Abstract
Microbicides based on nonnucleoside reverse transcriptase inhibitors (NNRTIs) are currently being developed to protect women from HIV acquisition through sexual contact. However, the large-scale introduction of these products raises two major concerns. First, when these microbicides are used by undiagnosed HIV-positive women, they could potentially select for viral resistance, which may compromise subsequent therapeutic options. Second, NNRTI-based microbicides that are inactive against NNRTI-resistant strains might promote the selective transmission of these viruses. In order to address these concerns, drug resistance was selected in vitro by the serial passage of three viral isolates from subtypes B and C and CRF02_AG (a circulating recombinant form) in activated peripheral blood mononuclear cells (PBMCs) under conditions of increasing concentrations of three NNRTIs (i.e., TMC120, UC781, and MIV-160) that are currently being developed as candidate microbicides. TMC120 and MIV-160 displayed a high genetic barrier to resistance development, whereas resistance to UC781 emerged rapidly, similarly to efavirenz and nevirapine. Phenotypically, the selected viruses appeared to be highly cross-resistant to current first-line therapeutic NNRTIs (i.e., delavirdine, nevirapine, and efavirenz), although they retained some susceptibility to the more recently developed NNRTIs lersivirine and etravirine. The ability of UC781, TMC120, and MIV-160 to inhibit the in vitro-selected NNRTI-resistant viruses was also limited, although residual activity could be observed for the candidate microbicide NNRTI MIV-170. Interestingly, only four p2/p7/p1/p6/PR/RT/INT recombinant NNRTI-resistant viruses (i.e., TMC120-resistant VI829, EFV-resistant VI829, MIV-160-resistant VI829, and EFV-resistant MP568) showed impairments in replicative fitness. Overall, these in vitro analyses demonstrate that due to potential cross-resistance, the large-scale introduction of single-NNRTI-based microbicides should be considered with caution.
As the number of people living with human immunodeficiency virus type 1 (HIV-1) continues to grow (73), prevention remains absolutely necessary to combat this pandemic. Most of the 3 million HIV infections each year occur in women who often have no control over condom use by their sexual partners (39, 42, 86). Vaginal microbicides that could empower women to protect themselves from sexual transmission are therefore urgently needed (68). However, the development of an effective and safe microbicide appears to be a challenging task. The first candidate microbicides, such as the detergent nonoxynol-9 (13, 40), the polyanion cellulose sulfate (70, 74), carrageenan (67), and PRO2000 (26), failed in clinical trials due to toxicity problems or a lack of efficacy. Therefore, new candidate microbicides based on more potent and specific anti-HIV agents that interfere with viral entry (binding or fusion) (76), reverse transcription, or integration steps of the HIV life cycle (6, 7) are currently being developed.
Reverse transcriptase (RT) inhibitors (RTIs) are the most advanced compounds used in candidate microbicides because they are very specific and potent and potentially have a long-term inhibitory effect (47). The FDA-approved nucleotide reverse transcriptase inhibitor (NtRTI) tenofovir (TFV) recently proved to be effective as a vaginal microbicide in a phase IIb clinical trial (35) and is currently being tested as preexposure prophylaxis in phase III clinical trials (1). On the other hand, the nonnucleoside reverse transcriptase inhibitor (NNRTI) dapivirine (TMC120) has shown potent antiviral activity against both wild-type (wt) and NNRTI-resistant HIV-1 strains (23, 75). This diarylpyrimidine (DAPY) is currently being evaluated as a microbicide in phase I clinical trials and has a favorable safety profile when administered intravaginally (1, 44, 61, 80). Also in phase I clinical trials is the thiocarboxanilide UC781, which is known to have a very high therapeutic index (9). Although this NNRTI shows decreased activity against some NNRTI-resistant viruses (30), it was proven to be safe as a vaginal microbicide in the form of once-daily dosing for 6 days (64). Other NNRTIs such as the phenylethylthioureathiazoles (PETTs) MIV-160 and MIV-170 are still in preclinical development as candidate microbicides. However, they show high potency against wt and certain drug-resistant strains (21, 62).
Both NRTIs and NNRTIs also constitute the backbone of highly active antiretroviral therapy (HAART) (15), and therefore, their use in HIV prevention raises two major concerns. First, it is likely that these microbicides will be used by undiagnosed HIV-infected women since they should be available over-the-counter to guarantee maximum accessibility. As such, significant systemic absorption of RTI-based microbicides could lead to suboptimal drug pressure and, hence, resistance development that could compromise first-line antiretroviral therapy (39, 42). Second, it is not known whether RTI-based microbicides will protect against RTI-resistant strains, which, as a consequence of therapy, are already circulating within the HIV-infected population (66). In this case, the large-scale use of RTI-based microbicides could potentially promote the selective transmission of RTI-resistant viruses (39, 42), contributing to an already increasing public health problem in developing countries (73).
Although the likelihood and severity of these potential problems remain uncertain, they need to be addressed, particularly when developing NNRTI-based microbicides, since a relatively small number of common mutations usually confer resistance to this drug class. Therefore, in this study we analyzed the in vitro resistance or cross-resistance profiles of three viral isolates from subtypes B, C, and CRF02_AG that are resistant to three NNRTIs currently under development as potential microbicides, namely, TMC120, UC781, and MIV-160. Subsequently, their susceptibilities to 26 antiretroviral drugs, including five NNRTIs in development (i.e., TMC120, UC781, MIV-160, MIV-170, etravirine [ETR], and lersivirine [LSV]), were evaluated. Finally, the replicative fitness of p2/p7/p1/p6/PR/RT/INT recombinant NNRTI-resistant viruses was determined by using viral growth kinetics and growth competition experiments.
MATERIALS AND METHODS
Antiretroviral compounds.
The nucleoside reverse transcriptase inhibitor (NRTI) zidovudine (AZT) and the NNRTIs nevirapine (NVP) and efavirenz (EFV) were obtained through the NIH AIDS Research and Reference Reagent Program, Germantown, MD. The nucleotide analogue tenofovir (TFV) and the thiocarboxanilide NNRTI UC781 were kindly provided by the Rega Institute, Leuven, Belgium, whereas the NNRTIs dapivirine (TMC120), and delavirdine (DLV) were donated by Tibotec BVBA, Beerse, Belgium. The PETT NNRTIs MIV-160 and MIV-170 were kindly provided by Medivir AB, Sweden, and the NNRTI lersivirine (LSV) was synthesized by the Medicinal Chemistry of the University of Antwerp, Antwerp, Belgium. Compounds were dissolved in dimethyl sulfoxide (DMSO) or water and used at noncytotoxic concentrations as determined by the WST-1 tetrazolium cytotoxicity assay (Roche Diagnostic, Mannheim, Germany) as described previously (24). The antiretroviral drugs used in the Viralarts HIV assay, i.e., AZT, didanosine (ddI), stavudine (d4T), lamivudine (3TC), abacavir (ABC), TFV, emtricitabine (FTC), NVP, DLV, EFV, etravirine (ETR), saquinavir (SQV), ritonavir (RTV), indinavir (IDV), nelfinavir (NFV), amprenavir (APV), lopinavir (LPV), atazanavir (ATV), tipranavir (TPV), darunavir (DRV), and raltegravir (RAL), were obtained from ENZO Life Sciences International, Inc., Plymouth Meeting, PA (formerly BioMol International, LP).
Primary cells and cell lines.
Human peripheral blood mononuclear cells (PBMCs) were used to grow wild-type (wt) and drug-resistant virus stocks. PBMCs were isolated from buffy coats of healthy blood donors (kindly provided by the Antwerp Blood Transfusion Centre) through Ficoll density gradient centrifugation and stimulated for 48 h in RPMI 1640 medium containing 10% heat-inactivated fetal bovine serum (FBS) and 50 μg/ml gentamicin (Lonza, Verviers, Belgium) supplemented with 2 μg/ml phytohemagglutinin (PHA) (Remel, Kent, United Kingdom) and 2 μg/ml Polybrene (Sigma-Aldrich, Bornem, Belgium). Subsequently, cells were activated for 24 h in RPMI 1640 medium with 15% FBS and 50 μg/ml gentamicin supplemented with 1 ng/ml interleukin-2 (IL-2) (Gentaur, Brussels, Belgium), 2 μg/ml Polybrene, and 5 μg/ml hydrocortisone (Calbiochem, Leuven, Belgium). The JC53-BL cell line (85), also known as the TZM-bl cell line (NIH AIDS Research and Reference Reagent Program, Germantown, MD), was used for the evaluation of drug susceptibilities of wt and drug-resistant virus strains. TZM-bl cells were cultured in Dulbecco's minimum essential medium (DMEM) (Lonza) containing 10% heat-inactivated FBS and 50 μg gentamicin/ml at 37°C in a humidified 5% CO2-95% air environment. MT-4 cells were maintained in RPMI 1640-2 mM l-glutamine medium (Cellgro; Mediatech, Herndon, VA) supplemented with 10% FBS (Cellgro), 10 mM HEPES (Sigma), 100 U of penicillin/ml, and 100 μg of streptomycin/ml (Gibco BRL).
In vitro selection of NNRTI-resistant viruses.
