Abstract
We identified GRL-1388 and -1398, potent nonpeptidic human immunodeficiency virus type 1 (HIV-1) protease inhibitors (PIs) containing a bicyclic P2 functional moiety, tetrahydropyrano-tetrahydrofuran (Tp-THF). GRL-1388 was as potent as darunavir (DRV) against various drug-resistant HIV-1 laboratory strains with 50% effective concentration (EC50s) of 2.6 to 32.6 nM. GRL-1398 was significantly more potent against such variants than DRV with EC50s of 0.1 to 5.7 nM. GRL-1388 and -1398 were also potent against multiple-PI-resistant clinical HIV-1 variants (CLHIV-1MDR) with EC50s ranging from 2.7 to 21.3 nM and from 0.3 to 4.8 nM, respectively. A highly DRV-resistant HIV-1 variant selected in vitro remained susceptible to GRL-1398 with the EC50 of 21.9 nM, while the EC50 of DRV was 214.1 nM. When HIV-1NL4-3 was selected with GRL-1398, four amino acid substitutions—leucine to phenylalanine at a position 10 (L10F), A28S, L33F, and M46I—emerged, ultimately enabling the virus to replicate in the presence of >1.0 μM the compound beyond 57 weeks of selection. When a mixture of 10 different CLHIV-1MDR strains was selected, the emergence of resistant variants was more substantially delayed with GRL-1398 than with GRL-1388 and DRV. Modeling analyses revealed that GRL-1398 had greater overall hydrogen bonding and hydrophobic interactions than GRL-1388 and DRV and that GRL-1388 and -1398 had hydrogen bonding interactions with the main chain of the active-site amino acids (Asp29 and Asp30) of protease. The present findings warrant that GRL-1398 be further developed as a potential drug for treating individuals with HIV-1 infection.
Currently available combination therapy or highly active antiretroviral therapy (HAART) has been shown to suppress the replication of human immunodeficiency virus type 1 (HIV-1) and significantly extend the life expectancy of HIV-1-infected individuals (20, 33). However, the ability to provide effective long-term antiretroviral therapy for HIV-1 infection remains a complex issue since the reverse transcriptase of HIV-1 is error-prone, and the replication rate of the virus is enormously rapid, thus enabling HIV-1 to eventually develop resistance to virtually any existing antiretroviral agents. Therefore, continuous efforts are required to develop more potent and safer therapeutics of a different structure(s), mechanism(s), and/or class(es) with a high genetic barrier against HIV-1's acquisition of drug resistance.
HIV-1 protease inhibitors (PIs) are one of the most often used classes of antiretroviral drugs in HAART. Darunavir (DRV), the latest addition among currently available PIs, contains a unique nonpeptidic P2 functional group, bis-tetrahydrofuranylurethane (bis-THF), exerts greatly potent activity against a wide spectrum of laboratory and clinical multidrug-resistant HIV-1 variants (HIV-1MDR). We have previously reported that the high efficacy and genetic barrier of DRV should be associated with its dual antiviral activity: (i) protease enzymatic inhibition and (ii) protease dimerization inhibition (18, 19).
In the present work, we designed, synthesized, and evaluated nonpeptidic HIV-1 protease inhibitors, GRL-1388 and -1398, which contain a novel bicyclic P2 functional group, tetrahydropyrano-tetrahydrofuran (Tp-THF), instead of the bis-THF moiety of DRV, and show highly potent antiretroviral activity against not only wild-type HIV-1 but also a variety of multi-PI-resistant laboratory HIV-1 strains, including a highly DRV-resistant HIV-1 variant, selected against DRV in vitro as recently described (17), and various multidrug-resistant clinical isolates. Structural modeling analyses revealed that the Tp-THF moiety of GRL-1388 and -1398 forms effective and strong hydrogen bond interactions with the main chain atoms of the protease active site amino acids, as does the bis-THF moiety of DRV. In particular, the additional polar and hydrophobic interactions between GRL-1398 and HIV-1 protease should be an explanation of its greater potency compared to that of GRL-1388.
MATERIALS AND METHODS
Cells and viruses.
CD4+ MT-2 and MT-4 cell lines were grown in RPMI 1640-based culture medium supplemented with 10% fetal calf serum (PAA Laboratories GmbH, Linz, Austria) plus 50 U of penicillin and 100 μg of kanamycin per ml. The following HIV-1 strains were used for the drug susceptibility assay: HIV-1LAI, HIV-1NL4-3, a clinical HIV-1 strain isolated from a treatment-naive AIDS patient (HIV-1ERS104pre) (28), and six HIV-1 clinical isolates that were originally isolated from patients with AIDS, who had received long-term antiretroviral therapy using 9 to 11 anti-HIV-1 drugs (without DRV) over 32 to 83 months and were genotypically and phenotypically characterized as clinical multiple-PI-resistant HIV-1 variants (CLHIV-1MDR) (36, 37). Ten such CLHIV-1MDR strains were used as follows: CLHIV-1MDR/A, CLHIV-1MDR/B, CLHIV-1MDR/C, CLHIV-1MDR/EV, CLHIV-1MDR/G, CLHIV-1MDR/TM, CLHIV-1MDR/MM, CLHIV-1MDR/JSL, CLHIV-1MDR/SS, and CLHIV-1MDR/13-52.
Antiviral agents.
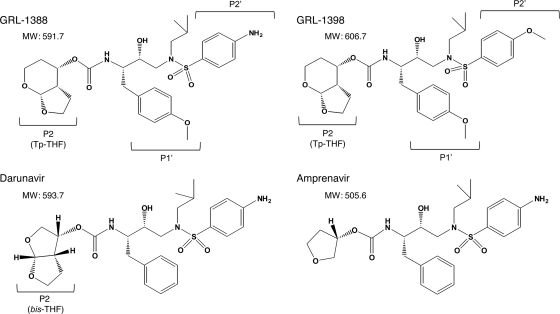
Nonpeptidic PIs, GRL-1388 and -1398 (Fig. 1) (molecular weights of 591.7 and 606.7, respectively), both of which contain a polycyclic ligand (Tp-THF) in place of bis-THF of DRV, were synthesized. The method of synthesis of GRL-1388 and -1398 will be published elsewhere by A. K. Ghosh et al. Saquinavir (SQV) and ritonavir (RTV) were kindly provided by Roche Products, Ltd. (Welwyn Garden City, United Kingdom), and Abbott Laboratories (Abbott Park, IL), respectively. Amprenavir (APV) was kindly provided by GlaxoSmithKline (Research Triangle Park, NC). Nelfinavir (NFV) and lopinavir (LPV) were kindly provided by Japan Energy, Inc., Tokyo, Japan. Atazanavir (ATV) was kindly provided by Bristol-Myers Squibb (New York, NY). Darunavir (DRV) was synthesized as previously described (16).
FIG. 1.
Structures of GRL-1388, GRL-1398, darunavir, and amprenavir.
Drug susceptibility assay.
The susceptibility of HIV-1LAI to various drugs was determined as previously described (36), with minor modifications. Briefly, MT-2 cells (2 × 104/ml) were exposed to 100 50% tissue culture infective doses (TCID50s) of HIV-1LAI in the presence or absence of various concentrations of drugs in 96-well microtiter culture plates, followed by incubation at 37°C for 7 days. After 100 μl of the culture medium was removed from each well, 3-(4,5-dimetylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (10 μl, 7.5 mg/ml in phosphate-buffered saline) was added to each well, followed by incubation at 37°C for 3 h. After incubation to dissolve the formazan crystals, 100 μl of acidified isopropanol containing 4% (vol/vol) Triton X-100 was added to each well, and the optical density was measured by using a kinetic microplate reader (Vmax; Molecular Devices, Sunnyvale, CA). All assays were performed in duplicate or triplicate. To determine the drug sensitivity of laboratory HIV-1 strains, MT-4 cells were used as target cells. In brief, MT-4 cells (105/ml) were exposed to 50 TCID50s of HIV-1NL4-3 and PI-resistant HIV-1 strains in the presence or absence of various concentrations of drugs, the supernatants were harvested on day 7 of culture, and the amount of p24 Gag protein were determined by using a fully automated chemiluminescent enzyme immunoassay system (Lumipulse F; Fujirebio, Inc., Tokyo, Japan) (22). To determine the susceptibility of primary HIV-1 isolates to drugs, phytohemagglutinin-activated peripheral blood mononuclear cells (PHA-PBMCs; 106/ml) were exposed to 50 TCID50s of HIV-1ERS104pre or each CLHIV-1MDR isolate in the presence or absence of various concentrations of drugs and incubated for 7 days. Upon the conclusion of the culture, the amounts of p24 Gag protein in the supernatants were quantified. The drug concentrations that suppressed the production of p24 Gag protein by 50% (50% effective concentrations [EC50s]) were determined. The p24 Gag amounts were compared to the amounts of those produced in drug-free control cell cultures. All assays were performed in duplicate or triplicate.
