remaining pair, thnP versus thnO. The ability of thnL, thnO, and thnN mutants to produce thienamycin in the neighborhood of thnP; thnL; and thnO, thnL, and thnP, respectively, demonstrates that the first mutant incorporates an intermediate secreted by the second one and converts it into thienamycin. As outlined in Fig. 5, these results might provide an indication of the possible order of the enzymatic steps in the pathway, as follows: ThnN, ThnO, ThnL, and ThnP.
FIG. 5.
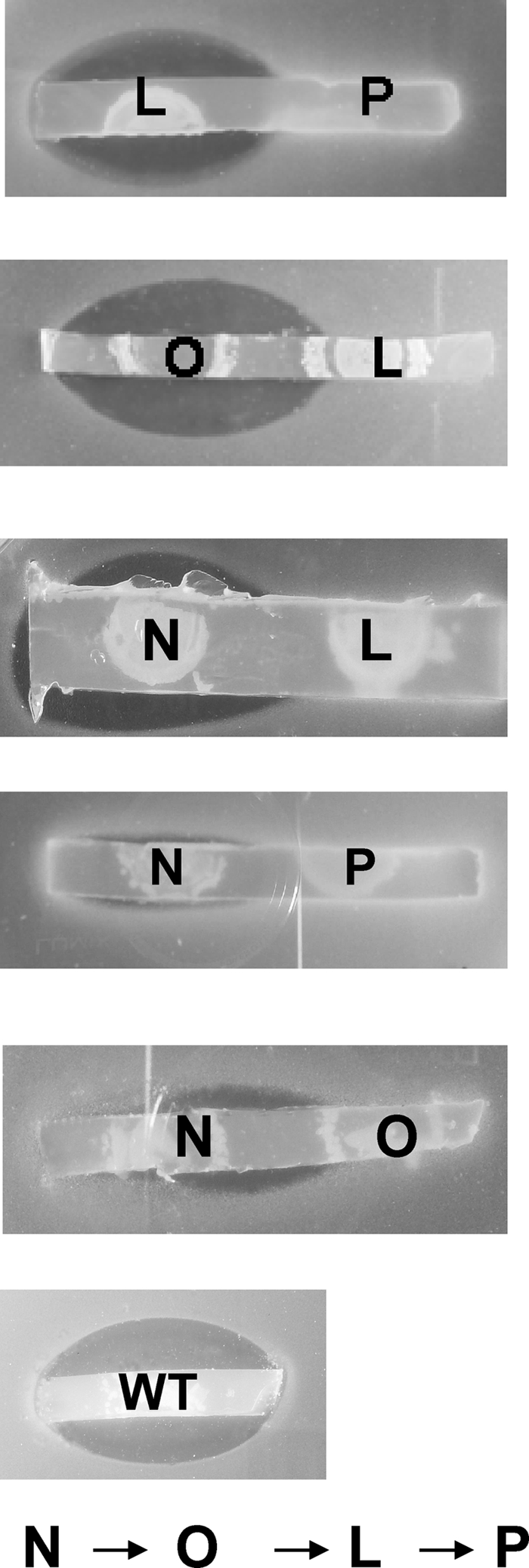
Cross-feeding relationship of thn structural mutants. Cosynthesis experiments were performed with thnN, thnL, thnO, and thnP non-thienamycin-producing structural mutants. Agar strips from R5A (minus sucrose) agar plates containing the mutants grown for 6 to 10 days at 28°C were used in the bioassays against thienamycin-sensitive S. aureus strain ATCC 6538P. The inhibition zones indicate a restoration of thienamycin biosynthesis due to the diffusional exchange of accumulated intermediates. No cross-feeding was detectable in the rest of the pairs. The wild-type strain was used as a positive control. L, N, O, and P indicate mutants generated in genes thnL, thnN, thnO, and thnP.
Identification of putative intermediates in thienamycin biosynthesis-blocked mutants by HPLC-MS.
