Abstract
The in vitro activities of modithromycin against Gram-positive and -negative respiratory pathogens, including macrolide-resistant cocci with different resistance mechanisms, were compared with those of other macrolide and ketolide agents. MICs were determined by the broth microdilution method. All 595 test strains used in this study were isolated from Japanese medical facilities. The erm (ribosome methylase) and/or mef (efflux pump) gene, which correlated with resistance to erythromycin as well as clarithromycin and azithromycin, was found in 81.8%, 21.3%, and 23.2% of Streptococcus pneumoniae, Streptococcus pyogenes, and methicillin-susceptible Staphylococcus aureus (MSSA) strains, respectively. Modithromycin showed MIC90s of 0.125 μg/ml against these three cocci, including macrolide-resistant strains. In particular, the MIC of modithromycin against ermB-carrying S. pyogenes was ≥32-fold lower than that of telithromycin. The activities of modithromycin as well as telithromycin were little affected by the presence of mefA or mefE in both streptococci. Against Gram-negative pathogens, modithromycin showed MIC90s of 0.5, 8, and 0.031 μg/ml against Moraxella catarrhalis, Haemophilus influenzae, and Legionella spp., respectively. The MICs of modithromycin against M. catarrhalis and H. influenzae were higher than those of telithromycin and azithromycin. However, modithromycin showed the most potent anti-Legionella activity among the macrolide and ketolide agents tested. These results suggested that the bicyclolide agent modithromycin is a novel class of macrolides with improved antibacterial activity against Gram-positive cocci, including telithromycin-resistant streptococci and intracellular Gram-negative bacteria of the Legionella species.
Macrolides such as clarithromycin and azithromycin are antimicrobial agents for the treatment of mild to moderate community-acquired pneumonia and are clinically useful for pediatric and penicillin-allergic patients (2, 19). However, the rate of macrolide resistance among Gram-positive cocci, particularly Streptococcus pneumoniae, has been increasing throughout the world (12, 16). Therefore, the choice of these agents for use in empirical treatment has become complicated. In addition, although the ketolide agent telithromycin was shown to be active even against macrolide-resistant clinical strains of S. pneumoniae, the emergence of high-level telithromycin-resistant strains has also been reported recently (1, 10, 29).
Target site modifications and active efflux pumps are known to be the predominant macrolide resistance mechanisms in streptococci and staphylococci (17, 27, 28). As for the target site modification, some methylase enzymes encoded by the erm genes induce dimethylation of an adenine residue in domain V, the peptidyltransferase site, of a 23S ribosomal subunit, which results in reduced binding affinity to macrolide, lincosamide (clindamycin), and streptogramin B (MLSB) antibiotics. The erm-induced resistance can be classified into inducible and constitutive phenotypes. The efflux pumps encoded by the mef and msr genes have been found in clinical strains of streptococci and staphylococci, respectively (17, 28). The strains carrying mef genes are resistant to low or moderate levels of 14- and 15-membered macrolides but are not resistant to 16-membered macrolides, clindamycin, streptogramin B, or the ketolide telithromycin. On the other hand, the Msr efflux pumps encoded by msr genes confer resistance to 14- and 15-membered macrolides and streptogramin B and also, in some cases, to clindamycin and telithromycin (24). In addition, mutations in L4 or L22 ribosomal proteins, either alone or in combination with the presence of erm genes or 23S rRNA mutations, have been reported to confer reduced susceptibility to macrolides and, to a lesser extent, telithromycin in streptococci (1, 28, 29).
Modithromycin (EDP-420, EP-013420, S-013420), a novel 6,11-bridged bicyclolide, was designed and synthesized by Enanta Pharmaceuticals, Inc., to potentiate the antibacterial activity against Gram-positive and -negative respiratory pathogens (Fig. 1) (28). In this study, we examined the in vitro activity of modithromycin against clinical strains of Gram-positive cocci, including macrolide-resistant strains with different macrolide resistance mechanisms, and Gram-negative organisms, including intracellular pathogens.
FIG. 1.