Generation of FRET-based HIV-1 expression system.
The intermolecular fluorescence resonance energy transfer (FRET)-based HIV-1 expression assay (FRET/HIV-1 assay) using cyan and yellow fluorescent protein-tagged protease monomers (CFP and YFP, respectively) was performed as previously described (18). In brief, CFP- and YFP-tagged HIV-1 protease constructs were generated by using BD Creator DNA cloning kits (BD Biosciences, San Jose, CA). For the generation of full-length molecular infectious clones containing CFP- or YFP-tagged protease, the PCR-mediated recombination method was used (7). A linker consisting of five alanines was inserted between the protease and the fluorescent proteins. The phenylalanine-proline site that HIV-1 protease cleaves was also introduced between the fluorescent protein and reverse transcriptase sites. The DNA fragments obtained were subsequently joined by using the PCR-mediated recombination reaction performed under the standard conditions for Ex Taq polymerase (Takara Bio, Inc., Otsu, Japan). The amplified PCR products were cloned into the pCR-XL-TOPO vector according to the manufacturer's instructions (Gateway cloning system; Invitrogen, Carlsbad, CA). PCR products were generated with the pCR-XL-TOPO vector and used as templates, followed by digestion by both ApaI and SmaI, and the ApaI-SmaI fragment was introduced into pHIV-1NLSma (10), generating pHIV-PRWTCFP and pHIV-PRWTYFP (where WT indicates wild type), respectively.
FRET procedure.
COS7 cells plated on an EZVIEW glass-bottom culture plate (Iwaki, Tokyo, Japan) were cotransfected with pHIV-PRWTCFP and pHIV-PRWTYFP using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions, in the presence of various concentrations of each test compound, cultured for 72 h, and analyzed under a Fluoview FV500 confocal laser scanning microscope (Olympus Optical Corp., Tokyo, Japan) at room temperature, as previously described (18). In the assay, each test compound was added to the culture, followed by cotransfection. The results were determined by measurement of the quenching of the CFP (donor) fluorescence and the increase in the YFP (acceptor) fluorescence (sensitized emission), since a part of CFP energy is transferred to YFP instead of being emitted. This phenomenon can be measured by bleaching YFP, which should result in an increase in CFP fluorescence (2, 3, 27, 29). The changes in the CFP and YFP fluorescence intensities in the images of the selected regions were examined and quantified by using the FV500 image software system (Olympus Optical Corp.). The ratios of the intensities of the CFP fluorescence after photobleaching to the CFP fluorescence before photobleaching (CFPA/B ratios) were determined. When the CFPA/B ratios were <1, it was indicated that the association of the two subunits did not occur, being interpreted that protease dimerization was inhibited.
Generation of PI-resistant HIV-1 using HIV-1NL4-3 in vitro.
In the experiments of the selection of drug-resistant variants, MT-4 cells were exploited as target cells, since HIV-1 in general replicates at greater levels in MT-4 cells than in MT-2 cells. MT-4 cells (105/ml) were exposed to HIV-1NL4-3 (500 TCID50s) and cultured in the presence of various PIs at an initial concentration of an EC50. Viral replication was monitored with the determination of the amount of p24 Gag produced by MT-4 cells. The culture supernatants were harvested on day 7 or by up to day 14 when the amount of p24 Gag was >250 ng/ml and used to infect fresh MT-4 cells for the next round of culture in the presence of increasing concentrations of each drug. When the virus began to propagate in the presence of the drug, the drug concentration of the following round of culture was generally increased 2- to 3-fold. To determine whether amino acid substitutions associated with HIV-1 acquisition of drug resistance occurred, high-molecular-weight DNA obtained from the lysates of the cells was subjected to nucleotide sequencing.
Generation of highly GRL-1388- and -1398-resistant HIV-1 variants using a mixture of CLHIV-1MDR isolates in vitro.
Ten CLHIV-1MDR strains (CLHIV-1MDR/A, CLHIV-1MDR/B, CLHIV-1MDR/C, CLHIV-1MDR/EV, CLHIV-1MDR/G, CLHIV-1MDR/TM, CLHIV-1MDR/MM, CLHIV-1MDR/JSL, CLHIV-1MDR/SS, and CLHIV-1MDR/13-52) were isolated from patients with AIDS who had failed existing anti-HIV regimens after receiving 9 to 12 anti-HIV-1 drugs, not including DRV (17, 30). These strains contained 9 to 21 amino acid substitutions in protease, which have reportedly been associated with HIV-1 resistance to various PIs. The mixture of the viruses was propagated initially in MT-4 cells and PHA-PBMCs as previously described (37). The mixture was transferred to the culture with fresh MT-4 cells on day 7, and the culture supernatant was harvested and used to infect fresh MT-4 cells to continue the selection. This cycle of cell-free transmission was repeated every 7 to 14 days, each time increasing the drug concentration by a factor of 2 or 3.
Determination of nucleotide sequences.
Molecular cloning and the determination of nucleotide sequences of HIV-1 passaged in the presence of each agent were performed as previously described (36, 37). In brief, high-molecular-weight DNA was extracted from HIV-1-infected MT-4 cells by using the InstaGene matrix (Bio-Rad Laboratories, Hercules, CA) and was subjected to molecular cloning, followed by sequence determination. The primers used for the first-round PCR amplification of the entire Gag- and protease-encoding regions of the HIV-1 genome were LTR F1 (5′-GAT GCT ACA TAT AAG CAG CTG C-3′) and PR12 (5′-CTC GTG ACA AAT TTC TAC TAA TGC-3′). The first-round PCR mixture consisted of 1 μl of proviral DNA solution, 2.0 U of premix Taq (Ex Taq version; Takara Bio, Inc., Otsu, Japan), and 12.5 pmol of each of the first-round PCR primers in a total volume of 50 μl. The PCR conditions used were an initial 2-min step at 94°C, followed by 30 cycles of 30 s at 94°C, 30 s at 58°C, and 3 min at 72°C, with a final 8 min of extension at 72°C. The first-round PCR products (1 μl) were used directly in the second round of PCR with primers LTR F2 (5′-GAG ACT CTG GTA ACT AGA GAT C-3′) and Ksma2.1 (5′-CCA TCC CGG GCT TTA ATT TTA CTG GTA C-3′) under the same PCR conditions described above. The second-round PCR products were purified with spin columns (MicroSpin S-400 HR; Amersham Biosciences Corp., Piscataway, NJ), cloned, and subjected to sequencing with a model 377 automated DNA sequencer (Applied Biosystems, Foster City, CA).
Determination of replication kinetics of GRL-1398-resistant HIV-1 variants and HIV-1NL4-3.
We determined the replication kinetics of two HIV-1 variant populations, which were selected in the presence of up to 1 μM GRL-1398. One population was selected for 53 weeks in experiment I (designated HIV-1GRL1398-1μMExp.I) and the other for 57 weeks in experiment II (HIV-1GRL1398-1μMExp.II) (see Fig. 4). MT-4 cells (3 × 105) were exposed to an HIV-1GRL1398-1μMExp.I, HIV-1GRL1398-1μMExp.II or wild-type HIV-1NL4-3 preparation that contained 30 ng of p24 in six-well culture plates for 3 h, and each MT-4 cell population was divided into three fractions. Each fraction was propagated in the presence of 0, 0.1, or 1 μM GRL-1398. The amounts of p24 were measured every 2 days in culture for up to 8 or 10 days.
Modeling and analysis of GRL-1388 and -1398 interactions with wild-type HIV-1 protease.
The structures of GRL-1388 and -1398 were modeled into the active site cavity of the wild-type HIV-1 protease using the extra precision Glide 5.5-ligand docking method from the Schrödinger suite of programs (8, 9, 26). Crystal structures of protease complexed with DRV (PDB code 2IEN [31]) and protease complexed with brecanavir (PDB code 2FDE [23]) obtained from the RCSB Protein Data Bank (http://www.rcsb.org) were used as template protease structures. All of the ions and glycerol-related atoms were deleted from the structures. The hydrogen atoms in both structures, 2IEN and 2FDE, were energy minimized (MacroModel, version 9.7) by using constraints on the heavy atoms of the structures. Crystallographic waters were deleted from both of the minimized structures (except the conserved water molecule that forms a tetracoordinated hydrogen bond interaction between the protease flaps and the inhibitor). These structures were then processed using the protein preparation wizard of MAESTRO (version 9.1), and the resultant structures were used to generate the receptor grid. Molecular models for GRL-1388 and -1398 were prepared by using the structure of DRV as a template. The correct stereochemistries of GRL-1388 and -1398 were assigned, and a set of diverse conformers was generated for the latter compound by using ConfGen (version 2.1). These conformers were then docked against the receptor grid. Among the docked conformers, the best binding conformations and the interactions with wild-type HIV-1 protease were analyzed. The structural model for GRL-1388 was generated by energy minimization from the optimized model of GRL-1398. Hydrogen bonds were calculated by using MAESTRO (with a 3.0-Å distance cutoff and angle constraints of 90° [donor] and 60° [acceptor], respectively), and the hydrophobic contacts of the inhibitor were obtained by probing all of the protease atoms within a radius of 4 Å using the CCP4 suite of programs (4). The programs MacroModel (version 9.7), ConfGen (version 2.1), and Glide (version 5.5) from Schrödinger LLC (New York, NY) were used in generating the final models described above. All of the graphics were prepared by using PyMOL molecular graphics program (version 0.99 DeLano Scientific LLC: http://pymol.org/).