Three different CCR5-tropic HIV-1 isolates were grown in activated PBMC cultures in the presence of increasing concentrations of three NNRTIs (i.e., TMC120, UC781, and MIV-160) currently being developed as candidate microbicides (also referred to as candidate microbicide NNRTIs) and two NNRTIs (i.e., NVP and EFV) commonly used in therapeutic regimens (also referred to as therapeutic NNRTIs) (Fig. 1 A). To this end, subtype B reference strain BaL and two primary clinical isolates, subtype C isolate VI829 and the CRF02_AG isolate MP568 (45, 71), were used. Viral cultures were initiated at a low multiplicity of infection (MOI) (MOIs of 0.001 for BaL and 0.01 for VI829 and MP568) in 10 million PHA-IL-2-stimulated PBMCs in the presence of the NNRTI at its 50% effective concentration (EC50). An identical culture without drug was started in parallel as a wt negative control (referred to as control virus). All cultures were incubated at 37°C in a humidified 5% CO2-95% air environment. IL-2 medium was replaced twice weekly, and viral replication was monitored every 3 to 4 days with an in-house p24 antigen capture enzyme-linked immunosorbent assay (ELISA) (8). At the end of each week, half of the cell culture was centrifuged (680 × g for 8 min) and resuspended in 10 ml IL-2 medium with 5 × 106 uninfected PHA-IL-2-stimulated PBMCs and exposed to a 5- or 2-fold-higher concentration of the NNRTI, only when the p24 level in the resistant culture was ≥50% of the p24 level in the control culture. Cultures with insufficient amounts of viral replication were maintained for another week at unchanged compound concentrations. Viral cultures were passaged for several weeks until the preferred resistance concentration was reached. The endpoint concentration for resistance selection against the microbicide NNRTIs was set to 1,000 nM (roughly 329 ng/g), based on TMC120 tissue concentrations measured previously in the macaque model and for humans (46, 61). The EFV resistance selection was also stopped at 1,000 nM, which was defined as a subtherapeutic concentration considering the expected EFV plasma levels of 1 to 4 mg/liter (18, 49, 56) in HIV-infected subjects. For NVP, the endpoint was set at 5,000 nM because NVP plasma levels range from 0.36 to 1.59 mg/liter 3 days after the receipt of a single-dose of 200 mg to prevent mother-to-child transmission (43). When the desired resistance was obtained, large stocks (30 ml) of cell-free virus were generated, aliquoted, and stored at −80°C. For each of these stocks, the tissue culture dose for 50% infectivity (TCID50) was determined. Briefly, six replicates of 10,000 TZM-bl cells (at 105 cells/ml in culture medium supplemented with 30 μg/ml DEAE-dextran) were incubated in a 96-well plate with serially diluted virus for 48 h at 37°C in 5% CO2. Subsequently, the luciferase activity was quantified through the addition of Steadylite HTS (Perkin-Elmer, Life Sciences, Zaventem, Belgium) and by using a TriStar LB941 luminometer (Berthold Technologies GmbH & Co. KG, Bad Wildbad, Germany). Viral titers were calculated according to the Reed and Muench method and are expressed as infectious units (IU) per milliliter (57).
FIG. 1.
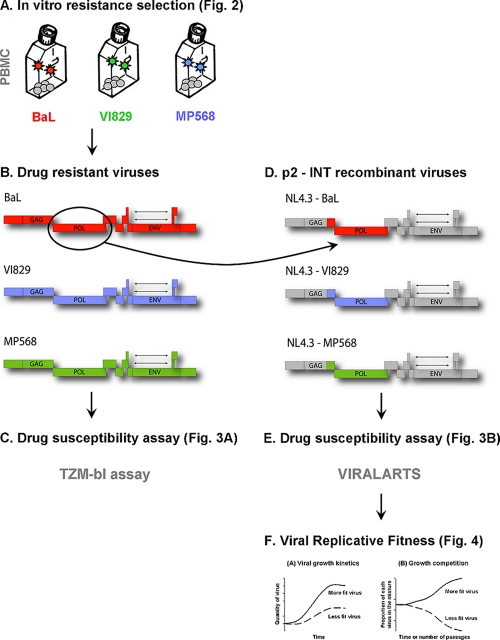
Experimental setup. (A and B) Viruses resistant to the candidate microbicide NNRTIs TMC120, MIV-160, and UC781 and the first-line therapeutic NNRTIs NVP and EFV were generated by the serial passage of reference strain BaL (subtype B) and the clinical isolates VI829 (subtype C) and MP568 (CRF02_AG) on mitogen-activated PBMCs with a gradual increase of drug pressure. (C) Subsequently, all 15 NNRTI-resistant viruses were tested in a TZM-bl assay for their susceptibility to four candidate microbicide RTIs and four therapeutic RTIs. (D) In order to confirm the data obtained with the TZM-bl assay and to correlate the phenotyping of the selected viruses to mutations within the pol gene, recombinant viruses were constructed by using p2/p7/p1/p6/PR/RT/INT genomic fragments as described in Materials and Methods. (E) The susceptibility of these p2-INT recombinant viruses to five candidate microbicide RTIs and 21 antiretroviral drugs was then tested. (F) Finally, the replicative fitness of the p2-INT recombinant NNRTI-resistant viruses was determined by using viral growth kinetics and growth competition experiments.
Genotyping of NNRTI-resistant viruses.
Mutations in the reverse transcriptase gene associated with resistance to RTI were determined by population sequencing (Fig. 1B). Viral RNA (vRNA) was extracted from culture supernatants by using the QIAamp viral RNA kit (Qiagen, Venlo, Netherlands) according to the manufacturer's protocol. Subsequently, vRNA was reverse transcribed into cDNA, and the RT region amplified by using the one-step Expand high-fidelity PCR system (Roche Applied Science, Lewes, United Kingdom). Primer SJH12a (5′-CCTAATGCATACTGTGAGTCTG-3′) was used for the reverse transcription of vRNA and in combination with SJH11A (5′-AAAAGGGCTGTTGGAAATGTGG-3′) for the amplification of a 2,042-bp Gag-Pol fragment (nucleotide positions 2,018 to 4,060 according the HxB2 reference sequence). Nested PCR was performed on 1 μl of the first-round product, using primers SJH13a (5′-GAGAGACAGGCTAATTTTTTAGGG-3′) and SJH14 (5′-CCTATTAGCTGCCCCATCTACATA-3′), as previously described (32, 33). Amplicons were purified by using Wizard SV gel and PCR Clean-Up Start-Up (Promega, Leiden, Netherlands). Sequencing was performed at the VIB Genetic Service Facility (Wilrijk, Belgium) using an Applied Biosystems 3730 DNA analyzer and ABI Prism BigDye terminator cycle sequencing reagents using primers SJH13a, H1P3339 (5′-TCCCATTCAGGAATCCAGGTGGC-3′), and H1P1697 (5′-AGAGCCAACAGCCCCACCAG-3′). The sequences were then assembled by using DNAsis software (Hitachi Software Engineering, Molecular Biology Insights, CO), translated and aligned with BioEdit Sequence Alignment Editor (Ibis Therapeutics, CA), and finally entered into the Stanford University Drug Resistance Database (version 6.0.8, last updated on 22 March 2010; http://hivdb.stanford.edu/index.htmL) to identify resistance-associated mutations (RAMs).
Drug susceptibility of NNRTI-resistant viruses using the TZM-bl assay.
The antiviral activities of four candidate microbicide RTIs (i.e., TMC120, MIV-160, UC781, and TFV) and four therapeutic RTIs (i.e., DLV, NVP, EFV, and AZT) was measured by preincubating 10,000 TZM-bl cells (at 105 cells/ml in culture medium supplemented with 30 μg/ml DEAE-dextran) in a 96-well plate for 30 min at 37°C in 5% CO2 in the presence or absence of serial dilutions of the respective compound (Fig. 1C). Subsequently, 200 TCID50 of the control or resistant HIV-1 strain BaL, VI829, or MP568 was added to each well, and cultures were incubated for 48 h before luciferase activity was quantified. Each condition was tested in duplicate or triplicate, and each experiment was independently repeated at least three times. Antiviral activity was expressed as the percentage of viral inhibition compared to that of the untreated controls and subsequently plotted against the compound concentration. Nonlinear regression analysis was used to calculate the EC50 based on at least three independent measurements and using GraphPad Prism software, version 5.03, for Windows (GraphPad Software, San Diego, CA). EC50 fold changes (FCs) were calculated relative to the EC50 determined for the control virus of each in vitro-selected NNRTI-resistant virus. Technical cutoff (TCO) values were used to assess significant fold changes due to the lack of a biological cutoff (BCO) or clinical cutoff (CCO) determined for these antiretroviral drugs (i.e., TMC120, MIV-160, UC781, TFV, DLV, NVP, EFV, and AZT) in this assay (TZM-bl). TCO values were defined for each RTI tested as the means and standard deviations (SD) of the EC50 values obtained for 15 wild-type viruses according to the following formula: TCO = 1 + 2 SD/mean.
Construction of 3′ Gag(p2/p7/p1/p6)/PR/RT/INT recombinant resistant viruses.