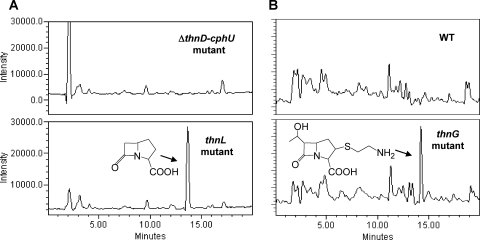
To investigate the biosynthetic steps of the thienamycin pathway, phenotypic analysis of the available thn mutants was performed by HPLC-MS analysis. We explored the possibility of detecting putative intermediates in the biosynthetic pathway by HPLC-MS analysis of culture supernatants of all available non-thienamycin-producing mutants. The production conditions were previously described for thienamycin production by growth in R5A liquid medium (minus glucose) (27). Samples were taken at various times (4, 5, 6, and 7 days) during growth. The mutants included in the analysis were the ones affecting the following genes: thnN, thnO, thnP, thnL, and thnG. The mutant blocked in the regulatory gene thnI, affected in the expression of the ThnI-dependent genes, was also analyzed, as it was still able to express the structural ThnI-independent genes. The S. cattleya wild-type strain was used as a positive control for the HPLC mass analysis. A ΔthnD-cphU deletion mutant (Fig. 3D) was used as a negative control. This mutant lacks most of the thn genes (from thnD up to thnT and the promoter region and first 13 cphU codons) and is affected in both thienamycin and cephamycin C production (L. E. Nuñez et al., unpublished data).
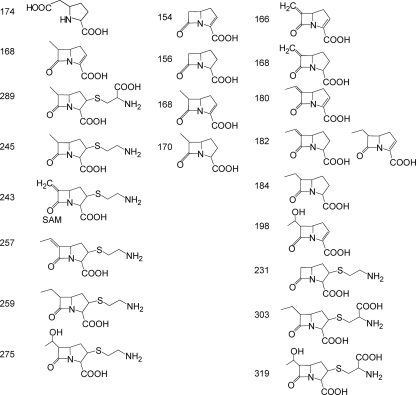
The molecular ions included in the analysis (Fig. 6) were selected according to different putative intermediates reported in the literature. SIR detection of protonated ions was performed at m/z 174, 168, 289, 245, 243, 257, 259, and 275 (25, 35) and at m/z 154, 156, 168, and 170 (30). In addition, novel combinations in the enzymatic step order corresponding to m/z 166, 168, 180, 182, 184, 198, 231, 303, and 319 were also analyzed (Fig. 6). One of the major difficulties of this approach was the low level of thienamycin production in the wild-type strain (about 1 μg/ml) and, consequently, of the putative intermediates. Despite this fact, two compounds of m/z 156 and 275 (Fig. 7) were detected by HPLC-MS analysis. Due to the low level of production and the hydrophilic nature of the compounds, which does not permit their solvent extraction from the culture medium, there was not sufficient yield to allow scale-up purification and nuclear magnetic resonance (NMR) analyses. Nevertheless, according to the mutants and masses of compounds detected, some of the peaks could be putatively ascribed to specific compounds. A peak with m/z 156 was detected in cultures of the thnL mutant (Fig. 7A) and at lower levels in the wild-type strain and the thnP and thnN mutants as well (however, it was absent in the ΔthnD-cphU, thnI, and thnO mutants). According to its mass, this compound could correspond to carbapenam-3-carboxylic acid, one of the putative intermediates proposed previously for the initial steps of thienamycin biosynthesis (30). Another peak corresponding to a product with m/z 275 was found to accumulate in the thnG mutant (Fig. 7B). The molecular mass could correspond to 2,3-dihydrothienamycin, one of the proposed late intermediates in the thienamycin pathway (25, 35). However, no such peak was detected for any of the remaining analyzed strains, including the rest of the mutants and the wild-type strain.
FIG. 6.
Chemical structures and molecular masses ([M + H]+) of putative intermediates included in the MS analysis.
FIG. 7.
Detection of putative intermediates by MS analysis. (A) SIR at m/z 156 ([M + H]+) of culture supernatants of S. cattleya ΔthnD-cphU and thnL mutants. The peak indicated by an arrow putatively corresponds to carbapenam-3-carboxylic acid. (B) SIR at m/z 275 ([M + H]+) of culture supernatants of the S. cattleya wild type and the thnG mutant. The peak indicated by an arrow putatively corresponds to 2,3-dihydrothienamycin.