RESULTS
Antiviral activity of GRL-1388 and -1398 against HIV-1LAI.
We designed and synthesized ∼100 novel PIs containing the bis-THF component, which we reported plays a major role in the potent antiviral activity of DRV (15, 19) and its related moieties, including polycyclic ligands (11-14). Among such novel PIs, we identified GRL-1388 and -1398 (Fig. 1) as potent anti-HIV-1 agents, which contain Tp-THF. As shown in Table 1, GRL-1388 and -1398 were highly potent in vitro against a laboratory wild-type HIV-1 strain, HIV-1LAI, with EC50s of 3.6 ± 1.8 and 0.2 ± 0.2 nM, respectively, as examined in the MTT assay with CD4+ MT-2 cells. Of note, the antiviral activity of GRL-1388 against HIV-1LAI was comparable to or more potent than four representative U.S. Food and Drug Administration (FDA)-approved PIs: SQV, APV, ATV, and DRV. GRL-1398 was significantly more potent against HIV-1LAI than these four PIs by a factor of 16.5 to 98. Both GRL-1388 and -1398 had favorable cytotoxicity profiles with 50% cytotoxicity concentrations [CC50s] of more than 100 and 37.7 μM, giving selectivity index (SI) values of more than 27,800 and 188,500, respectively (Table 1).
TABLE 1.
Antiviral activities of GRL-1388 and -1398 against HIV-1LAI and their cytotoxicities in vitroa
| Drug | Mean ± SD |
Selectivity indexb | |
|---|---|---|---|
| EC50 (nM) | CC50 (μM) | ||
| SQV | 6.2 ± 1.9 | 19.7 ± 6.1 | 3,200 |
| APV | 19.6 ± 3.9 | >100 | >5,100 |
| ATV | 5.0 ± 1.9 | 27.6 ± 0.7 | 5,500 |
| DRV | 3.3 ± 0.9 | >100 | >30,300 |
| GRL-1388 | 3.6 ± 1.8 | >100 | >27,800 |
| GRL-1398 | 0.2 ± 0.2 | 37.7 ± 2.7 | 188,500 |
All assays were conducted in duplicate, and the data shown represent mean values (±1 standard deviation) derived from the results of three independent experiments.
Each selectivity index denotes a ratio of CC50 to EC50 against HIV-1LAI.
GRL-1388 and -1398 are potent against various PI-selected laboratory HIV-1 variants.
We also examined whether GRL-1388 and -1398 were active against a variety of HIV-1 variants that had been selected in vitro with each of six FDA-approved PIs: SQV, RTV, NFV, LPV, ATV, and APV (Table 2). Each HIV-1 variant was selected in vitro by propagating HIV-1NL4-3 in the presence of increasing concentrations of each PI (up to 5 μM) in MT-4 cells and was confirmed to have acquired multiple amino acid substitutions in protease of the virus, which have reportedly been associated with viral resistance to PIs (see footnote a of Table 2). Each of the variants (HIV-1SQV-5μM, HIV-1RTV-5μM, HIV-1NFV-5μM, HIV-1LPV-5μM, and HIV-1ATV-5μM) was highly resistant to the corresponding PI, with which the variant was selected, having the EC50s of >1 μM, and the fold differences in the EC50s relative to the EC50 of each drug against HIV-1NL4-3 ranged from >25 to >400 (Table 2). The activity of GRL-1388 against all five variants was well maintained, with the fold changes being 1 to 11. GRL-1398 was significantly more potent against five such HIV-1 variants than DRV with an EC50 of 0.1 to 5.7 nM. Of note, GRL-1388 and -1398 were moderately active against HIV-1APV-5μM, with EC50s of 475.7 and 49.0 nM, presumably due to the structural resemblance of GRL-1388 and -1398 to APV (Fig. 1).
TABLE 2.
Antiviral activities of GRL-1388 and -1398 against laboratory PI-selected HIV-1variantsa
| Virus | Mean EC50 in nM ± SD (fold change) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| SQV | RTV | NFV | LPV | ATV | APV | DRV | GRL-1388 | GRL-1398 | |
| HIV-1NL4-3 | 5.2 ± 1.7 | 40.4 ± 4.9 | 30.2 ± 3.0 | 27.8 ± 5.3 | 2.5 ± 0.8 | 34.3 ± 1.3 | 3.2 ± 0.3 | 3.1 ± 0.2 | 0.2 ± 0.1 |
| HIV-1SQV-5μM | >1,000 (192) | >1,000 (>25) | >1,000 (>33) | >1,000 (>36) | 415.0 ± 15.1 (166) | 353.7 ± 12.4 (10) | 30.4 ± 0.5 (10) | 32.6 ± 7.5 (11) | 5.7 ± 1.3 (29) |
| HIV-1RTV-5μM | 66.3 ± 8.9 (13) | >1,000 (>25) | 386.4 ± 84.7 (13) | 698.3 ± 87.4 (25) | 32.5 ± 5.9 (13) | 334.5 ± 42.5 (10) | 19.9 ± 13.5 (6) | 28.8 ± 1.8 (9) | 3.1 ± 0.2 (16) |
| HIV-1NFV-5μM | 24.1 ± 6.3 (5) | 46.5 ± 9.4 (1) | >1,000 (>33) | 37.2 ± 3.3 (1) | 12.3 ± 2.2 (5) | 71.3 ± 10.5 (2) | 2.7 ± 0.4 (1) | 2.6 ± 0.4 (1) | 0.1 ± 0.1 (1) |
| HIV-1LPV-5μM | 33.5 ± 0.9 (6) | >1,000 (>25) | 401.6 ± 11.0 (13) | >1,000 (>36) | 17.0 ± 7.7 (7) | 320.7 ± 23.2 (9) | 4.0 ± 0.9 (1) | 29.9 ± 12.5 (10) | 3.5 ± 0.4 (18) |
| HIV-1ATV-5μM | 134.3 ± 22.8 (26) | >1,000 (>25) | >1,000 (>33) | >1,000 (>36) | >1,000 (>400) | 521.0 ± 124.1 (15) | 6.8 ± 0.6 (2) | 21.9 ± 7.7 (7) | 3.1 ± 0.4 (16) |
| HIV-1APV-5μM | 47.3 ± 2.1 (9) | >1,000 (>25) | >1,000 (>33) | 633.2 ± 21.0 (23) | 453.6 ± 12.2 (181) | >1,000 (>29) | 423.9 ± 6.0 (132) | 475.7 ± 38.1 (153) | 49.0 ± 1.1 (245) |
The amino acid substitutions identified in protease of HIV-1SQV-5μM, HIV-1RTV-5μM, HIV-1NFV-5μM, HIV-1LPV-5μM, HIV-1ATV-5μM, and HIV-1APV-5μM compared to the consensus type B sequence cited from the Los Alamos database include L10I/N37D/G48V/I54V/L63P/G73C/I84V/L90M, L10I/M46L/I54V/V82A, L10F/K20T/D30N/K45I/A71V/V77I, L10F/M46I/I54V/V82A, L23I/E34Q/K43I/M46I/I50L/G51A/L63P/A71V/V82A/T91A, and L10F/V32I/L33F/M46L/I54M/A71V, respectively. Numbers in parentheses represent fold changes in EC50s for each isolate compared to the EC50s for wild-type HIV-1NL4-3. All assays were conducted in duplicate or triplicate, and the data shown represent mean values (±1 standard deviation) derived from the results of three independent experiments.
GRL-1388 and -1398 exert potent activity against multiple-PI-resistant clinical HIV-1 strains (CLHIV-1MDR).