Thirty viruses passaged in vitro in the presence or absence of a compound were used to construct infectious chimeric viruses in an HIV-1 subtype B NL4-3 backbone as previously described (19, 82) (Fig. 1D). Briefly, a single fragment, p2-INT single (3,428 nucleotides [nt]), or two overlapping fragments, p2-half RT (1,657 nt) and half RT-INT (2,002 nt), were introduced via yeast-based homologous recombination into an NL4-3 acceptor vector containing a near-full-length HIV-1 genome with the yeast uracil biosynthesis (URA3) gene replacing the p2-INT HIV-1 coding sequence and expressing the human Renilla luciferase (hRluc) gene. The HIV-1 recombinant vector was then transfected into HEK293T cells. At 2 days posttransfection, chimeric HIV-1 viruses were harvested, and viral titers (TCID50) were determined by quantifying hRluc expression in MT-4 cells.
Drug susceptibility of p2-INT recombinant NNRTI-resistant viruses using the Viralarts HIV assay.
The drug susceptibilities of 30 p2-INT recombinant viruses were measured by determining the extents to which five candidate microbicide RTIs (i.e., MIV-160, MIV-170, UC781, TMC120, and TFV) and an additional panel of 21 antiretroviral drugs (i.e., LSV, AZT, ddI, d4T, 3TC, ABC, FTC, NVP, DLV, EFV, ETR, SQV, RTV, IDV, NFV, APV, LPV, ATV, TPV, DRV, and RAL) inhibited viral replication in MT-4 cells (82) (Fig. 1E). Briefly, serial dilutions of drugs were added in triplicate to a 96-well plate in RPMI medium with l-glutamine (Cellgro; Mediatech) supplemented with 10% fetal bovine serum, 100 U of penicillin/ml, 100 μg of streptomycin/ml (all reagents from Mediatech), and 10 mM HEPES (VWR, Radnor, PA). MT-4 cells were infected in bulk with a virus mixture of the reference virus expressing the firefly luciferase gene (HIV-1 NL4-3-fluc2) and the corresponding query virus tagged with hRluc, at a total multiplicity of infection of 0.01 IU/cell (both viruses at an equal ratio of 0.005 IU/cell) for 1 h at 37°C in 5% CO2. Subsequently, MT-4 cells were resuspended in RPMI medium, and 30,000 cells were added to each well with the preplated antiretroviral drugs. After 3 days of incubation at 37°C in 5% CO2, Dual-Glo luciferase reagent (Promega, Madison, WI) was added to the wells, and firefly luciferase activity was measured with a multiwell plate reader (Victor V multilabel reader; Perkin-Elmer, Waltham, MA). hRluc luminescence was measured after the addition of Dual-Glo Stop & Glo reagent. EC50 values were calculated and graphs were constructed by using nonlinear regression analysis (GraphPad Prism 5.03). Fold resistance was calculated based on the EC50 determined for the HIV-1 subtype B NL4-3-fluc2 reference virus used as an internal control for each dual infection. Although BCOs have been calculated for all 21 FDA-approved antiretroviral drugs using the Viralarts HIV assay, these cutoffs have yet to be determined for the four NNRTIs (i.e., MIV-160, MIV-170, UC781, and TMC120) that are being developed as candidate microbicides. Thus, we decided to use TCOs to be able to compare the results. As described above, TCO values were defined for each drug as the means and SD of the EC50 values obtained by analyzing 24 replicates of HIV-1 NL4-3 according to the following formula: TCO = 1 + 2 SD/mean.
Viral growth kinetics of p2-INT recombinant NNRTI-resistant viruses.
The replication of all 30 p2-INT viruses, plus the HIV-1 subtype B NL4-3 wt control, was measured by using growth kinetics as described previously (83) (Fig. 1F). Briefly, 3 × 106 MT-4 cells were infected at an MOI of 0.01 IU/cell in 1 ml of culture medium and incubated for 2 h at 37°C in 5% CO2. HIV-infected cells were washed twice with 1× phosphate-buffered saline (PBS) and split to be cultured in triplicate wells of a 24-well plate (106 cells/well). The culture supernatant was assayed by using a virus yield assay on days 0, 3, 4, 5, 6, 7, and 10 postinfection (p.i.) as described previously (53). Viral replication was quantified during the exponential growth phase (days 0 to 6) using the slope of the growth curves and by performing linear regression analysis derived from the equation log(y) = mt + log(h), where y is virus quantity (RT activity), m is the slope, t is time in days, and h is the intercept (day 0).
Growth competition of p2-INT recombinant NNRTI-resistant viruses.
Dual-infection/competition experiments were carried out as previously described (53, 84) (Fig. 1F). Briefly, in vitro-passaged and control (NL4-3) viruses tagged with hRluc and fluc2, respectively, were competed in a 1:1 initial proportion using an MOI of 0.01 IU/cell. One milliliter of the virus mixture was incubated with 6 × 105 MT-4 cells for 2 h at 37°C in 5% CO2 in the absence of drug pressure. Cells were subsequently washed three times with 1× PBS and cultured in triplicate in a 96-well plate (2 × 105 cells/well). At day 5 p.i., the final proportion of the two viruses in each competition mixture was quantified by using the Dual-Glo luciferase assay system (Promega, Madison, WI) in a multiwell plate reader (Victor V multilabel reader; Perkin-Elmer) after normalization to viral production in HIV-1 monoinfections as described previously (53, 81, 84). The replicative fitness for each virus was calculated and expressed as a percentage of the replicative fitness of the reference virus HIV-1 NL4-3.
RESULTS
In vitro selection of NNRTI-resistant viruses.
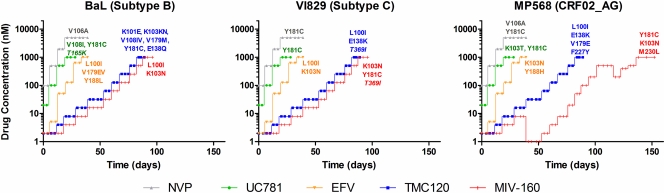
Viruses resistant to the candidate microbicide NNRTIs TMC120, MIV-160, and UC781 and the first-line therapeutic NNRTIs NVP and EFV were generated by the serial passage of reference strain BaL (subtype B) and the clinical isolates VI829 (subtype C) and MP568 (CRF02_AG) on mitogen-activated PBMCs with a gradual increase of drug pressure. In total, 15 NNRTI-resistant and as many control (passed in the absence of drug) viruses were selected (Fig. 2). Reduced susceptibility to the NNRTI UC781 and to the therapeutic NNRTIs EFV and NVP was detected in vitro within 5 weeks (Fig. 2). On the other hand, viruses that were fully resistant to the NNRTIs TMC120 and MIV-160 emerged only after 12 weeks. The CRF02_AG strain MP568 even required 20 weeks to develop resistance against MIV-160 (Fig. 2).
FIG. 2.
In vitro selection of NNRTI-resistant viruses. Reference strain BaL (subtype B) and two clinical isolates, VI829 (subtype C) and MP568 (CRF02_AG), were grown in the presence of increasing concentrations of three candidate microbicide NNRTIs (i.e., TMC120, UC781, and MIV-160) and two therapeutic NNRTIs (i.e., NVP and EFV) in activated PBMC cultures. Viral cultures were initiated at the 50% effective concentration (EC50) of the drug, while identical cultures without drug were started in parallel as a wild-type negative control. p24 concentrations were measured every 3 to 4 days by ELISA, and drug concentrations were raised only at the end of each week when the p24 level of the resistant culture was ≥50% of the p24 level of the control culture. Viral cultures were passaged for several days until the preferred resistance concentration of 1,000 nM (for TMC120, UC781, MIV-160, and EFV) or 5,000 nM (for NVP) was reached. Finally, resistance-associated mutations (RAMs) were determined by population sequencing of the viral Pol RNA after reverse transcription. Mutations that are not associated with therapeutic NNRTI resistance are shown in italics.
Genotyping of NNRTI-resistant viruses.
A series of amino acid substitutions (ranging from 1 to 6 substitutions for each individual virus) were identified in the RT gene of the NNRTI-resistant viruses (Fig. 2). Most of these mutations have been described as being resistance-associated mutations (RAMs) (http://hivdb.stanford.edu/). One of the most prevalent NNRTI RAMs, the Y181C mutation, was selected in all three UC781-resistant viruses. This confirms previous work which reported the emergence of the Y181C mutation during the in vitro selection of UC781 resistance together with other mutations such as the V108I mutation (10, 30). In agreement with these findings, the V108I mutation was also selected in UC781-resistant BaL. The K103T mutation, which emerged in the UC781-resistant MP568 virus, has not been associated with UC781 resistance, although it is known to be associated with intermediate- to high-level resistance to NVP, DLV, and EFV (59, 65).
All three MIV-160-resistant viruses harbored the common K103N mutation, which is known to cause major resistance to most NNRTIs (59, 65). In MIV-160-resistant BaL, the K103N mutation was accompanied by the L100I mutation, which can act synergistically with the K103N mutation, causing high-level resistance to the newer NNRTI ETR (TMC125) (78), while in MIV-160-resistant MP568 and MIV-160-resistant VI829, the K103N mutation occurred together with the Y181 mutation. MIV-160-resistant MP568 also contained the M230L mutation, which is known to emerge under ETR pressure in vitro and to cause intermediate- to high-level resistance to all NNRTIs (78, 87).