DISCUSSION
The carbapenem group of β-lactams is one of the most therapeutically potent antibiotics currently available (24). Although carbapenems are natural products, the clinically used antibiotics are thienamycin derivatives produced by chemical synthesis, since no fermentation procedures have been developed yet. This is due mainly to the low yields of thienamycin production by the producing strains and to molecule instability. Deciphering the biosynthetic pathway in order to facilitate its metabolic manipulation might enable more efficient production and the possibility of obtaining more stable derivatives. This might also lead to the generation of novel carbapenems, which are needed for the fight against emergent antibiotic-resistant pathogens.
Thienamycin and its derivatives have the same carbapenem nucleus as the simplest member of carbapenems, Car, whose biosynthetic pathway has been extensively studied by both genetics and biochemistry (4). Although not clinically useful itself, knowledge of Car biosynthesis has been of great help as a model system for thienamycin biosynthesis, as they share precursors, glutamate (pyrrolidine ring) and acetate (β-lactam carbons), as well as core biosynthetic enzymes for bicyclic ring assembly. However, the pathways to the C-2,C-6-substituted carbapenems as thienamycin involve multiple enzymes. The essential steps in the biosynthesis of Car in P. carotovorum are catalyzed directly by three enzymes, CarABC (18, 23). Sequencing analysis of the thn gene cluster revealed the close relationship of ThnE and ThnM to CarB and CarA, respectively (25). The CarB and ThnE carboxymethylproline synthases are members of the crotonase superfamily, which catalyze pyrroline ring formation by the conversion of glutamate semialdehyde (l-GSA) into trans-carboxymethylproline using malonyl-CoA as a cosubstrate. There is evidence supporting that ThnE catalyzes the formation of (2S,5S)-trans-carboxymethylproline from precursors with an efficiency similar to that of CarB (13). The β-lactam ring was proposed to be formed by the β-lactam synthetase ThnM in a way similar to that of CarA, generating a carbapenam bicyclic ring (25). The oxidation of carbapenam to carbapenem, and ring inversions, must take place at some later point, as occurs in Car by the action of the 2-oxoglutarate-dependent dioxygenase CarC. Despite the lack of a convincing CarC homologue in the thn gene cluster, it was proposed that two members of the same family, ThnG and ThnQ, might catalyze analogous reactions. These enzymes have been proposed to be responsible for the coupled C-5 epimerization and C-2/C-3 desaturation of (2S,5S)-carbapenam to (5R)-carbapenem-3-carboxylate (13, 30). Recent studies of the genetics of thienamycin regulation in S. cattleya have revealed that the thn genes display a different expression pattern (27, 28). Interestingly, genes encoding the carbapenem ring assembly enzymes (ThnE, ThnM, and ThnG/ThnQ) show a completely different organization than those of their homologues in P. carotovorum, as they are not adjacent and not transcriptionally coupled (Fig. 1). Furthermore, they are differentially regulated, since only thnM and thnQ are ThnI dependent, whereas thnE and thnG display a ThnI-independent regulation pattern.