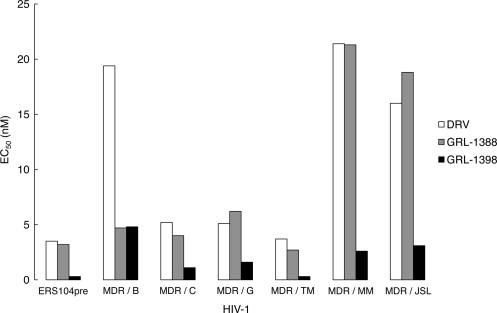
We also sought to determine whether GRL-1388 and -1398 were active against CLHIV-1MDR isolates, including CLHIV-1MDR/B, CLHIV-1MDR/C, CLHIV-1MDR/G, CLHIV-1MDR/TM, CLHIV-1MDR/MM, and CLHIV-1MDR/JSL, which contained 9 to 14 PI-resistance-associated amino acid substitutions in protease (see footnote a of Table 3). The EC50s of LPV against these multi-PI-resistant clinical HIV-1 isolates were mostly >1 μM, and the activity of other three PIs (SQV, ATV, and APV) had also been significantly compromised, as determined using PHA-PBMCs as the target cells and p24 production inhibition as the endpoint (Table 3). Both GRL-1388 and DRV remained active against all of the clinical variants examined and the fold differences between the EC50s against HIV-1ERS104pre and those against each clinical variant ranged from as low as 1 to 7. The fold changes seen with GRL-1398 ranged from 1 to 16; however, the absolute EC50s remained substantially lower, ranging from 0.3 to 4.8 nM, than those of DRV (Fig. 2).
TABLE 3.
Antiviral activity of GRL-1388 and -1398 against multidrug-resistant clinical isolates in PHA-PBMCsa
| Virus (syncytium formation) | Mean EC50 in nM ± SD (fold change) |
||||||
|---|---|---|---|---|---|---|---|
| SQV | LPV | ATV | APV | DRV | GRL-1388 | GRL-1398 | |
| CLHIV-1ERS104pre (SI) | 4.4 ± 1.8 | 41.0 ± 6.0 | 1.9 ± 1.2 | 35.1 ± 4.3 | 3.5 ± 0.6 | 3.2 ± 0.3 | 0.3 ± 0.1 |
| CLHIV-1MDR/B (SI) | 206.9 ± 81.2 (47) | >1,000 (>24) | 228.0 ± 82.6 (120) | 328.8 ± 138.7 (9) | 19.4 ± 3.6 (6) | 4.7 ± 3.6 (1) | 4.8 ± 0.3 (16) |
| CLHIV-1MDR/C (SI) | 38.8 ± 16.2 (9) | >1,000 (>24) | 25.4 ± 7.5 (13) | 265.2 ± 79.8 (8) | 5.2 ± 1.5 (1) | 4.0 ± 1.0 (1) | 1.1 ± 0.1 (4) |
| CLHIV-1MDR/G (SI) | 26.3 ± 2.4 (6) | 319.3 ± 31.0 (8) | 16.3 ± 4.5 (9) | 260.1 ± 55.8 (7) | 5.1 ± 2.3 (1) | 6.2 ± 0.6 (2) | 1.6 ± 1.1 (5) |
| CLHIV-1MDR/TM (SI) | 110.6 ± 48.6 (25) | 658.1 ± 115.9 (16) | 22.4 ± 3.8 (12) | 197.7 ± 65.0 (6) | 3.7 ± 1.3 (1) | 2.7 ± 0.7 (1) | 0.3 ± 0.1 (1) |
| CLHIV-1MDR/MM (NSI) | 190.7 ± 75.9 (43) | >1,000 (>24) | 68.9 ± 27.8 (36) | 402.2 ± 193.9 (11) | 21.4 ± 1.7 (6) | 21.3 ± 5.0 (7) | 2.6 ± 1.4 (9) |
| CLHIV-1MDR/JSL (NSI) | 281.8 ± 35.7 (64) | >1,000 (>24) | 347.0 ± 56.9 (183) | 306.6 ± 124.4 (9) | 16.0 ± 4.4 (5) | 18.8 ± 9.0 (6) | 3.1 ± 0.1 (10) |
Amino acid substitutions identified in protease compared to the consensus type B sequence cited from the Los Alamos database include L63P in CLHIV-1ERS104pre; L10I, K14R, L33I, M36I, M46I, F53I, K55R, I62V, L63P, A71V, G73S, V82A, L90M, and I93L in CLHIV-1MDR/B; L10I, I15V, K20R, L24I, M36I, M46L, I54V, I62V, L63P, K70Q, V82A, and L89M in CLHIV-1MDR/C; L10I, V11I, T12E, I15V, L19I, R41K, M46L, L63P, A71T, V82A, and L90M in CLHIV-1MDR/G; L10I, K14R, R41K, M46L, I54V, L63P, A71V, V82A, L90M, and I93L in CLHIV-1MDR/TM; L10I, K43T, M46L, I54V, L63P, A71V, V82A, L90M, and Q92K in CLHIV-1MDR/MM; L10I, L24I, I33F, E35D, M36I, N37S, M46L, I54V, R57K, I62V, L63P, A71V, G73S, and V82A in CLHIV-1MDR/JSL. CLHIV-1ERS104pre served as a source of wild-type HIV-1. Numbers in parentheses represent the fold changes of EC50s against each isolate compared to the EC50s against wild-type CLHIV-1ERS104pre. All assays were conducted in duplicate or triplicate, and the data shown represent mean values (±1 standard deviation) derived from results of three independent experiments. PHA-PBMCs were derived from a single donor in each experiment.
FIG. 2.
Antiviral activity of GRL-1388, GRL-1398, and darunavir against multidrug-resistant clinical HIV-1 isolates. The EC50s of darunavir (white), GRL-1388 (gray), and GRL-1398 (black) against CLHIV-1ERS104pre, which served as a wild-type HIV-1, and six CLHIV-1MDR isolates were determined as described in the legend to Table 3 and Materials and Methods. Note that the EC50s of GRL-1398 against all of these isolates were substantially lower than those of DRV.
GRL-1398 is active against DRV-resistant variants.
We determined antiviral activity against DRV-resistant HIV-1 variants that had been selected in vitro by propagating a mixture of eight CLHIV-1MDR strains in the presence of increasing concentrations of DRV in MT-4 cells (17). Two such variants, HIV-1DRVRP10 and HIV-1DRVRP20, containing a set of amino acid substitutions at passages 10 and 20, respectively (Table 4), were resistant to DRV with EC50s of 29.1 and 214.1 nM and to GRL-1388 with EC50s of 24.6 and 150.8 nM. In contrast, GRL-1398 had substantially lower absolute EC50s—3.3 and 21.9 nM against HIV-1DRVRP10 and HIV-1DRVRP20, respectively(Table 4).
TABLE 4.
Antiviral activity of GRL-1388 and -1398 against laboratory darunavir-resistant HIV-1a
| Virus | Amino acid substitution(s) in protease | Mean EC50 in nM ± SD (fold change) |
||
|---|---|---|---|---|
| DRV | GRL-1388 | GRL-1398 | ||
| cHIV-1ERS104pre | L63P | 4.0 ± 1.0 | 2.5 ± 0.4 | 0.4 ± 0.1 |
| HIV-1DRVRP10 | L10I, I15V, K20R, L24I, V32I, M36I, M46L, I54V, I62V, L63P, K70Q, V82A, L88M | 29.1 ± 0.9 (7) | 24.6 ± 6.9 (10) | 3.3 ± 0.3 (8) |
| HIV-1DRVRP20 | L10I, I15V, K20R, L24I, V32I, M36I, M46L, L63P, A71T, V82A, L88M | 214.1 ± 47.9 (54) | 150.8 ± 46.7 (60) | 21.9 ± 5.7 (54) |
DRV-resistant HIV-1 variants were selected in vitro by propagating a mixture of eight CLHIV-1MDR isolates in the presence of increasing concentrations of DRV in MT-4 cells. Six of the eight isolates were the same as those used for drug susceptibility assay (Table 3). Amino acid substitutions identified in protease of the other two isolates compared to the consensus type B sequence cited from the Los Alamos database include L10I, I15V, E35D, N37E, K45R, I54V, L63P, A71V, V82T, L90M, I93L, and C95F in CLHIV-1MDR/A and L10R, N37D, M46I, I62V, L63P, A71V, G73S, V74I, V82T, L90M, and I93L in CLHIV-1MDR/SS. Numbers in parentheses represent the fold changes of EC50s against each isolate compared to the EC50s against HIV-1ERS104pre. All assays were conducted in duplicate or triplicate, and the data shown represent mean values (±1 standard deviation) derived from the results of three independent experiments. PHA-PBMCs were derived from a single donor.
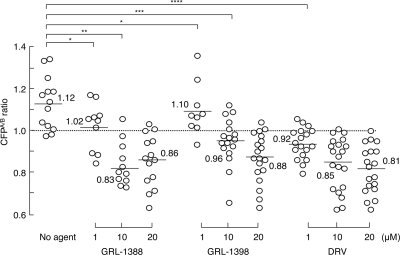
GRL-1388 and -1398 moderately disrupt the dimerization of HIV-1 protease.