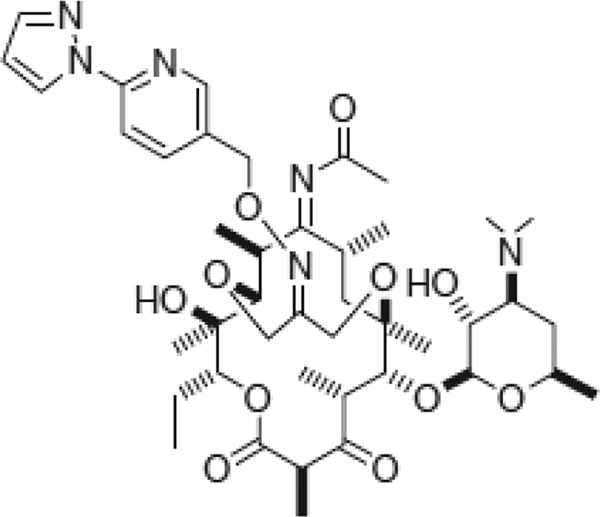
Chemical structure of modithromycin (EDP-420, EP-013420, S-013420).
(This work was presented in part at the 46th Interscience Conference on Antimicrobial Agents and Chemotherapy, abstr. E-1856, San Francisco, CA, 2006, and the 47th Interscience Conference on Antimicrobial Agents and Chemotherapy, abstr. E-1628, Chicago, IL, 2007.)
MATERIALS AND METHODS
Antimicrobial agents.
Modithromycin and telithromycin were kindly supplied from Shionogi & Co., Ltd. (Osaka, Japan). Clarithromycin, azithromycin, erythromycin, and clindamycin were purchased from U.S. Pharmacopeia (Rockville, MD). Levofloxacin was kindly supplied from Daiichi Sankyo Co., Ltd. (Tokyo, Japan). The solvent for modithromycin was 1 M acetate buffer (pH 4.5).
Bacteria.
A total of 534 clinical strains, including S. pneumoniae (148 strains), Streptococcus pyogenes (94 strains), methicillin-susceptible Staphylococcus aureus (MSSA; 99 strains), Haemophilus influenzae (93 strains), and Moraxella catarrhalis (100 strains), were isolated at Toho University Omori Hospital in 2006. Sixty-one clinical strains of Legionella spp., including Legionella pneumophila (56 strains), Legionella longbeachae (2 strains), Legionella sainthelensi (1 strain), Legionella micdadei (1 strain), and an unidentified Legionella species (1 strain), were collected from 34 Japanese medical facilities from 1994 to 2006.
Determination of MICs.
MICs were determined by the broth microdilution method recommended by the Clinical and Laboratory Standards Institute (CLSI) (7). Cation-adjusted Mueller-Hinton broth (Difco) was used for MSSA and M. catarrhalis. For streptococci, cation-adjusted Mueller-Hinton broth supplemented with 5% lysed horse blood was used. For H. influenzae, Haemophilus test medium (Nissui Pharmaceutical Co., Ltd., Japan) was used. For Legionella spp., N-(2-acetamido)-2-aminoethanesulfonic acid-buffered yeast extract broth supplemented with 0.1% α-ketoglutarate, 0.04% l-cysteine, and 0.025% iron(III) diphosphate (final pH, 6.9) was used (22, 26). After inoculation of the bacteria into the broth medium containing serial dilutions of the antimicrobial agents in a 96-well microplate (inoculum, approximately 5 × 105 CFU/ml), incubation was performed for 20 h at 35°C for all organisms except Legionella species. For Legionella spp., incubation was performed for 67 h at 35°C. The MIC was defined as the lowest concentration of antimicrobial agent which inhibited visible growth of bacteria. Interpretation of MICs as susceptible, intermediate resistant, or resistant was determined on the basis of the CLSI breakpoints (8).
Determination of macrolide-resistant genotypes and phenotypes.
Macrolide resistance genes were detected by using a conventional PCR protocol. For streptococci, three sets of primers for ermB, ermTR, and mefA (mefE) were used (13, 15). For MSSA, four sets of primers for ermA, ermB, ermC, and msrA (msrB) were used (21, 23). Among erythromycin-resistant strains of MSSA carrying the erm genes, the strains resistant to clindamycin were judged to have the constitutive resistance phenotype of methylase expression. Other erythromycin-resistant erm-carrying MSSA and ermTR-carrying streptococci, which were susceptible to clindamycin, were judged to have the inducible resistance phenotype of methylase expression because a D-shaped inhibition zone around the clindamycin disk was observed by the double-disk diffusion test on Todd-Hewitt agar with clindamycin and erythromycin disks (17).
RESULTS
Antibacterial activity of modithromycin against clinical strains.