Interestingly, and unlike UC781 and MIV-160, TMC120 did not select for mutations common to all three isolates, although the subtype C strain VI829 and the CRF02_AG strain MP568 viruses contained the L100I and E138K mutations (Fig. 2). This supports previous work which associated the L100I mutation with resistance to the structurally related (48) DAPY compound ETR in non-B HIV-1 subtypes (41, 79), whereas the E138K mutation is commonly selected during ETR pressure in vitro (4, 69). Apart from the E138K and L100I mutations, other mutations were also selected under TMC120 pressure, namely, the E138Q, K101E, V108I, K103N, Y181C, V179M/E, and F227Y mutations (Fig. 2). These TMC120 resistance-associated mutations occur at exactly the same position as many of the mutations associated with ETR resistance, such as the E138K, E138A, F227C, K101E/H/P, Y181C/I/V, and V179D/F/T mutations (2, 41, 79).
Finally, the selection of resistance to EFV and NVP was associated with the presence of well-known RAMs that are commonly selected during NNRTI therapy. EFV selected for its key resistance mutations, Y188L/H and K103N, and two accessory mutations, V179E and L100I (Fig. 2) (60). As is the case for the EFV-resistant VI829 mutation, the L100I mutation usually occurs in combination with the K103N mutation (59, 65). NVP selected for two key mutations, V106A and Y181C, which are both known to increase NVP resistance by >50-fold (36, 60) and to emerge after single-dose NVP treatment to prevent mother-to-child transmission (46).
Through comparisons with the wt control virus, additional amino acid substitutions could be identified which are currently not associated with therapeutic NNRTI resistance, as determined by the Stanford University HIV Drug Resistance Database (http://hivdb.stanford.edu/), namely, the T369I (selected in TMC120-resistant VI829 and MIV-160-resistant VI829) and T165K (selected in UC781-resistant BaL) mutations (Fig. 2). However, the T369I mutation was previously identified as a novel mutation in the RT connection domain in patient-derived viruses with reduced NNRTI and AZT susceptibility (28) and in viruses selected in vitro against the novel NNRTI VRX-480773 (89). The T165K mutation, on the other hand, was previously reported to emerge in vitro during the selection of viruses resistant to the novel NNRTI MK-4965 (38).
Drug susceptibility of the NNRTI-resistant viruses determined by use of the TZM-bl assay.
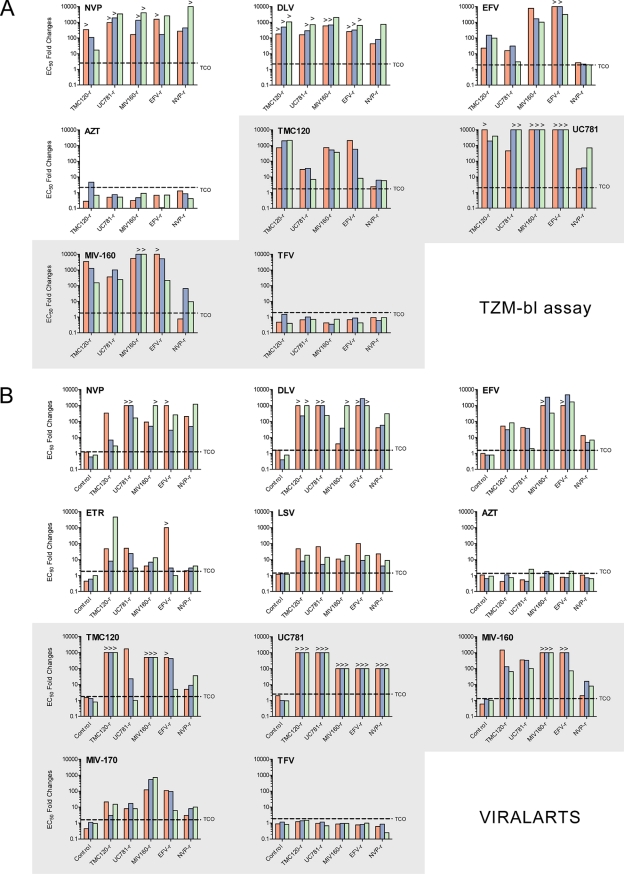
All 15 NNRTI-resistant viruses (subtype B, C, and CRF02_AG TMC120-resistant, UC781-resistant, MIV-160-resistant, EFV-resistant, and NVP-resistant strains) were tested in a TZM-bl assay for their susceptibility to four candidate microbicide RTIs (i.e., TMC120, UC781, MIV-160, and TFV) and four therapeutic RTIs (i.e., DLV, NVP, EFV, and AZT). Although at different levels, all NNRTI-resistant viruses showed a decrease in susceptibility to the different NNRTIs tested (i.e., TMC120, UC781, MIV-160, DLV, NVP, and EFV) while remaining fully sensitive to the NRTIs AZT and TFV (Fig. 3 A). The therapeutically used DLV and NVP lost activity against all resistant viruses, as reflected by the large EC50 fold changes (FCs) (>100-fold). EFV, however, retained some level of potency against TMC120-resistant BaL and all three UC781-resistant and NVP-resistant viruses, with an FC range of 2 to 31 (see the supplemental material). The ability of the microbicide NNRTIs TMC120, UC781, and MIV-160 to inhibit the in vitro-selected NNRTI-resistant viruses was also markedly reduced (Fig. 3A). Broad and severe cross-resistance against UC781 was observed (FC range of 32 to >100), while the other two candidates showed a similar pattern although with a more diverse range of EC50 fold changes (FC range of 0.77 to >100). For example, TMC120 retained some potency against the NVP-resistant viruses and the UC781-resistant viruses (FC ranges of 2.4 to 6.3 and 6.9 to 35, respectively), while MIV-160 displayed some residual activity against the NVP-resistant BaL and MP568 viruses (0.77- and 9.6-fold, respectively) (Fig. 3A). Moreover, in contrast with UC781, MIV-160 and TMC120 were still able to fully inhibit most (9/15 and 11/15 viruses, respectively) of the NNRTI-resistant viruses at submicromolar concentrations (see the supplemental material). Finally, a statistical analysis comparing the EC50 fold changes obtained for all the drugs did not reveal a significant difference between the three strains of different HIV-1 subtypes (data not shown).
FIG. 3.
Drug susceptibility of NNRTI-resistant viruses. (A) Fifteen viruses passaged in vitro in the presence and absence of TMC120, UC781, MIV160, EFV, and NVP were tested in a TZM-bl assay for their susceptibilities to four candidate microbicide RTIs (i.e., TMC120, UC781, MIV-160, and TFV) and four therapeutic RTIs (i.e., DLV, NVP, EFV, and AZT). Drug EC50s were determined by nonlinear regression analysis on data from at least three independent experiments. EC50 fold changes were calculated relative to the EC50 determined for the control virus of each in vitro-selected NNRTI-resistant virus. Technical cutoff (TCO) values were used to define the susceptibility or resistance of each virus to a given antiretroviral drug. TCOs were defined as the means and standard deviations (SD) of the EC50 values obtained for the 15 wild-type viruses according to the following formula: TCO = 1 + 2 SD/mean. (B) The same 15 NNRTI-resistant viruses described above (A) were used to construct p2-INT recombinant viruses containing the p2/p7/p1/p6/PR/RT/RT fragment in an HIV-1 subtype B NL4-3 backbone. Viral susceptibility to five candidate microbicide RTIs (i.e., TMC120, UC781, MIV-160, MIV-170, and TFV) and six therapeutic RTIs (i.e., EFV, NVP, DLV, ETR, LSV, and AZT) was subsequently tested with MT-4 cells using the Viralarts HIV antiviral resistance test system. EC50 fold changes were calculated relative to the value for the HIV-1 NL4-3 control virus. TCO values for each drug were defined as described above but using the means and SD of the EC50 values obtained by analyzing 24 replicates of HIV-1 NL4-3. EC50 fold changes are shown for TMC120-resistant (TMC120-r), UC781-resistant, MIV160-resistant, EFV-resistant, and NVP-resistant viruses. Viral strains BaL (subtype B), VI829 (subtype C), and MP568 (CRF02_AG) are represented by red, blue, and green, respectively. > symbols indicate EC50 fold changes that are larger than could be measured in the range of drug concentrations tested. Plots of microbicide NNRTIs are depicted with a gray background.
Drug susceptibility of p2-INT recombinant NNRTI-resistant viruses by using the Viralarts HIV assay.