We previously reported that DRV effectively disrupts the dimerization of HIV-1 protease monomer subunits as determined with the FRET/HIV-1 expression assay (18). We therefore sought to determine whether GRL-1388 and -1398 exerted such protease dimerization inhibition activity. In the absence of agents, the mean CFPA/B ratio obtained in the assay was 1.12, indicating that protease dimerization clearly occurred (Fig. 3). However, when COS7 cells were cotransfected in the presence of 1 μM GRL-1388 and -1398, protease dimerization still occurred, with the mean CFPA/B ratios being 1.02 and 1.10, respectively. However, the mean CFPA/B ratios obtained in the presence of 10 and 20 μM GRL-1388 and -1398 were all <1.0, indicating that both compounds blocked the dimerization. Of note, DRV blocked the dimerization at as low as 1.0 μM under the same conditions, suggesting that GRL-1388 and -1398 had relatively moderate activity to disrupt the dimerization of HIV-1 protease compared to DRV.
FIG. 3.
Inhibition of HIV-1 protease dimerization. COS7 cells were exposed to each of the agents (GRL-1388, GRL-1398, and DRV) in various concentrations (1, 10, and 20 μM) and subsequently cotransfected with plasmids encoding full-length molecular infectious HIV-1 (HIVNL4-3) clones producing CFP- or YFP-tagged protease. After 72 h, cultured cells were examined in the FRET-based HIV-1 expression assay and the CFPA/B ratios (y axis) were determined. The mean values of the ratios obtained are shown as horizontal bars. A CFPA/B ratio that is >1 signifies that protease dimerization occurred, whereas a ratio that is <1 signifies the disruption of protease dimerization. All of the experiments were conducted in a blind fashion. *, Not significant; **, P < 0.0001; ***, P = 0.0005; ****, P = 0.0001.
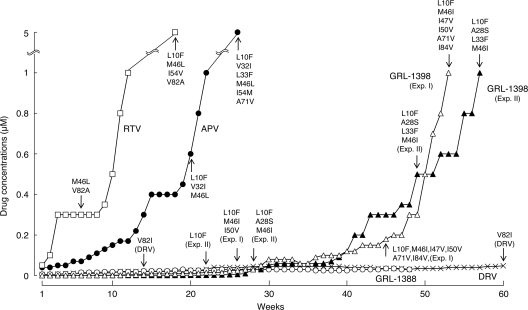
In vitro selection of HIV-1 variants resistant to GRL-1388 and -1398.
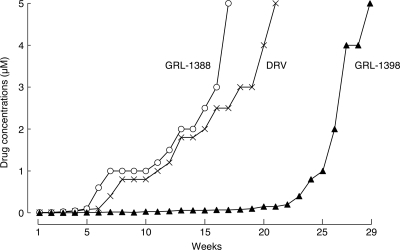
We attempted to select HIV-1 variants resistant to GRL-1388 and -1398 by propagating HIV-1NL4-3 in MT-4 cells in the presence of increasing concentrations of each compound. Considering that drug resistance-associated amino acid substitutions occur stoichiometrically at random, we conducted the selection with GRL-1398 on two different occasions. When selected in the presence of increasing concentrations of RTV and APV, the virus quickly became resistant to the drugs and started replicating in the presence of 5 μM at 18 and 26 weeks of selection (Fig. 4) and had acquired L10I/M46L/I54V/V82A and L10F/V32I/L33F/M46L/I54M/A71V, respectively (Fig. 4). In contrast, the acquisition of resistance to GRL-1398 was significantly delayed and it took 53 and 57 weeks in the selection until the virus started replicating in the presence of 1 μM in experiments I and II, respectively (Fig. 4). The virus preparations selected with GRL-1398 in experiments I and II had acquired no amino acid substitutions in protease by 20 and 16 weeks of culture; however, they acquired apparently resistance-associated substitutions by 26 and 22 weeks with L10F/M46I/I50V and L10F, respectively. The virus had acquired A28S by 28 weeks of culture, along with L10F and M46M/I in experiment II (HIV1398II-Wk28); thereafter, HIV1398II-Wk28 appeared to start developing resistance at a faster rate, although the virus in experiment I did not acquire A28S but had L10F/M46I/I50V and developed resistance. In contrast, when HIV-1NL4-3 was selected with GRL-1388 under the same conditions, the virus remained wild-type over 45 weeks of selection, whereas HIV-1NL4-3, selected with DRV, had acquired V82I by 14 weeks. All amino acid substitutions identified during the selection are illustrated in Fig. S1 in the supplemental material.
FIG. 4.
In vitro selection of HIV-1 variants against GRL-1388 and -1398. HIV-1NL4-3 was propagated in MT-4 cells in the presence of increasing concentrations of ritonavir (□), amprenavir (•), darunavir (×), GRL-1388 (○), or GRL-1398 (experiment I ▵ and experiment II ▴). Each passage of the virus was carried out in a cell-free manner. Amino acid substitutions identified in the protease of each HIV-1 at each indicated time of the selection are shown. Note that, by week 48, no amino acid mutations were detected in the protease of GRL-1388-resistant isolate. All amino acid substitutions identified during the selection are illustrated in Fig. S1 in the supplemental material.
Selection with GRL-1388 and -1398 of a mixture of 10 CLHIV-1MDR variants.
Considering that amino acid substitutions in HIV-1 stoichiometrically and randomly occur and the resulting amino acid substitutions can vary from one experiment to another as seen above (Fig. 4), we performed an additional selection experiment with MT4 cells and a mixture of 10 CLHIV-1MDR variants as a starting virus population with DRV, GRL-1388, and -1398. In line with our previous observations that a highly DRV-resistant HIV-1 variants emerged when selected with DRV using a mixture of eight CLHIV-1MDR variants (17), HIV-1 started replicating in the presence of 5 μM DRV and GRL-1388 by 21 and 17 weeks of selection, respectively (Fig. 5). In contrast to the results of selection experiments using MT-4 cells and HIV-1NL4-3, in which HIV-1 started replicating with GRL-1398, but not with GRL-1388 or DRV (Fig. 4), the emergence of HIV-1 capable of replicating in the presence of GRL-1398 was significantly delayed compared to the cases with GRL-1388 and DRV (Fig. 5). Sequence analyses of the protease-encoding gene of the viruses in these selection experiments revealed that the HIV-1 replicating in the presence of GRL-1398 had acquired A28S by 24 weeks of selection (see Fig. S2 in the supplemental material), when the virus appeared to obtain robust replication fitness despite the presence of GRL-1398. There was no A28S substitution in those selected with GRL-1388 or DRV, as seen in the selection experiments discussed above in Fig. 4. All amino acid substitutions identified during the selection are illustrated in Fig. S2 in the supplemental material.
FIG. 5.
In vitro selection multidrug-resistant clinical isolates against GRL compounds and DRV. A mixture of 10 CLHIV-1MDR variants were propagated in MT-4 cells in the presence of increasing concentrations of darunavir (×), GRL-1388 (○), or GRL-1398 (▴). Each selection was conducted in a cell-free fashion. The amino acid substitutions in protease of eight CLHIV-1MDR isolates are given in the footnotes for Tables 3 and 4. The amino acid substitutions identified in protease were L10V, T12E, G16A, L19I, K20R, L33F, E35D, M36I, N37S, M46I, I50V, F53L, I54V, K55R, R57K, D60E, I62V, L63P, A71V, V82A, and L90M in CLHIV-1MDR/EV and N47D, I54V, D60E, L63P, A71V, V74I, V82A, L90M, and I93L in CLHIV-1MDR/13-52. HIV-1 strains, capable of replicating in the presence of each agent at 5 μM, had acquired L10V, T12E, G16A, L19I, K20R, V32I, L33F, E34K, E35D, M36I, N37S, M46I, I50V, F53L, I54V, K55R, R57K, D60E, I62V, L63P, A71V, G73S,V82A, and L90M in HIVMIX-DRVWK21; L10V, T12E, G16A, L19I, K20R, V32I, L33F, E35D, M36I, N37S, M46I, I50V, F53L, I54V, K55R, R57K, D60E, I62V, L63P, A71V, V82A, and L90M in HIVMIX-1388WK17; and L10I, I15V, A28S, L33I, M36I, M46I, I50V, F53L, K55R, I62V, L63P, A71V, G73S, L90M, and I93L in HIVMIX-1398WK29. The amino acid substitutions identified in each HIV-1 during the selection are illustrated in Fig. S2 in the supplemental material.
Effects of amino acid substitutions in the protease of GRL-1398-resistant variants on the antiviral activity of GRL-1388 and DRV.
We also examined whether GRL-1398-resistant variants had cross-resistance with GRL-1388 and DRV. As described above, we selected two HIV-1 variant populations in the presence of up to 1 μM GRL-1398 (Fig. 4). One population was selected for 53 weeks in experiment I (designated as HIV-1GRL1398-1μMExp.I) and the other for 57 weeks in experiment II (HIV-1GRL1398-1μMExp.II). These two populations were resistant to GRL-1398 with EC50s of 505.1 and 552.8 nM (Table 5). GRL-1388 and DRV were moderately active against HIV-1GRL1398-1μMExp.I, whereas both GRL-1388 and DRV essentially lost their activity against HIV-1GRL1398-1μMExp.II, having EC50s of >1,000 nM (Table 5).