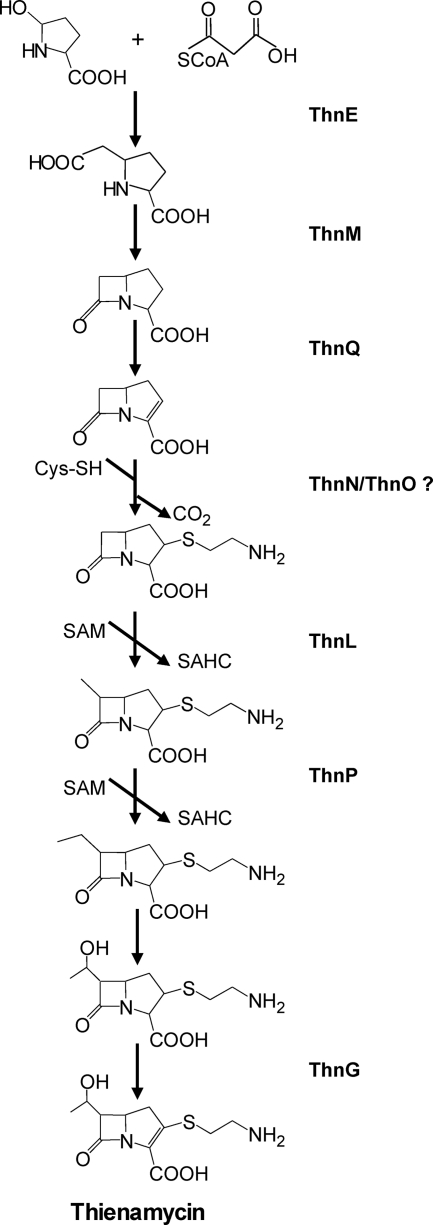
Based on the results provided here for the characterization of several thn mutants together with latest findings in the literature for this area, we propose some enzymatic steps in the thienamycin biosynthetic pathway (Fig. 8). As shown by HPLC-MS analysis, a compound with m/z 156 ([M + H]+), which might correspond to carbapenam-3-carboxilic acid, has been detected in some of the mutants (thnL, thnP, and thnN mutants) as well as in the S. cattleya wild-type strain. Although further investigations will be needed to address this question, these results suggest that carbapenam-3-carboxilic acid might be an intermediate in the biosynthetic pathway. Thus, bicyclic nucleus formation would occur in the same order as that for Car assembly, being the β-lactam ring formed immediately after the five-member ring. This contrasts with the initially proposed pathway, in which a methylation reaction at C-6 would take place, immediately after the five-member ring is formed and before the generation of the β-lactam one (25, 35). However, this result is in agreement with a proposed alternative pathway in which ThnE and ThnM catalyze sequential steps, like CarB and CarA in P. carotovorum (30). The absence of this compound in the thnI deletion mutant reinforces this hypothesis, since it expresses thnE and not thnM. In fact, this regulatory mutant behaves as a structural mutant blocked in the enzymatic step that follows the ThnE reaction.
FIG. 8.
Thienamycin biosynthetic pathway in S. cattleya and assignment of functions to different genes.
On the other hand, if carbapenam-3-carboxylic acid is an intermediate in thienamycin assembly, the incorporation of the first methyl group at C-6 to generate the hydroxyethyl side chain of thienamycin would probably occur after ThnM synthetizes the β-lactam ring. The accumulation of a compound with an m/z corresponding to carbapenam-3-carboxilic acid in the thnL- and thnP-disrupted mutants is in agreement with this hypothesis, although it should be pointed out that another putative methyltransferase, ThnK, might contribute to this process (25). The results of cosynthesis analysis of thienamycin-nonproducing mutants have provided an indication of the possible order of the ThnL and ThnP enzymatic reactions in the pathway: since the thnP mutant restores thienamycin production in the thnL mutant when growing in close proximity, the thnP mutant appears to be blocked later in the pathway.
The phenotype of the thnG mutant is quite remarkable and difficult to understand, as besides accumulating a putative intermediate in the pathway, the mutant also overproduces thienamycin. First, the detection of a compound with m/z 275 ([M + H]+), corresponding to 2,3-dihydrothienamycin, in this mutant suggests that ThnG might be responsible for the oxidation reaction that converts the carbapenam to the carbapenem ring (Fig. 8). This is in agreement with one of the two possibilities proposed previously for the last step in the thienamycin pathway (25, 35). Second, the thienamycin production in this mutant might be explained by the similarity between ThnG and ThnQ, as they share 30% amino acid identity. As mentioned above, the encoding genes display a different regulation pattern, and the thnQ monocistronic transcripts were found to be five times more abundant than the monocistronic transcripts for thnG (27). The function of both enzymes in the thienamycin biosynthetic pathway has been the subject of multiple hypotheses (2, 13, 25, 30). A prominent feature of the role of the 2-oxoglutarate-dependent oxygenases in β-lactam biosynthesis pathways is their ability to catalyze more than one reaction (15). One of the proposed roles of ThnG/ThnQ is to catalyze the C-2/C-3 desaturation of the carbapenam nucleus to carbapenem (13, 30), which takes place in two independent enzymatic steps (35). In agreement with this, we can speculate that the restoration of thienamycin production in the thnG mutant might be the result of a partial complementation by ThnQ, resulting in 2,3-dihydrothienamycin accumulation in the mutant. This role for ThnQ indirectly suggests that the early oxidation step converting the carbapenam ring to carbapenem might be carried out by ThnQ earlier on in the pathway. In addition, ThnG was recently found to give rise to oxidative carbapenem diversity being able to hydroxylate the C-6 carbapenem moiety in vitro (2). Third, the 2.5-fold increase in thienamycin production observed for the thnG deletion mutant, in comparison with the S. cattleya wild-type strain, might be explained by a putative lack of expression of the downstream thnF gene as a consequence of the mutation. ThnF is an acetyltransferase proposed previously to give rise to the S. cattleya cometabolite N-acetylthienamycin through the acetylation of the reactive primary amine in thienamycin (12). Thus, the lack of N-acetylthienamycin formation might explain the thienamycin accumulation in this mutant in relation to the wild-type strain.