In order to confirm the data obtained with the TZM-bl assay and to correlate the phenotyping of the selected viruses to mutations within the pol gene, recombinant viruses were constructed by using p2/p7/p1/p6/PR/RT/INT genomic fragments as described above. The replication of these p2-INT recombinant viruses was then tested against five candidate microbicide RTIs (i.e., TMC120, UC781, MIV-160, MIV-170, and TFV) and six therapeutic RTIs (i.e., DLV, NVP, EFV, ETR, LSV, and AZT). As expected, a statistically significant positive correlation was observed between the phenotypic data obtained with both drug susceptibility assays, i.e., TZM-bl and Viralarts HIV (Spearman rank [r] = 0.77; P < 0.0001). In general, all 15 p2-INT recombinant NNRTI-resistant viruses showed broad cross-resistance to the nine NNRTIs (i.e., EFV, NVP, DLV, ETR, TMC120, UC781, MIV-160, MIV170, and LSV) while remaining fully sensitive to the two NRTIs AZT and TFV (Fig. 3B). However, and similarly to results obtained with the TZM-bl assay, the therapeutic NNRTIs EFV and ETR and the NNRTIs TMC120, MIV-160, and MIV-170 retained some level of activity against the NVP-resistant viruses (FC ranges of 5 to 12, 1.5 to 3.6, 5 to 36, 1.6 to 16, and 3 to 10, respectively) (Fig. 3B). Some NNRTIs also remained active against particular viruses, for example, NVP and MIV-170 against TMC120-resistant VI829 (6.5- and 3-fold, respectively), NVP against TMC120-resistant MP568 (3.1-fold), EFV against UC781-resistant MP568 (2.5-fold), and DLV against MIV-160-resistant BaL (3.9-fold) (Fig. 3B). Interestingly, although UC781 was not able to inhibit the replication of any of the 15 p2-INT recombinant viruses carrying NNRTI resistance mutations (i.e., EC50 fold changes above the limit of detection), ETR, MIV-170, and LSV showed the best inhibitory profile, defined as the mean of the FCs of all 15 viruses for each NNRTI (i.e., 387-, 113-, and 24-fold, respectively, compared to a range of 479- to 828-fold for the other six NNRTIs). Finally, the susceptibilities of all 18 p2-INT recombinant (3 wt control and 15 NNRTI-resistant) viruses to nine protease inhibitors (i.e., SQV, RTV, IDV, NFV, APV, LPV, ATV, TPV, and DRV), five additional NRTIs (i.e., ddI, d4T, 3TC, ABC, and FTC), and one integrase strand transfer inhibitor (RAL) were assayed. All 18 viruses were susceptible to all 15 antiretroviral drugs (data not shown).
Replicative fitness of p2-INT recombinant NNRTI-resistant viruses.
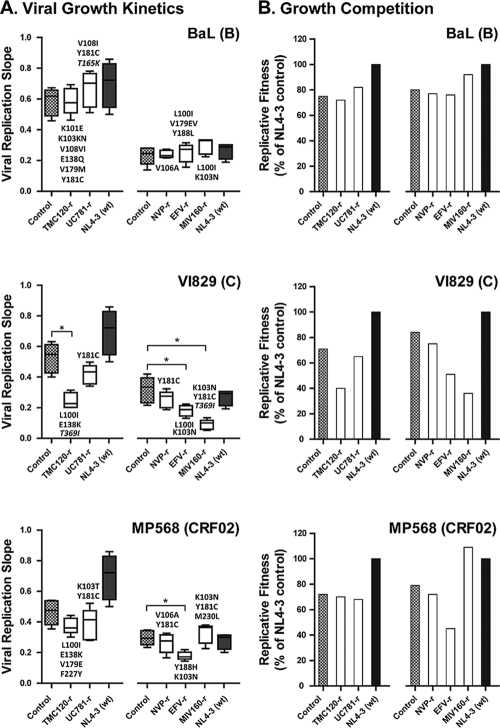
In general, mutations associated with drug resistance reduce the ability of the virus to replicate (viral fitness) (54, 55). Consequently, the presence of these impaired strains instead of wt viruses could be of some benefit to HIV-infected individuals (51, 52). Here, the replicative fitness of the NNRTI-resistant viruses was assayed by using two different but complementary approaches. A first glimpse of the replicative fitness was determined for the p2-INT recombinant viruses constructed from the 15 NNRTI-resistant viruses, 3 wt control strains, and HIV-1 subtype B strain NL4-3 using viral growth kinetics in MT-4 cells. Only four viruses, i.e., three subtype C strains (TMC120-resistant VI829, EFV-resistant VI829, and MIV-160-resistant VI829) and one CRF02_AG strain (EFV-resistant MP568) showed a statistically significant impairment in replicative fitness compared to their respective wt controls passaged in the absence of drug pressure (Fig. 4A). Interestingly, some of the amino acid substitutions observed for these four viruses, e.g., L100I plus E138K (TMC120-resistant VI829), L100I plus K103N (EFV-resistant VI829), K103N plus Y181C (MIV-160-resistant VI829), and K103N plus Y188H (EFV-resistant MP568), were also detected in other NNRTI-resistant viruses although not exactly in the same combination and/or genetic background (Fig. 4A). These results were corroborated using growth competition experiments in the absence of drug pressure. The replicative fitness values for the TMC120-resistant VI829, EFV-resistant VI829, MIV-160-resistant VI829, and EFV-resistant MP568 viruses, relative to that of the HIV-1 NL4-3 control, were 40%, 51%, 36%, and 45%, respectively (Fig. 4B), in contrast to a range in replicative fitness from 65% to 109% for the other NNRTI-resistant viruses.
FIG. 4.
Replicative fitness of p2-INT recombinant NNRTI-resistant viruses. (A) Viruses were propagated in MT-4 cells in the absence of drug pressure, and virus replication was measured by using luciferase expression, quantified as relative light units (RLU). Viral replication slopes were calculated by using the slopes between RLU values at days 0 and 3, 0 and 4, 0 and 5, and 0 and 6. All four slope values for each virus were used to calculate the means, standard deviations, and 10th and 90th percentiles, as indicated by the box plots. Differences in the mean values were evaluated by using a one-way analysis of variance test, and significant differences from HIV-1 subtype B NL4-3 were calculated by using Bonferroni's multiple-comparison test (GraphPad Prism). The replication kinetics of viruses marked with an asterisk are significantly different from their respective parental controls (P < 0.05; 95% confidence interval). Mutations that were not associated with therapeutic NNRTI resistance are shown in italics. (B) Each p2-INT recombinant virus was competed in a single experiment against the wt HIV-1 NL4-3 control in the absence of drug pressure, and their replicative fitnesses were calculated and expressed as a percentage of the replicative fitness of the reference virus NL4-3.
DISCUSSION
With the success of the CAPRISA 004 trial using TFV gel (35), RTI-based microbicides are back in the picture as a promising HIV prevention strategy. However, the large-scale introduction of these products raises two major concerns: first, when these microbicides are used by undiagnosed HIV-positive women, they could potentially select for viral resistance, which will compromise subsequent therapeutic options, and second, when NNRTI-based microbicides are inactive against resistant strains, they might promote the selective transmission of these viruses.
The first concern of developing microbicide resistance could emerge if the use of RTI-based microbicides by HIV-infected women leads to systemic drug absorption and subsequent viral resistance development. Thus far, only low-level systemic absorption has been observed for the three most advanced RTI-based microbicide candidates, namely, TFV (12, 39, 63), TMC120 (46, 61), and UC781 (64), although it remains unclear whether these low plasma levels are sufficient to select for resistance (7). However, even if the level of systemic absorption is low, local diffusion of the RTIs in the genital mucosa (47, 61, 63) could possibly result in the development of RTI-resistant genital reservoirs. During therapy, systemic drug exposure may select for resistant viruses from these reservoirs, thereby severely limiting the number of therapeutic options. Local resistance selection is especially a concern for NNRTI-based microbicides, as they are more likely to accumulate in the vaginal tissue due to their hydrophobic nature (14, 23, 46). In this study, we addressed this issue by simulating resistance selection in vitro against NNRTIs that are currently being developed as potential microbicides (Fig. 2). Three HIV-1 isolates were serially passaged in activated PBMC cultures in the presence of increasing concentrations of the microbicide NNRTIs UC781, TMC120, and MIV-160. Under increasing UC781 pressure, the viruses rapidly acquired one (Y181C) or two (V108I or K103T in addition) amino acid substitutions, which were sufficient to confer high-level resistance. This confirms previous work which has shown similar rates of resistance development (10) and clearly indicates that UC781 has a low genetic barrier to resistance (defined as the combination of the number of mutations and the time required to overcome drug-selective pressure). Moreover, the emergence of the Y181C mutation in all three UC781-resistant viruses is also in accordance with data from previously reported in vitro studies (10, 30), suggesting that this mutation is key for the development of UC781 resistance. In contrast to UC781, the selection of MIV-160 resistance was more difficult, resulting in a combination of at least two or three RAMs (L100I, K103N, Y181C, and/or M230L), suggesting a higher genetic barrier. This is consistent with studies of the related compound MIV-150, which also selected two to three mutations at a 3-fold-lower rate than that of EFV (14, 22, 29). Here MIV-160 resistance developed after 100 days of serial passage, compared to EFV resistance, which emerged after only 35 days. A similar rate was recently observed during in vitro selection experiments with MIV-160 (Bo Öberg, Medivir, personal communication). As the K103N mutation emerged in all three MIV-160-resistant viruses, this mutation might be crucial for the development of MIV-160 resistance. Finally, a series of mutations was observed for all three TMC120-resistant viruses (L100I, K101E, K103N, V108I, E138K/Q, V179M/E, Y181C, and/or F227Y). Of interest, an amino acid change at position 138 was present in each virus. Because mutations of the E138 residue have been associated with ETR (2, 79) and rilpivirine (TMC278) resistance (5), our data suggest an important role for amino acid position 138 in the development of resistance to TMC120 and other DAPY compounds. Furthermore, the delayed resistance selection and the high number of RAMs (two to six mutations) emerging under TMC120 pressure provide further evidence that TMC120 has a high genetic barrier to resistance development. The higher genetic barrier of MIV-160 and TMC120 could imply a lower risk of resistance selection during the potential use of these drugs as microbicides. However, their long-term use in sub-Saharan Africa might overcome this barrier, resulting in the emergence of microbicide resistance. To investigate the consequences of such a scenario, we assessed the susceptibilities of UC781-, MIV-160-, and TMC120-resistant viruses to FDA-approved and novel antiretroviral drugs, such as ETR and LSV (Fig. 3). As expected, the microbicide NNRTI-resistant viruses were susceptible to all protease inhibitors, NRTIs, and the integrase inhibitor raltegravir. On the other hand, the microbicide NNRTI-resistant viruses showed high-level resistance to all NNRTIs tested, which could limit the number of therapeutic options in sub-Saharan Africa, where NNRTIs are commonly used. Although newer NNRTIs such as LSV and ETR showed residual antiviral activity in the presence of NNRTI RAMs, it is possible that their use could also be compromised. For instance, the Y181C mutation, which provides the foundation for high-level ETR resistance (58), was selected under UC781 pressure. Furthermore, TMC120 selects for RAMs that are known to cause resistance to related drugs such as ETR and rilpivirine.