TABLE 5.
Antiviral activity of darunavir and GRL-1388 against GRL-1398-resistant HIV-1a
| Virus | Amino acid substitutions in protease | Mean EC50 (nM) ± SD |
||
|---|---|---|---|---|
| DRV | GRL-1388 | GRL-1398 | ||
| HIV-1NL4-3 | 3.1 ± 0.4 | 3.0 ± 0.3 | 0.2 ± 0.1 | |
| HIV-1GRL1398-1μMExp.I | L10F, M46I, I47V, I50V, A71V, I84V | 67.3 ± 12.0 | 151.9 ± 10.5 | 505.1 ± 168.0 |
| HIV-1GRL1398-1μMExp.II | L10F, A28S, L33F, M46I | >1,000 | >1,000 | 552.8 ± 213.3 |
GRL-1398-resistant HIV-1 was selected in vitro by propagating HIV-1NL4-3 in the presence of increasing concentrations of GRL-1398 in MT-4 cells (Fig. 4). All assays to determine the EC50s were conducted in duplicate or triplicate, and the data shown represent mean values (±1 standard deviation) derived from the results of three independent assays.
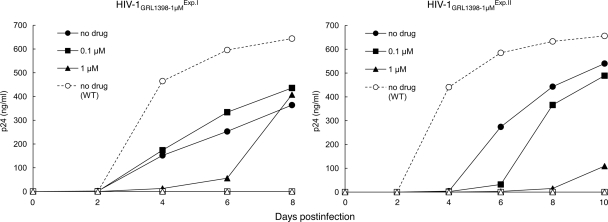
Moderately compromised replication fitness of GRL-1398-resistant HIV-1 variants.
We have conducted a replication kinetics assay and determined the fitness of HIV-1GRL1398-1μMExp.I, HIV-1GRL1398-1μMExp.II, and HIV-1NL4-3 with or without GRL-1398 (0, 0.1, 1 μM). It is of note that the resistant viruses were selected and adapted in MT-4 cells, and MT-4 cells were used in the assay. Although HIV-1NL4-3 failed to replicate in the presence of 0.1 and 1 μM GRL-1398, both HIV-1GRL1398-1μMExp.I and HIV-1GRL1398-1μMExp.II replicated despite the presence of GRL-1398 (Fig. 6). It is also noteworthy that when HIV-1GRL1398-1μMExp.I and HIV-1GRL1398-1μMExp.II were propagated in the presence or absence of GRL-1398, their replication activities were found to be moderately compromised compared to that of HIV-1NL4-3 without GRL-1398 (Fig. 6).
FIG. 6.
Replication kinetics of GRL-1398-resistant HIV-1 variants and HIV-1NL4-3. MT-4 cells were exposed to HIV-1GRL1398-1μMExp.I (left panel, closed symbols), HIV-1GRL1398-1μMExp.II (right panel, closed symbols), or wild-type HIV-1NL4-3 (WT, ○) for 3 h, and each MT-4 cell population was cultured in the presence of 0.1 μM (▪) or 1 μM (▴) of GRL-1398 or without the agent (•). The amounts of p24 were measured every 2 days for up to 8 or 10 days. Note that HIV-1NL4-3 failed to replicate in the presence of 0.1 and 1 μM GRL-1398 (□ and ▵, respectively), both HIV-1GRL1398-1μMExp.I and HIV-1GRL1398-1μMExp.II replicated in the presence of GRL-1398.
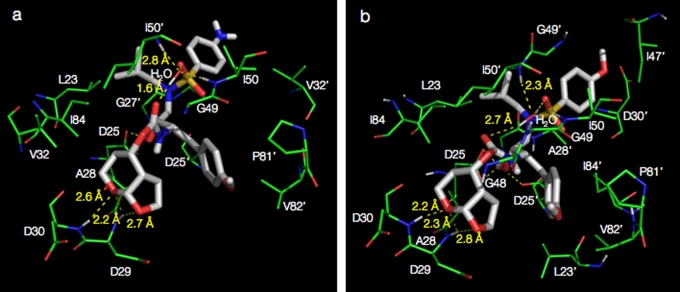
GRL-1398 forms greater interactions with protease than GRL-1388.
Analysis of the molecular complexes of GRL-1388 and -1398 with wild-type HIV-1 protease, generated by docking simulations, revealed that GRL-1398 has a greater number of hydrophobic interactions with protease compared to GRL-1388 and DRV (Fig. 7). The bicyclic structure of Tp-THF representing the P2 group in both GRL-1388 and -1398 shows hydrogen bond interactions with the backbone amide nitrogen atoms of Asp29 and Asp30, similar to that of the bis-THF of DRV. The oxygen atoms of the pyran and the furan rings showed two and one hydrogen bonds with the amide nitrogen atoms of Asp29 and Asp30, respectively. The transition state mimetic hydroxyl group in both compounds shows two hydrogen bonds with the catalytic Asp25, in addition, GRL-1398 shows a hydrogen bond with the catalytic Asp25′ residue of protease. The P2′ amine group in GRL-1388 has a potential to form contacts with the delta oxygen atoms on the side chains of Asp29′ and Asp30′, respectively. The oxygen atom from the O-methoxy (P2′) functional group of GRL-1398 has a potential to form hydrogen bonds with the two backbone amide nitrogen atoms of Asp29′ and Asp30′, respectively. Both GRL-1388 and -1398 form hydrogen bonds with the conserved bridging water molecule that connects to the protease flaps via hydrogen bonds with the amide nitrogen atoms of I50 and I50′. As shown in Table 6, the hydrophobic contacts were examined for both compounds and were compared to each other as well as to those of DRV. GRL-1388 has a similar interaction profile as DRV, whereas GRL-1398 has an overall greater number of interactions.
FIG. 7.
Structural analysis of GRL-1388 and -1398 modeled into the active-site cavity of wild-type HIV-1 protease. GRL-1388 (a) and GRL-1398 (b) are shown as stick models (white color). Protease residues involved in either hydrogen bonding or hydrophobic interactions are highlighted in green. The distances for selected hydrogen bonds are shown. The novel Tp-THF functional group in both the compounds shows strong hydrogen bonding interactions with the backbone of residues Asp29 and Asp30.
TABLE 6.
Interactions between wild type HIV-1 protease and GRL-1388 or -1398a
| Compound | Amino acid residues and a bridging water involved in H bonding (no. of H bonds) | Total no. of H bonds | Amino acid residues involved in hydrophobic interactions (no. of hydrophobic interactions) | Total no. of hydrophobic interactions |
|---|---|---|---|---|
| DRV | D25 (1), G27 (1), D29 (2), D30 (1), D29′ (1), D30′ (1), water (2) | 9 | L23 (1), G27 (1), A28 (2), V32 (2), P81 (5), V82 (1), I84 (4), L23′ (1), D29′ (1), G49′ (3), G50′ (3), V82′ (1), I84′ (1) | 26 |
| GRL-1388 | D25 (2), D29 (2), D30 (1), water (2) | 7 | L23 (1), A28 (4), D29 (1), V32 (2), G49 (5), I50 (1), I84 (2), G27′ (1), A28′ (6), V32′ (1), I50′ (1), P81′ (4), V82′ (2) | 31 |
| GRL-1398 | D25 (2), D29 (2), D30 (1), D25′ (2), water (2) | 8 | L23 (1), D25 (1), A28 (1), D29 (2), D30 (3), G48 (1), G49 (3), I84 (2), L23′ (1), D25′ (1), G27′ (2), A28′ (3), D30′ (1), I47′ (1), G49′ (1), P81′ (4), V82′ (4), I84′ (2) | 34 |
The hydrogen bonding, as well as the hydrophobic interaction profiles of GRL-1388 and -1398 formed within the hydrophobic cavity of wild type HIV-1 protease, are shown along with those of DRV. The protease amino acid residues are listed, along with the numbers (in parentheses) of corresponding interactions. Note that GRL-1398 has relatively more hydrophobic interactions than GRL-1388, which should be one explanation for the greater potency of GRL-1398 against HIV-1. The two identical subunits that HIV-1 protease consists of were distinguished from each other by the use of a prime sign (′) at the upper right of each amino acid number.