Another controversial point in thienamycin biosynthesis is the incorporation of the cysteaminyl side chain at C-2. It has been long assumed that cysteine is the source of cysteamine and is incorporated directly into the antibiotic (35). The authors of that study also suggested an alternative branch pathway leading to thienamycin after the incorporation of pantetheine (through the formation of OA-6129 carbapenems). Some recent reports on the enzymatic activities of recombinant ThnT, ThnR, and ThnH have shown that they process in vitro CoA through pantetheine to yield cysteamine (12). Gene expression analyses have shown that the genes encoding these proteins are differentially regulated in S. cattleya, as only thnH is ThnI dependent, while thnT and thnR are ThnI independent (27). As deduced from the phenotypic analysis of the respective mutants of thnT and thnR, we provide evidence here supporting that under our standard culture conditions, ThnT and ThnR are not essential for thienamycin biosynthesis. Recombinant ThnT, shown previously in vitro to hydrolyze pantetheine to cysteamine (12), appears to be required for cephamycin biosynthesis, as deduced from the HPLC-MS analysis of the thnT deletion mutant. ThnR, reported previously to cleave CoA to 4-phosphopantetheine (12), is a member of the Nudix hydrolase superfamily whose general role is to hydrolyze nucleotide pools and to maintain cell viability, serving as surveillance and “house-cleaning” enzymes. In this regard, thnR might represent the physical boundary of this side of the thienamycin gene cluster. Mutational analyses of two other genes in the thn cluster, thnN and thnO, have revealed that they are absolutely required for thienamycin biosynthesis (25), although their role is still unknown. Based on its similarity to GriC-GriD, a carboxylic acid reductase complex essential for grixazone biosynthesis in Streptomyces griseus, it was proposed previously that ThnN-ThnO may be responsible for the reduction of a carboxylic acid to the corresponding aldehyde in the thienamycin pathway (33). A decarboxylation step occurs during the generation of the cysteaminyl side chain after cysteine incorporation in thienamycin biosynthesis (25, 35). Cross-feeding relationships suggest that ThnN catalyzes an earlier step than ThnO and that both ThnN and ThnO carry out reactions previous to those of the ThnL and ThnP putative methyltransferases. In addition, the presence of the putative carbapenam-3-carboxilic acid in the thnN-disrupted mutant suggests that ThnN acts after the bicyclic ring is formed (although its absence in the thnO mutant is not in agreement with the cross-feeding result). Conserved domain analysis of ThnN has shown the presence of an amino acid adenylation domain (TIGR01733), which can activate amino acids in nonribosomal peptide synthetases. These enzymes are involved in the peptide biosynthesis of antibiotics and other pharmacological molecules of microbial origin (17). ThnO presents a conserved domain belonging to the NAD(P)-dependent aldehyde dehydrogenase superfamily (cl11961). Although further investigations will be needed to address this question, and it is still a matter of speculation, the involvement of ThnN-ThnO in generating the thienamycin cysteaminyl side chain from cysteine might be a possibility.