Besides the emergence of microbicide resistance, there is also a second concern that RTI-based microbicides could potentially promote the selective transmission of RTI-resistant viruses circulating within the infected population (66). Indeed, the suboptimal implementation of national HIV treatment programs has already been shown to result in high rates of resistance (77), and also, the single-dose administration of nevirapine, commonly used to prevent mother-to-child transmission, is known to select for resistance in the treated mothers (27, 34). If candidate microbicides show a decreased activity against these viruses while inhibiting the wild-type viruses, they might promote the selective transmission of these RTI-resistant strains. To address this issue, the activities of four NNRTIs (i.e., UC781, TMC120, MIV-160, and MIV-170) and one NRTI (i.e., TFV) that are currently being developed as candidate microbicides were evaluated against different NNRTI-resistant viruses (Fig. 3). Our results show that the potency of the NNRTIs was markedly reduced, while, as expected, the NRTI TFV retained its activity. The presence of key mutations associated with UC781 resistance (i.e., L100I, Y181C, or K103N) explains the failure of UC781 to protect against the tested NNRTI-resistant viruses (10). As the Y181C and K103N mutations also belong to the most prevalent mutations in vivo, this clearly raises concerns about the use of UC781 as a microbicide. The high therapeutic index (>62,000) reported for this compound was suggested to compensate for this reduced susceptibility (10). However, here we show that most of the tested viruses (9/15) could not be inhibited by UC781, even at concentrations of ≥50,000 nM. The activities of MIV-160 and TMC120, on the other hand, were also severely impacted, although they remained potent against the NVP-resistant viruses. More importantly, of all tested microbicide NNRTIs, MIV-170 displayed the highest level of residual activity against our set of NNRTI-resistant viruses, providing further support for the development of MIV-170 as a microbicide. Although the potency of the microbicide NNRTIs against NNRTI-resistant strains was very limited, the question remains regarding whether these resistant viruses are transmitted at a similar frequency as that of wild-type viruses. For instance, some NRTI-resistant strains have shown a lower rate of transmission than wild-type viruses, which was attributed to a decreased viral fitness as a consequence of NRTI RAMs (16, 72, 88). Conversely, many major NNRTI mutations are not expected to impact viral fitness (16, 72), which might pave the path for the selective transmission of NNRTI-resistant strains. We therefore assessed the replicative fitnesses of 15 NNRTI-resistant viruses using viral growth kinetics and growth competition experiments (Fig. 4). As determined by these two complementary assays, only four NNRTI-resistant viruses showed a statistically significant decrease in replicative fitness. These viruses harbored different combinations of mutations, i.e., the L100I, K103N, E138K, Y181C, and Y188H mutations, some of which were previously associated with a decreased viral replicative fitness (54, 55). Original studies showed that single mutations such as the K103N and Y181C mutations, despite conferring high-level resistance to NNRTIs, have limited effects on viral fitness (11, 17, 20, 25). However, contradictory results described a decrease (3) or an increase (31) in the replication capacity of viruses harboring the Y181C mutation, which could be due to differences in the methodologies used to quantify viral fitness (50). Similar results were reported for viruses carrying the K103N mutation (52). Viruses harboring the L100I mutation have been shown to be impaired compared to the wt control, while viruses with the L100I and K103N mutations had a greater reduction in fitness (37). Interestingly, in this study, two viruses (MIV160-resistant BaL and EFV-resistant VI829) shared the same two mutations but in different genomic contexts (subtypes B and C, respectively). However, only EFV-resistant VI829 showed an impairment in replicative fitness compared to those of its wt VI829 parental virus (P < 0.05) and the HIV-1 subtype B NL4-3 control (51%). Moreover, none of the NNRTI-resistant BaL viruses showed a reduction of their replication capacity. It is possible that the laboratory-adapted BaL strain has evolved to replicate well in vitro and that the p2-INT fragment introduced into the NL4-3 backbone allows for strong replication in MT-4 cells. On the other hand, the replication of 3 out of 5 NNRTI-resistant viruses based on a subtype C clinical isolate was impaired. Finally, scarce information is available about the role of the E138K and Y188H mutations in viral fitness. However, it is possible that these amino acid substitutions are selected to compensate for the reduction in fitness attributed to the L100I and K103N mutations in the TMC120-resistant VI829 and EFV-resistant MP568 viruses, respectively.
In summary, we have shown that broad cross-resistance among NNRTIs that are currently under development as microbicides warrants caution against the large-scale introduction of single-NNRTI-based microbicides. If these microbicides select for drug resistance in the female genital tract, major cross-resistance against first-line NNRTIs could be expected, which, as a consequence, may increase therapy failure in sub-Saharan Africa. However, our data suggest that although resistance to UC781 can rapidly emerge, a higher genetic barrier for resistance to microbicides based on TMC120 and MIV-160 is expected. Additional data on animal models are thus urgently needed to assess the likelihood of microbicide resistance upon long-term application. Finally, we have shown that the activity of UC781, TMC120, and MIV-160 to inhibit in vitro-selected NNRTI-resistant viruses was markedly reduced, which might promote the selective transmission of circulating NNRTI-resistant viruses. In analogy to HAART, the combination of different antiretroviral drugs in one microbicide might provide a solution to these problems, as this could not only diminish the possibility of drug resistance selection but also increase the residual activity of these microbicides against circulating resistant viruses.
Supplementary Material
Acknowledgments
We are grateful to Bo Öberg for his generous gift of reagents and for the proofreading of the manuscript. We also thank Paul Lewi for his assistance in processing the data and the Antwerp Red Cross Blood Transfusion Center for providing us with buffy coats. We thank the EUROPRISE Network of Excellence for their support.
Philippe Selhorst is a predoctoral fellow of the Research Foundation-Flanders (FWO), Flanders, Belgium, and the research leading to these results has received funding from the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 242135 (CHAARM) and from the FWO (grant G.0125.06). Diagnostic Hybrids, Inc., acknowledges the contribution of the State of Ohio, Department of Development and Third Frontier Commission, which provided funding in support of the Platform for Antiviral Resistance Testing and Vaccine Development project. This publication was prepared with financial support from the State of Ohio.
Footnotes
Published ahead of print on 31 January 2011.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Alliance for Microbicide Development. 2009. Microbicide and PreP candidates in ongoing clinical trials as of September 2009. Alliance for Microbicide Development, Silver Spring, MD. www.microbicide.org.