DISCUSSION
We have previously designed and synthesized a series of PIs possessing a bis-THF moiety that interacts with the backbone atoms of the catalytic site amino acids, Asp29 and Asp30, of HIV-1 protease (1, 19, 32). In the present study, we report two newly generated PIs, GRL-1388 and -1398 (Fig. 1), containing a polycyclic ligand, Tp-THF, in place of the bis-THF moiety, which displayed potent anti-HIV-1 activity against wild-type HIV-1 and a wide spectrum of laboratory and primary multi-PI-resistant HIV-1 strains with favorable cytotoxicity profiles in vitro. Structurally, GRL-1388 has a methoxybenzene moiety and an aminobenzene moiety at the P1′ and P2′ sites, respectively, while GRL-1398 has a methoxybenzene moiety at each of the P1′ and P2′ sites, respectively. The activity of GRL-1388 against HIV-1LAI was comparable to or more potent than four representative FDA-approved PIs, SQV, APV, ATV, and DRV, whereas that of GRL-1398 was significantly more potent than these four PIs by factors of 16.5 to 98 (Table 1). It should be noted that the cytotoxicity of GRL-1398 is relatively greater, with a CC50 value of 37.7 μM and a SI of 188,500, compared to GRL-1388 with a CC50 value of >100 μM and an SI of >27,800. The mechanism of the relatively greater cytotoxicity of GRL-1398 compared to that of GRL-1388 is unknown at this time. Nevertheless, the CC50 values of two currently widely used FDA-approved PIs, SQV and ATV, were 19.7 and 27.6 μM, with their SI values of 3,200 and 5,500, respectively, when assessed under the same conditions together with GRL-1388 and -1398 (Table 1). Thus, even if GRL-1398 is relatively more cytotoxic than GRL-1388, one can assume that the level of toxicity of GRL-1398 is likely to be reasonably favorable, although the actual safety issue of GRL-1388 and -1398 has to be carefully examined in the setting of preclinical and clinical trials as needed.
GRL-1388 was also potently active against HIV-1 variants, which were selected to be resistant in vitro to each of five FDA-approved PIs, SQV, RTV, NFV, LPV, and ATV. GRL-1398 was significantly more potent against such five HIV-1variants with EC50s of as low as 0.1 to 5.7 nM (Table 2). It is noteworthy that both GRL-1388 and -1398 were moderately active against HIV-1APV-5μM, with EC50s of 475.7 and 49.0 nM, which is explained by the fact that the two compounds have a resemblance to APV, as does DRV (Fig. 1). Moreover, GRL-1388 was also potent against all six highly multi-PI-resistant clinical HIV-1 isolates examined with the observed fold differences between EC50s against a wild-type clinical isolate HIV-1ERS104pre and those against each clinical variant, ranging from as low as 1 to 7. The fold changes seen with GRL-1398 similarly ranged from 1 to 16; however, the absolute EC50s of GRL-1398 remained substantially low, ranging from 0.3 to 4.8 nM, compared to 3.7 to 21.4 nM for DRV (Table 3 and Fig. 2). These data strongly suggest that these two new PIs could serve as good candidates for further development as potential anti-HIV-1 therapeutics, but it was noted that, of the two compounds, GRL-1398 could be more promising since it is such a potent PI against both wild-type HIV-1 and various multi-PI-resistant HIV-1 variants.
We previously demonstrated that DRV effectively disrupts the dimerization of HIV-1 protease monomer subunits, as determined with a FRET/HIV-1 expression assay (18); this might explain the reason for the highly favorable clinical efficacy and high-level genetic barrier against HIV-1 acquisition of resistance to DRV in clinical settings (6, 21, 25). However, the protease dimerization inhibition activity of both GRL-1388 and -1398 was modest compared to that of DRV, suggesting that the protease dimerization activity of GRL-1388 and -1398 does not appear to significantly contribute to the potency of the two compounds. Indeed, none or only one of the set of four amino acid substitutions (V32I, L33F, I54M, and I84V), which appears to reduce the activity of DRV to disrupt HIV-1 protease dimerization (17), was seen in the protease of HIV-1 that replicated in the presence of high concentrations of GRL-1398, although both V32I and L33F were seen in HIVMIX-DRVWK19 and HIVMIX-DRVWK21 (see Fig. S2 in the supplemental material). Of interest, when HIV-1NL4-3 was propagated in the presence of GRL-1398, the A28S substitution was seen in two sets of selection experiments (Fig. 4 and 5; see Fig. S1 and S2 in the supplemental material), whereas A28S did not appear throughout the selection experiments with GRL-1388 or DRV. Although A28S substitution has been seen in HIV-1 selected with TMC-126 (36), GRL-98065 (1), and brecanavir (GW640385) (35), all of which contain the bis-THF moiety, the appearance of A28S has not been reported in the case of HIV-1 exposed to DRV in vitro (17) or in vivo (6, 24). Considering that both TMC-126 and GRL-1398 contain a methoxybenzene moiety but both DRV and GRL-1388 contain an aminobenzene moiety in the P2′ site, the presence of the methoxybenzene might be associated with the development of A28 substitution (Fig. 7b).
Amino acid substitutions in HIV-1 stoichiometrically and randomly occur, and the resulting amino acid substitutions in in vitro selection can vary from one experiment to another (34, 36). Thus, the results of a single selection experiment have to be confirmed with repeated selection experiments. In the present study, the selection experiment using HIV-1NL4-3 as a starting HIV-1 strain was conducted twice, and different profiles of amino acid substitutions were observed: the A28S substitution developed in the experiment II with GRL-1398, while it did not in the experiment I of the same compound (Fig. 4). Another selection experiment with GRL-1398 was planned; however, the selection experiment is, in general, labor-intensive and time-consuming. In fact, the selection experiment in the present study illustrated in Fig. 4 took more than 50 weeks each time. We thus performed the third selection experiment with a mixture of 10 CLHIV-1MDR variants as a starting virus population, expecting that highly GRL-1398-resistant HIV-1 variants would develop much sooner than when a single HIV-1 strain was used as a starting virus population since multiple CLHIV-1MDR variants would undergo homologous recombination and also acquire amino acid substations de novo under the pressure of GRL-1398, resulting in quicker development of GRL-1398 resistance. In fact, using a mixture of eight multi-PI resistant clinical isolates, we successfully selected a highly DRV-resistant HIV-1 variant by 51 passages (68 weeks), although our group (17) and other groups (5) had failed to select such a variant using a single strain as a starting virus population. Indeed, in the third selection experiment, HIV-1 variants that replicated in the presence of 5 μM GRL-1388, DRV, and GRL-1398 appeared in 17, 21, and 29 weeks of selection periods, respectively.
One can assume that the amino acid positions, where secondary and further mutations occur, are affected under the influence of primary mutations. In the selection assay starting with 10 CLHIV-1MDR isolates, certain HIV-1 isolates most likely had already possessed various sets of amino acid substitutions in protease, some of which might have readily given replication advantages to the resulting GRL-1388- and DRV-resistant variants but not to GRL-1398-resistant variant. This should explain why the emergence of GRL-1398 resistance-associated mutations was delayed compared to the viruses selected with GRL-1388 or DRV. Moreover, homologous recombination among these 10 CLHIV-1MDR isolates might have given certain advantages in the speed for gaining resistance.
HIV-1GRL1398-1μMExp.I was more susceptible to DRV and GRL-1388 than was HIV-1GRL1398-1μMExp.II (Table 5). This susceptibility difference should stem from the difference in the amino acid substitutions obtained differently by the two variant populations. Both populations contained L10F and M46I substitutions; however, the former additionally had four amino acid substitutions and the latter had two substitutions. Both populations had compromised replication fitness compared to wild-type HIVNL4-3, whereas both of them had a similarly significant advantages in replication in the presence of GRL-1398 (Fig. 6), proving that each set of amino acid substitutions conferred on the population significant levels of resistance to GRL-1398.
Modeling studies of GRL-1388 and -1398 docked against wild-type HIV-1 protease showed that the overall binding conformation of both compounds is similar (albeit not identical) to that of DRV. GRL-1398 shows more hydrogen bonds and hydrophobic interactions compared to GRL-1388. Structural analysis of GRL-1398 suggested that the potential extra hydrogen bonds with amide nitrogen atoms of Asp29′ and Asp30′ residues of the protease backbone could contribute to its greater potency. Based on this analysis, we postulate that these two hydrogen bonds with the backbone should be tolerant against mutations in the side chains from a wide spectrum of multidrug-resistant HIV-1 variants that we examined in comparison to either GRL-1388 or DRV. Due to the strong hydrogen bonds formed by GRL-1398 with the protease backbone, the overall binding profile of this compound is different from that of GRL-1388. This unique binding conformation of GRL-1398 is explained by the difference in the profiles of both GRL-1388 and -1398 with respect to their hydrophobic interactions in the protease active site.
Supplementary Material
Acknowledgments
We thank Matthew L. Danish for helpful discussion and carefully reading the manuscript.
This study was supported in part by the Intramural Research Program of Center for Cancer Research, National Cancer Institute, National Institutes of Health (D.D. and H.M.); a grant from the National Institutes of Health (GM53386 to A.K.G.); a Grant-in-Aid for Scientific Research (Priority Areas to H.M.) from the Ministry of Education, Culture, Sports, Science, and Technology (Monbu-Kagakusho) of Japan (H.M.); and a Grant for Promotion of AIDS Research from the Ministry of Health, Labor, and Welfare (Kosei-Rodosho) of Japan (H.M.). This study utilized the computational resources of the Biowulf cluster at the National Institutes of Health.