Acknowledgments
Part of this research was supported by grants of the Plan Regional de Investigación del Principado de Asturias (grants GE-MEDO1-05 and FC-02-PC-REC01-20).
We are very grateful to Luis A. García for his continuous support and fruitful discussions. We also thank Leire Peña for technical assistance.
Footnotes
Published ahead of print on 24 January 2011.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipmann. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bodner, J. J., R. M. Phelan, M. F. Freeman, R. Li, and C. A. Townsend. 2010. Non-heme iron oxygenases generate natural structural diversity in carbapenem antibiotics. J. Am. Chem. Soc. 132:12-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchan, T., et al. 1994. Mutants of Streptomyces cattleya defective in the synthesis of a factor required for thienamycin production. J. Antibiot. 47:992-1000. [DOI] [PubMed] [Google Scholar]
- 4.Coulthurst, S. J., A. M. L. Barnard, and G. P. C. Salmond. 2005. Regulation and biosynthesis of carbapenem antibiotics in bacteria. Nat. Rev. Microbiol. 3:295-306. [DOI] [PubMed] [Google Scholar]
- 5.Cox, A. R., et al. 1998. A pheromone-independent CarR protein controls carbapenem antibiotic synthesis in the opportunistic human pathogen Serratia marcescens. Microbiology 144:201-209. [DOI] [PubMed] [Google Scholar]
- 6.Demain, A. L. 2009. Antibiotics: natural products essential to human health. Med. Res. Rev. 29:821-842. [DOI] [PubMed] [Google Scholar]
- 7.Derzelle, S., E. Duchaud, F. Kunst, A. Danchin, and P. Bertin. 2002. Identification, characterization, and regulation of a cluster of genes involved in carbapenem biosynthesis in Photorhabdus luminescens. Appl. Environ. Microbiol. 68:3780-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ducho, C., et al. 2009. Synthesis of regio- and stereoselectively deuterium-labelled derivatives of L-glutamate semialdehyde for studies on carbapenem biosynthesis. Org. Biomol. Chem. 7:2770-2779. [DOI] [PubMed] [Google Scholar]
- 9.Elander, R. P. 2003. Industrial production of beta-lactam antibiotics. Appl. Microbiol. Biotechnol. 61:385-392. [DOI] [PubMed] [Google Scholar]
- 10.Fernández Lozano, M. J., et al. 2000. Characterization of two polyketide methyltransferases involved in the biosynthesis of the antitumor drug mithramycin by Streptomyces argillaceus. J. Biol. Chem. 275:3065-3074. [DOI] [PubMed] [Google Scholar]
- 11.Flett, F., V. Mersinias, and C. P. Smith. 1997. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol. Lett. 155:223-229. [DOI] [PubMed] [Google Scholar]
- 12.Freeman, M. F., K. A. Moshos, M. J. Bodner, R. Li, and C. A. Townsed. 2008. Four enzymes define the incorporation of coenzyme A in thienamycin biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 105:11128-11133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamed, R. B., et al. 2009. Evidence that thienamycin biosynthesis proceeds via C-5 epimerization: ThnE catalyzes the formation of (2S,5S)-trans-carboxymethylproline. Chembiochem 10:246-250. [DOI] [PubMed] [Google Scholar]
- 14.Kahan, J. S., et al. 1979. Thienamycin, a new β-lactam antibiotic. I. Discovery, taxonomy, isolation and physical properties. J. Antibiot. 23:1255-1265. [DOI] [PubMed] [Google Scholar]
- 15.Kershaw, N. J., M. E. C. Caines, M. C. Sleeman, and C. J. Schofield. 2005. The enzymology of clavam and carbapenem biosynthesis. Chem. Commun. (Camb.) 34:4251-4263. [DOI] [PubMed] [Google Scholar]
- 16.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, United Kingdom.