- 2.Andries, K., et al. 2004. TMC125, a novel next-generation nonnucleoside reverse transcriptase inhibitor active against nonnucleoside reverse transcriptase inhibitor-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 48:4680-4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archer, R. H., et al. 2000. Mutants of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase resistant to nonnucleoside reverse transcriptase inhibitors demonstrate altered rates of RNase H cleavage that correlate with HIV-1 replication fitness in cell culture. J. Virol. 74:8390-8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asahchop, E. L., et al. 2010. In vitro selection of novel etravirine-associated resistance mutations in B and non-B HIV-1 subtypes. Antivir. Ther. 15:A134. [Google Scholar]
- 5.Azijn, H., et al. 2010. TMC278, a next-generation nonnucleoside reverse transcriptase inhibitor (NNRTI), active against wild-type and NNRTI-resistant HIV-1. Antimicrob. Agents Chemother. 54:718-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balzarini, J., and L. Van Damme. 2005. Intravaginal and intrarectal microbicides to prevent HIV infection. CMAJ 172:461-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balzarini, J., and L. Van Damme. 2007. Microbicide drug candidates to prevent HIV infection. Lancet 369:787-797. [DOI] [PubMed] [Google Scholar]
- 8.Beirnaert, E., et al. 1998. Design and evaluation of an in-house HIV-1 (group M and O), SIVmnd and SIVcpz antigen capture assay. J. Virol. Methods 73:65-70. [DOI] [PubMed] [Google Scholar]
- 9.Buckheit, R. W., M. Hollingshead, S. Stinson, and J. P. Bader. 1997. Efficacy, pharmacokinetics and in vivo anti-HIV activity of UC781. Antiviral Res. 34:62. [DOI] [PubMed] [Google Scholar]
- 10.Buckheit, R. W., Jr., et al. 1997. Highly potent oxathiin carboxanilide derivatives with efficacy against nonnucleoside reverse transcriptase inhibitor-resistant human immunodeficiency virus isolates. Antimicrob. Agents Chemother. 41:831-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins, J. A., et al. 2004. Competitive fitness of nevirapine-resistant human immunodeficiency virus type 1 mutants. J. Virol. 78:603-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cranage, M., et al. 2008. Prevention of SIV rectal transmission and priming of T cell responses in macaques after local pre-exposure application of tenofovir gel. PLoS Med. 5:e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cutler, B., and J. Justman. 2008. Vaginal microbicides and the prevention of HIV transmission. Lancet Infect. Dis. 8:685-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Cruz, O. J., and F. M. Uckun. 2006. Dawn of non-nucleoside inhibitor-based anti-HIV microbicides. J. Antimicrob. Chemother. 57:411-423. [DOI] [PubMed] [Google Scholar]
- 15.de Bethune, M.-P. 2009. Non-nucleoside reverse transcriptase inhibitors (NNRTIs), their discovery, development, and use in the treatment of HIV-1 infection: a review of the last 20 years (1989-2009). Antiviral Res. 85:75-90. [DOI] [PubMed] [Google Scholar]
- 16.de Mendoza, C., et al. 2004. Evidence for differences in the sexual transmission efficiency of HIV strains with distinct drug resistance genotypes. Clin. Infect. Dis. 39:1231-1238. [DOI] [PubMed] [Google Scholar]
- 17.Devereux, H. L., V. C. Emery, M. A. Johnson, and C. Loveday. 2001. Replicative fitness in vivo of HIV-1 variants with multiple drug resistance-associated mutations. J. Med. Virol. 65:218-224. [DOI] [PubMed] [Google Scholar]
- 18.Djabarouti, S., et al. 2006. Intracellular and plasma efavirenz concentrations in HIV-infected patients switching from successful protease inhibitor-based highly active antiretroviral therapy (HAART) to efavirenz-based HAART (SUSTIPHAR Study). J. Antimicrob. Chemother. 58:1090-1093. [DOI] [PubMed] [Google Scholar]
- 19.Dudley, D. M., et al. 2009. A novel yeast-based recombination method to clone and propagate diverse HIV-1 isolates. Biotechniques 46:458-467. [DOI] [PubMed] [Google Scholar]
- 20.Dykes, C., et al. 2001. Impact of clinical reverse transcriptase sequences on the replication capacity of HIV-1 drug-resistant mutants. Virology 285:193-203. [DOI] [PubMed] [Google Scholar]
- 21.Elinder, M., et al. 2010. Inhibition of HIV-1 by non-nucleoside reverse transcriptase inhibitors via an induced fit mechanism—importance of slow dissociation and relaxation rates for antiviral efficacy. Biochem. Pharmacol. 80:1133-1140. [DOI] [PubMed] [Google Scholar]
- 22.Fernández-Romero, J. A., et al. 2007. Carrageenan/MIV-150 (PC-815), a combination microbicide. Sex. Transm. Dis. 34:9-14. [DOI] [PubMed] [Google Scholar]
- 23.Fletcher, P., et al. 2009. Inhibition of human immunodeficiency virus type 1 infection by the candidate microbicide dapivirine, a nonnucleoside reverse transcriptase inhibitor. Antimicrob. Agents Chemother. 53:487-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gali, Y., et al. 2010. Development of an in vitro dual-chamber model of the female genital tract as a screening tool for epithelial toxicity. J. Virol. Methods 165:186-197. [DOI] [PubMed] [Google Scholar]
- 25.Gatanaga, H., A. Hachiya, S. Kimura, and S. Oka. 2006. Mutations other than 103N in human immunodeficiency virus type 1 reverse transcriptase (RT) emerge from K103R polymorphism under non-nucleoside RT inhibitor pressure. Virology 344:354-362. [DOI] [PubMed] [Google Scholar]
- 26.Global Campaign for Microbicides. 2009. Microbicide trial results signal end of one chapter, focus turns to promising ARV-based candidates. Global Campaign for Microbicides, Washington, DC. http://www.global-campaign.org/MDP301.htm.
- 27.Guay, L. A., et al. 1999. Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-child transmission of HIV-1 in Kampala, Uganda: HIVNET 012 randomised trial. Lancet 354:795-802. [DOI] [PubMed] [Google Scholar]
- 28.Gupta, S., et al. 2010. Combinations of mutations in the connection domain of human immunodeficiency virus type 1 reverse transcriptase: assessing the impact on nucleoside and nonnucleoside reverse transcriptase inhibitor resistance. Antimicrob. Agents Chemother. 54:1973-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Högberg, M., et al. 1999. Urea-PETT compounds as a new class of HIV-1 reverse transcriptase inhibitors. 3. Synthesis and further structure-activity relationship studies of PETT analogues. J. Med. Chem. 43:304. [DOI] [PubMed] [Google Scholar]
- 30.Hossain, M. M., and M. A. Parniak. 2006. In vitro microbicidal activity of the nonnucleoside reverse transcriptase inhibitor (NNRTI) UC781 against NNRTI-resistant human immunodeficiency virus type 1. J. Virol. 80:4440-4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iglesias-Ussel, M. D., C. Casado, E. Yuste, I. Olivares, and C. Lopez-Galindez. 2002. In vitro analysis of human immunodeficiency virus type 1 resistance to nevirapine and fitness determination of resistant variants. J. Gen. Virol. 83:93-101. [DOI] [PubMed] [Google Scholar]
- 32.Jallow, S., et al. 2009. Virological response to highly active antiretroviral therapy in patients infected with human immunodeficiency virus type 2 (HIV-2) and in patients dually infected with HIV-1 and HIV-2 in the Gambia and emergence of drug-resistant variants. J. Clin. Microbiol. 47:2200-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jallow, S., et al. 2007. Development and evaluation of an oligonucleotide ligation assay for detection of drug resistance-associated mutations in the human immunodeficiency virus type 2 pol gene. J. Clin. Microbiol. 45:1565-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson, J. A., et al. 2005. Emergence of drug-resistant HIV-1 after intrapartum administration of single-dose nevirapine is substantially underestimated. J. Infect. Dis. 192:16-23. [DOI] [PubMed] [Google Scholar]
- 35.Karim, Q. A., et al. 2010. Effectiveness and safety of tenofovir gel, an antiretroviral microbicide, for the prevention of HIV infection in women. Science 329:1168-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kiptoo, M., et al. 2008. Prevalence of nevirapine-associated resistance mutations after single dose prophylactic treatment among antenatal clinic attendees in North Rift Kenya. AIDS Res. Hum. Retroviruses 24:1555-1559. [DOI] [PubMed] [Google Scholar]
- 37.Koval, C. E., C. Dykes, J. Wang, and L. M. Demeter. 2006. Relative replication fitness of efavirenz-resistant mutants of HIV-1: correlation with frequency during clinical therapy and evidence of compensation for the reduced fitness of K103N + L100I by the nucleoside resistance mutation L74V. Virology 353:184-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai, M.-T., et al. 2010. Distinct mutation pathways of non-subtype B HIV-1 during in vitro resistance selection with nonnucleoside reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 54:4812-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lederman, M. M., R. E. Offord, and O. Hartley. 2006. Microbicides and other topical strategies to prevent vaginal transmission of HIV. Nat. Rev. Immunol. 6:371-382. [DOI] [PubMed] [Google Scholar]
- 40.Madan, R. P., M. J. Keller, and B. C. Herold. 2006. Prioritizing prevention of HIV and sexually transmitted infections: first-generation vaginal microbicides. Curr. Opin. Infect. Dis. 19:49-54. [DOI] [PubMed] [Google Scholar]
- 41.Maiga, A. I., et al. 2009. Resistance-associated mutations to etravirine (TMC-125) in Antiretroviral-naive patients infected with non-B HIV-1 subtypes. Antimicrob. Agents Chemother. 54:728-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinez, J., P. Coplan, and M. A. Wainberg. 2006. Is HIV drug resistance a limiting factor in the development of anti-HIV NNRTI and NRTI-based vaginal microbicide strategies? Antiviral Res. 71:343-350. [DOI] [PubMed] [Google Scholar]
- 43.Muro, E. P., et al. 2005. Nevirapine plasma concentrations are still detectable after more than 2 weeks in the majority of women receiving single-dose nevirapine: implications for intervention studies. J. Acquir. Immune Defic. Syndr. 39:419-421. [DOI] [PubMed] [Google Scholar]
- 44.Nel, A., et al. 2009. Safety and pharmacokinetics of dapivirine delivery from matrix and reservoir intravaginal rings to HIV-negative women. J. Acquir. Immune Defic. Syndr. 51:416-423. [DOI] [PubMed] [Google Scholar]
- 45.Njai, H. F., et al. 2006. The predominance of human immunodeficiency virus type 1 (HIV-1) circulating recombinant form 02 (CRF02_AG) in West Central Africa may be related to its replicative fitness. Retrovirology 3:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nuttall, J. P., et al. 2008. Concentrations of dapivirine in the rhesus macaque and rabbit following once daily intravaginal administration of a gel formulation of [14C]dapivirine for 7 days. Antimicrob. Agents Chemother. 52:909-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patton, D. L., et al. 2007. Preclinical safety assessments of UC781 anti-human immunodeficiency virus topical microbicide formulations. Antimicrob. Agents Chemother. 51:1608-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prajapati, D. G., R. Ramajayam, M. R. Yadav, and R. Giridhar. 2009. The search for potent, small molecule NNRTIs: a review. Bioorg. Med. Chem. 17:5744-5762. [DOI] [PubMed] [Google Scholar]
- 49.Puthanakit, T., P. Tanpaiboon, L. Aurpibul, T. R. Cressey, and V. Sirisanthana. 2009. Plasma efavirenz concentrations and the association with CYP2B6-516G>T polymorphism in HIV-infected Thai children. Antivir. Ther. 14:315-320. [PubMed] [Google Scholar]
- 50.Quinones-Mateu, M. E., and E. J. Arts. 2002. Fitness of drug resistant HIV-1: methodology and clinical implications. Drug Resist. Updat. 5:224-233. [DOI] [PubMed] [Google Scholar]
- 51.Quinones-Mateu, M. E., and E. J. Arts. 2006. Virus fitness: concept, quantification, and application to HIV population dynamics. Curr. Top. Microbiol. Immunol. 299:83-140. [DOI] [PubMed] [Google Scholar]
- 52.Quiñones-Mateu, M. E., and E. J. Arts. 2001. HIV-1 fitness: implications for drug resistance, disease progression, and global epidemic evolution, p. 134-170. In C. Kuiken et al. (ed.), HIV sequence compendium. Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM.