Footnotes
Published ahead of print on 31 January 2011.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Amano, M., et al. 2007. A novel bis-tetrahydrofuranylurethane-containing nonpeptidic protease inhibitor (PI), GRL-98065, is potent against multiple-PI-resistant human immunodeficiency virus in vitro. Antimicrob. Agents Chemother. 51:2143-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bastiaens, P. I., and T. M. Jovin. 1996. Microspectroscopic imaging tracks the intracellular processing of a signal transduction protein: fluorescent-labeled protein kinase C beta I. Proc. Natl. Acad. Sci. U. S. A. 93:8407-8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bastiaens, P. I., I. V. Majoul, P. J. Verveer, H. D. Soling, and T. M. Jovin. 1996. Imaging the intracellular trafficking and state of the AB5 quaternary structure of cholera toxin. EMBO J. 15:4246-4253. [PMC free article] [PubMed] [Google Scholar]
- 4.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D50:760-763. [DOI] [PubMed] [Google Scholar]
- 5.De Meyer, S., et al. 2005. TMC114, a novel human immunodeficiency virus type 1 protease inhibitor active against protease inhibitor-resistant viruses, including a broad range of clinical isolates. Antimicrob. Agents Chemother. 49:2314-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Meyer, S., et al. 2008. Resistance profile of darunavir: combined 24-week results from the POWER trials. AIDS Res. Hum. Retrovir. 24:379-388. [DOI] [PubMed] [Google Scholar]
- 7.Fang, G., B. Weiser, A. Visosky, T. Moran, and H. Burger. 1999. PCR-mediated recombination: a general method applied to construct chimeric infectious molecular clones of plasma-derived HIV-1 RNA. Nat. Med. 5:239-242. [DOI] [PubMed] [Google Scholar]
- 8.Friesner, R. A., et al. 2004. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 47:1739-1749. [DOI] [PubMed] [Google Scholar]
- 9.Friesner, R. A., et al. 2006. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 49:6177-6196. [DOI] [PubMed] [Google Scholar]
- 10.Gatanaga, H., et al. 2002. Amino acid substitutions in Gag protein at non-cleavage sites are indispensable for the development of a high multitude of HIV-1 resistance against protease inhibitors. J. Biol. Chem. 277:5952-5961. [DOI] [PubMed] [Google Scholar]
- 11.Ghosh, A. K., et al. 2008. Design and synthesis of stereochemically defined novel spirocyclic P2-ligands for HIV-1 protease inhibitors. Org. Lett. 10:5135-5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh, A. K., et al. 2008. Flexible cyclic ethers/polyethers as novel P2-ligands for HIV-1 protease inhibitors: design, synthesis, biological evaluation, and protein-ligand X-ray studies. J. Med. Chem. 51:6021-6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghosh, A. K., et al. 2010. Synthesis and biological evaluation of novel allophenylnorstatine-based HIV-1 protease inhibitors incorporating high-affinity P2-ligands. Bioorg. Med. Chem. Lett. 20:1241-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh, A. K., et al. 2008. Potent HIV-1 protease inhibitors incorporating meso-bicyclic urethanes as P2-ligands: structure-based design, synthesis, biological evaluation and protein-ligand X-ray studies. Org. Biomol. Chem. 6:3703-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh, A. K., et al. 1998. Potent HIV protease inhibitors incorporating high-affinity P2-ligands and (R)-(hydroxyethylamino)sulfonamide isostere. Bioorg. Med. Chem. Lett. 8:687-690. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh, A. K., S. Leshchenko, and M. Noetzel. 2004. Stereoselective photochemical 1,3-dioxolane addition to 5-alkoxymethyl-2(5H)-furanone: synthesis of bis-tetrahydrofuranyl ligand for HIV protease inhibitor UIC-94017 (TMC-114). J. Org. Chem. 69:7822-7829. [DOI] [PubMed] [Google Scholar]
- 17.Koh, Y., et al. 2010. In vitro selection of highly darunavir-resistant and replication-competent HIV-1 variants by using a mixture of clinical HIV-1 isolates resistant to multiple conventional protease inhibitors. J. Virol. 84:11961-11969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koh, Y., et al. 2007. Potent inhibition of HIV-1 replication by novel non-peptidyl small molecule inhibitors of protease dimerization. J. Biol. Chem. 282:28709-28720. [DOI] [PubMed] [Google Scholar]
- 19.Koh, Y., et al. 2003. Novel bis-tetrahydrofuranylurethane-containing nonpeptidic protease inhibitor (PI) UIC-94017 (TMC114) with potent activity against multi-PI-resistant human immunodeficiency virus in vitro. Antimicrob. Agents Chemother. 47:3123-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lohse, N., et al. 2007. Survival of persons with or without HIV infection in Denmark, 1995-2005. Ann. Intern. Med. 146:87-95. [DOI] [PubMed] [Google Scholar]
- 21.Madruga, J. V., et al. 2007. Efficacy and safety of darunavir-ritonavir compared with that of lopinavir-ritonavir at 48 weeks in treatment-experienced, HIV-infected patients in TITAN: a randomised controlled phase III trial. Lancet 370:49-58. [DOI] [PubMed] [Google Scholar]
- 22.Maeda, K., et al. 2001. Novel low-molecular-weight spirodiketopiperazine derivatives potently inhibit R5 HIV-1 infection through their antagonistic effects on CCR5. J. Biol. Chem. 276:35194-35200. [DOI] [PubMed] [Google Scholar]
- 23.Miller, J. F., et al. 2006. Ultra-potent P1 modified arylsulfonamide HIV protease inhibitors: the discovery of GW0385. Bioorg. Med. Chem. Lett. 16:1788-1794. [DOI] [PubMed] [Google Scholar]
- 24.Mitsuya, Y., T. F. Liu, S. Y. Rhee, W. J. Fessel, and R. W. Shafer. 2007. Prevalence of darunavir resistance-associated mutations: patterns of occurrence and association with past treatment. J. Infect. Dis. 196:1177-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ortiz, R., et al. 2008. Efficacy and safety of once-daily darunavir/ritonavir versus lopinavir/ritonavir in treatment-naive HIV-1-infected patients at week 48. AIDS 22:1389-1397. [DOI] [PubMed] [Google Scholar]
- 26.Reference deleted.
- 27.Sekar, R. B., and A. Periasamy. 2003. Fluorescence resonance energy transfer (FRET) microscopy imaging of live cell protein localizations. J. Cell Biol. 160:629-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shirasaka, T., et al. 1995. Emergence of human immunodeficiency virus type 1 variants with resistance to multiple dideoxynucleosides in patients receiving therapy with dideoxynucleosides. Proc. Natl. Acad. Sci. U. S. A. 92:2398-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szczesna-Skorupa, E., B. Mallah, and B. Kemper. 2003. Fluorescence resonance energy transfer analysis of cytochromes P450 2C2 and 2E1 molecular interactions in living cells. J. Biol. Chem. 278:31269-31276. [DOI] [PubMed] [Google Scholar]
- 30.Tamiya, S., S. Mardy, M. F. Kavlick, K. Yoshimura, and H. Mistuya. 2004. Amino acid insertions near Gag cleavage sites restore the otherwise compromised replication of human immunodeficiency virus type 1 variants resistant to protease inhibitors. J. Virol. 78:12030-12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tie, Y., et al. 2004. High resolution crystal structures of HIV-1 protease with a potent non-peptide inhibitor (UIC-94017) active against multidrug-resistant clinical strains. J. Mol. Biol. 338:341-352. [DOI] [PubMed] [Google Scholar]
- 32.Tojo, Y., et al. 2010. Novel protease inhibitors (PIs) containing macrocyclic components and 3(R),3a(S),6a(R)-bis-tetrahydrofuranylurethane that are potent against multi-PI-resistant HIV-1 variants in vitro. Antimicrob. Agents Chemother. 54:3460-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walensky, R. P., et al. 2006. The survival benefits of AIDS treatment in the United States. J. Infect. Dis. 194:11-19. [DOI] [PubMed] [Google Scholar]
- 34.Watkins, T., W. Resch, D. Irlbeck, and R. Swanstrom. 2003. Selection of high-level resistance to human immunodeficiency virus type 1 protease inhibitors. Antimicrob. Agents Chemother. 47:759-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yates, P. J., et al. 2006. In vitro development of resistance to human immunodeficiency virus protease inhibitor GW640385. Antimicrob. Agents Chemother. 50:1092-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshimura, K., et al. 2002. A potent human immunodeficiency virus type 1 protease inhibitor, UIC-94003 (TMC-126), and selection of a novel (A28S) mutation in the protease active site. J. Virol. 76:1349-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshimura, K., et al. 1999. JE-2147: a dipeptide protease inhibitor (PI) that potently inhibits multi-PI-resistant HIV-1. Proc. Natl. Acad. Sci. U. S. A. 96:8675-8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.