- 17.Konz, D., and M. A. Marahiel. 1999. How do peptide synthetases generate structural diversity? Chem. Biol. 6:R34-R48. [DOI] [PubMed] [Google Scholar]
- 18.Li, R., A. Stapon, J. T. Blanchfield, and C. A. Towsend. 2000. Three unusual reactions mediate carbapenem and carbapenam biosynthesis. J. Am. Chem. Soc. 122:9296-9297. [Google Scholar]
- 19.Liras, P., and J. F. Martín. 2006. Gene cluster for beta-lactam antibiotics and control of their expression: why have clusters evolved, and from where did they originate? Int. Microbiol. 9:9-19. [PubMed] [Google Scholar]
- 20.MacNeil, D. J., et al. 1992. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111:61-68. [DOI] [PubMed] [Google Scholar]
- 21.Mazodier, P., R. Petter, and C. J. Thompson. 1989. Intergeneric conjugation between Escherichia coli and Streptomyces species. J. Bacteriol. 171:3583-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGowan, S. J., et al. 1996. Analysis of bacterial carbapenem antibiotic production genes reveals a novel β-lactam biosynthesis pathway. Mol. Microbiol. 22:415-426. [DOI] [PubMed] [Google Scholar]
- 23.McGowan, S. J., et al. 1997. Analysis of the carbapenem gene cluster of Erwinia carotovora: definition of the antibiotic biosynthetic genes and evidence for a novel beta-lactam resistance mechanism. Mol. Microbiol. 26:545-556. [DOI] [PubMed] [Google Scholar]
- 24.Nicolau, D. P. 2008. Carbapenems: a potent class of antibiotics. Expert Opin. Invest. Drugs 9:23-37. [DOI] [PubMed] [Google Scholar]
- 25.Nuñez, L. E., C. Méndez, A. F. Braña, G. Blanco, and J. A. Salas. 2003. The biosynthetic gene cluster for the β-lactam carbapenem thienamycin in Streptomyces cattleya. Chem. Biol. 10:301-311. [DOI] [PubMed] [Google Scholar]
- 26.Puk, O., et al. 2004. Biosynthesis of chloro-β-hydroxytyrosine, a nonproteinogenic amino acid of the peptidic backbone of glycopeptide antibiotics. J. Bacteriol. 186:6093-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodríguez, M., et al. 2008. Identification of transcriptional activators for thienamycin and cephamycin C biosynthetic genes within the thienamycin gene cluster from Streptomyces cattleya. Mol. Microbiol. 69:633-645. [DOI] [PubMed] [Google Scholar]
- 28.Rodríguez, M., C. Méndez, J. A. Salas, and G. Blanco. 2010. Transcriptional organization of ThnI-regulated thienamycin biosynthetic genes in Streptomyces cattleya. J. Antibiot. 63:135-138. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Sleeman, M. C., and C. J. Schofield. 2004. Carboxymethylproline synthase (CarB), an unusual carbon-carbon bond-forming enzyme of the crotonase superfamily involved in carbapenem biosynthesis. J. Biol. Chem. 279:6730-6736. [DOI] [PubMed] [Google Scholar]
- 31.Stapon, A., R. F. Li, and C. A. Townsend. 2003. Synthesis of (3S,5R)-carbapenam-3-carboxylic acid and its role in carbapenem biosynthesis and the stereoinversion problem. J. Am. Chem. Soc. 125:15746-15747. [DOI] [PubMed] [Google Scholar]
- 32.Sun, Y., et al. 2002. ‘Streptomyces nanchangensis’, a producer of the insecticidal polyether antibiotic nanchangmycin and the antiparasitic macrolide meilingmycin, contains multiple polyketide gene clusters. Microbiology 148:361-371. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki, H., Y. Ohnishi, and S. Horinouchi. 2007. GriC and GriD constitute a carboxylic acid reductase involved in grixazone biosynthesis in Streptomyces griseus. J. Antibiot. 60:380-387. [DOI] [PubMed] [Google Scholar]
- 34.Vieira, J., and J. Messing. 1991. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene 100:189-194. [DOI] [PubMed] [Google Scholar]
- 35.Williamson, J. M., E. Inamine, K. E. Wilson, A. W. Douglas, J. M. Liesch, and G. Albers-Schönberg. 1985. Biosynthesis of the β-lactam antibiotic, thienamycin, by Streptomyces cattleya. J. Biol. Chem. 260:4637-4647. [PubMed] [Google Scholar]