- 53.Quinones-Mateu, M. E., et al. 2000. A dual infection/competition assay shows a correlation between ex vivo human immunodeficiency virus type 1 fitness and disease progression. J. Virol. 74:9222-9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quiñones-Mateu, M. E., D. M. Moore-Dudley, O. Jegede, and J. Weber. 2008. Viral drug resistance and fitness. Adv. Pharmacol. 56:257-296. [DOI] [PubMed] [Google Scholar]
- 55.Quiñones-Mateu, M. E., J. Weber, H. R. Rangel, and B. Chakraborty. 2001. HIV-1 fitness and antiretroviral drug resistance. AIDS Rev. 3:223-242. [Google Scholar]
- 56.Read, T. R. H., et al. 2009. Efavirenz plasma concentrations did not predict cessation of therapy due to neuropsychiatric symptoms in a large randomized trial. AIDS 23:2222-2223. [DOI] [PubMed] [Google Scholar]
- 57.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. (Lond.) 27:493-497. [Google Scholar]
- 58.Reuman, E. C., S.-Y. Rhee, S. P. Holmes, and R. W. Shafer. 2010. Constrained patterns of covariation and clustering of HIV-1 non-nucleoside reverse transcriptase inhibitor resistance mutations. J. Antimicrob. Chemother. 65:1477-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rhee, S.-Y., et al. 2003. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 31:298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rhee, S. Y., et al. 2006. Genotypic predictors of human immunodeficiency virus type 1 drug resistance. Proc. Natl. Acad. Sci. U. S. A. 103:17355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Romano, J., et al. 2009. Safety and availability of dapivirine (TMC120) delivered from an intravaginal ring. AIDS Res. Hum. Retroviruses 25:483-488. [DOI] [PubMed] [Google Scholar]
- 62.Sahlberg, C., and X.-X. Zhou. 2008. Development of non-nucleoside reverse transcriptase inhibitors for anti-HIV therapy. Antiinfect. Agents Med. Chem. 7:101-117. [Google Scholar]
- 63.Schwartz, J., et al. 2008. Preliminary results from a pharmacokinetic study of the candidate vaginal microbicide agent 1% tenofovir gel, abstr. BO11-610. Microbicides 2008 Conference, New Dehli, India.
- 64.Schwartz, J. L., et al. 2008. A randomized six-day safety study of an antiretroviral microbicide candidate UC781, a non-nucleoside reverse transcriptase inhibitor. Sex. Transm. Dis. 35:414-419. [DOI] [PubMed] [Google Scholar]
- 65.Shafer, R. W. 2006. Rationale and uses of a public HIV drug-resistance database. J. Infect. Dis. 194:S51-S58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shekelle, P., et al. 2007. Antiretroviral (ARV) drug resistance in the developing world. Evid. Rep. Technol. Assess. (Full Rep.) 2007:1-74. [PMC free article] [PubMed] [Google Scholar]
- 67.Skoler-Karpoff, S., et al. 2008. Efficacy of Carraguard for prevention of HIV infection in women in South Africa: a randomised, double-blind, placebo-controlled trial. Lancet 372:1977-1987. [DOI] [PubMed] [Google Scholar]
- 68.Smith, R. J., E. N. Bodine, D. P. Wilson, and S. M. Blower. 2005. Evaluating the potential impact of vaginal microbicides to reduce the risk of acquiring HIV in female sex workers. AIDS 19:413-421. [DOI] [PubMed] [Google Scholar]
- 69.Su, G., et al. 2007. In vitro selection and characterization of viruses resistant to R1206, a novel non-nucleoside reverse transcriptase inhibitor. Antivir. Ther. 12:S35. [Google Scholar]
- 70.Tao, W., C. Richards, and D. Hamer. 2008. Enhancement of HIV infection by cellulose sulfate. AIDS Res. Hum. Retroviruses 24:925-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Terrazas-Aranda, K., Y. Van Herrewege, P. J. Lewi, J. Van Roey, and G. Vanham. 2007. In vitro pre- and post-exposure prophylaxis using HIV inhibitors as microbicides against cell-free or cell-associated HIV-1 infection. Antivir. Chem. Chemother. 18:141-151. [DOI] [PubMed] [Google Scholar]
- 72.Turner, D. M. D., et al. 2004. Diminished representation of HIV-1 variants containing select drug resistance-conferring mutations in primary HIV-1 infection. J. Acquir. Immune Defic. Syndr. 37:1627-1631. [DOI] [PubMed] [Google Scholar]
- 73.UNAIDS. 2009. AIDS epidemic update: November 2009. UNAIDS/09.36E/JC1700E. UNAIDS, Geneva, Switzerland.
- 74.Van Damme, L., et al. 2008. Lack of effectiveness of cellulose sulfate gel for the prevention of vaginal HIV transmission. N. Engl. J. Med. 359:463-472. [DOI] [PubMed] [Google Scholar]
- 75.Van Herrewege, Y., et al. 2004. In vitro evaluation of nonnucleoside reverse transcriptase inhibitors UC-781 and TMC120-R147681 as human immunodeficiency virus microbicides. Antimicrob. Agents Chemother. 48:337-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Veazey, R. S., et al. 2005. Protection of macaques from vaginal SHIV challenge by vaginally delivered inhibitors of virus-cell fusion. Nature 438:99-102. [DOI] [PubMed] [Google Scholar]
- 77.Vergne, L., et al. 2002. Resistance to antiretroviral treatment in Gabon: need for implementation of guidelines on antiretroviral therapy use and HIV-1 drug resistance monitoring in developing countries. J. Acquir. Immune Defic. Syndr. 29:165-168. [DOI] [PubMed] [Google Scholar]
- 78.Vingerhoets, J., et al. 2005. TMC125 displays a high genetic barrier to the development of resistance: evidence from in vitro selection experiments. J. Virol. 79:12773-12782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vingerhoets, J., et al. 2010. Resistance profile of etravirine: combined analysis of baseline genotypic and phenotypic data from the randomized, controlled phase III clinical studies. AIDS 24:503. [DOI] [PubMed] [Google Scholar]
- 80.Vyankandondera, J., et al. 2009. Safety, tolerability, and systemic absorption of dapivirine vaginal microbicide gel in healthy, HIV-negative women. AIDS 23:1531-1538. [DOI] [PubMed] [Google Scholar]
- 81.Weber, J., et al. 2003. A novel TaqMan real-time PCR assay to estimate ex vivo human immunodeficiency virus type 1 fitness in the era of multi-target (pol and env) antiretroviral therapy. J. Gen. Virol. 84:2217-2228. [DOI] [PubMed] [Google Scholar]
- 82.Weber, J., et al. 2010. Resistance analysis to maturation, protease, reverse transcriptase and integrase inhibitors based on 3′Gag(p2/p7/p1/p6)/PR/RT/INT-recombinant viruses: a useful tool in the multi-target era of antiretroviral drugs. Antivir. Ther. 14:A127. [Google Scholar]
- 83.Weber, J., et al. 2010. Resistance mutations in protease, reverse transcriptase, and integrase genes: do they have an epistatic effect on drug susceptibility and/or HIV-1 replicative fitness? Antivir. Ther. 15:A94. [Google Scholar]
- 84.Weber, J., et al. 2006. Use of a novel assay based on intact recombinant viruses expressing green (EGFP) or red (DsRed2) fluorescent proteins to examine the contribution of pol and env genes to overall HIV-1 replicative fitness. J. Virol. Methods 136:102-117. [DOI] [PubMed] [Google Scholar]
- 85.Wei, X., et al. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 86.Wilson, D. P., P. M. Coplan, M. A. Wainberg, and S. M. Blower. 2008. The paradoxical effects of using antiretroviral-based microbicides to control HIV epidemics. Proc. Natl. Acad. Sci. U. S. A. 105:9835-9840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xu, H.-T., et al. 2010. The M230L nonnucleoside reverse transcriptase inhibitor resistance mutation in HIV-1 reverse transcriptase impairs enzymatic function and viral replicative capacity. Antimicrob. Agents Chemother. 54:2401-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yerly, S., et al. 2004. Infrequent transmission of HIV-1 drug-resistant variants. Antivir. Ther. 9:375-384. [PubMed] [Google Scholar]
- 89.Zhang, Z., et al. 2007. A novel nonnucleoside analogue that inhibits human immunodeficiency virus type 1 isolates resistant to current nonnucleoside reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 51:429-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.