The activities of modithromycin and the reference antimicrobial agents against clinical strains are shown in Table 1. The MIC90 of modithromycin was 0.125 μg/ml against S. pneumoniae, which was comparable to that of telithromycin. On the other hand, against S. pneumoniae, clarithromycin and azithromycin showed MIC50s of 32 and >64 μg/ml, respectively. Modithromycin showed a MIC90 of 0.125 μg/ml against S. pyogenes, which was 4-fold lower than that of telithromycin and 64-fold lower than the MIC90s of clarithromycin and azithromycin. Modithromycin inhibited the growth of all strains of these streptococci at a concentration of 0.5 μg/ml, whereas telithromycin did so at a concentration of 8 μg/ml. Against MSSA, the activity of modithromycin was comparable to that of telithromycin, with a MIC90 of 0.125 μg/ml, although some strains showed high-level resistance to modithromycin and telithromycin, with MICs of >64 μg/ml. However, modithromycin was much more active against MSSA than clarithromycin and azithromycin, which showed MIC90s of >64 μg/ml. Compared to levofloxacin, the MIC90s of modithromycin were 4- to 16-fold lower against these Gram-positive cocci.
TABLE 1.
Antibacterial activities of modithromycin and reference antimicrobial agents against clinical strains of Gram-positive and -negative organisms
| Organism (no. of strains) | Test agent | MIC (μg/ml) |
||
|---|---|---|---|---|
| Range | 50% | 90% | ||
| Gram-positive organisms | ||||
| Streptococcus pneumoniae (148) | Modithromycin | ≤0.063-0.5 | ≤0.063 | 0.125 |
| Telithromycin | ≤0.063-2 | ≤0.063 | 0.125 | |
| Clarithromycin | ≤0.063->64 | 32 | >64 | |
| Azithromycin | ≤0.063->64 | >64 | >64 | |
| Erythromycin | ≤0.063->64 | >64 | >64 | |
| Levofloxacin | 0.5-1 | 0.5 | 1 | |
| Streptococcus pyogenes (94) | Modithromycin | ≤0.063-0.25 | ≤0.063 | 0.125 |
| Telithromycin | ≤0.063-8 | ≤0.063 | 0.5 | |
| Clarithromycin | ≤0.063->64 | ≤0.063 | 8 | |
| Azithromycin | ≤0.063->64 | ≤0.063 | 8 | |
| Erythromycin | ≤0.063->64 | ≤0.063 | 16 | |
| Levofloxacin | 0.25-2 | 0.5 | 2 | |
| MSSA (99) | Modithromycin | ≤0.063->64 | 0.125 | 0.125 |
| Telithromycin | ≤0.063->64 | ≤0.063 | 0.125 | |
| Clarithromycin | ≤0.063->64 | 0.25 | >64 | |
| Azithromycin | 0.25->64 | 0.5 | >64 | |
| Erythromycin | 0.125->64 | 0.25 | >64 | |
| Levofloxacin | ≤0.063-16 | 0.25 | 0.5 | |
| Gram-negative organisms | ||||
| Moraxella catarrhalis (100) | Modithromycin | ≤0.063-1 | 0.25 | 0.5 |
| Telithromycin | ≤0.063-0.25 | 0.125 | 0.25 | |
| Clarithromycin | ≤0.063-0.5 | 0.125 | 0.25 | |
| Azithromycin | ≤0.063-0.125 | ≤0.063 | ≤0.063 | |
| Erythromycin | ≤0.063-1 | 0.125 | 0.5 | |
| Levofloxacin | ≤0.063-1 | ≤0.063 | ≤0.063 | |
| Haemophilus influenzae (93) | Modithromycin | 1-16 | 4 | 8 |
| Telithromycin | 0.5-4 | 1 | 2 | |
| Clarithromycin | 2-32 | 4 | 16 | |
| Azithromycin | 0.125-4 | 1 | 2 | |
| Erythromycin | 1-16 | 4 | 8 | |
| Levofloxacin | ≤0.063 | ≤0.063 | ≤0.063 | |
| Legionella pneumophila (56) and other Legionella spp. (5) | Modithromycin | 0.016-0.25 | 0.031 | 0.031 |
| Telithromycin | 0.031-0.25 | 0.125 | 0.125 | |
| Clarithromycin | 0.016-0.125 | 0.031 | 0.063 | |
| Azithromycin | 0.063-2 | 0.125 | 1 | |
| Erythromycin | 0.125-2 | 0.25 | 1 | |
| Levofloxacin | 0.016-0.063 | 0.031 | 0.031 | |
Against M. catarrhalis and H. influenzae, modithromycin showed MIC90s of 0.5 and 8 μg/ml, respectively, which were higher than those of telithromycin and azithromycin but comparable to the MIC90 of clarithromycin. On the other hand, modithromycin showed the most potent anti-Legionella activity, with a MIC90 of 0.031 μg/ml. The MIC90 of modithromycin was 4- and 32-fold lower than the MIC90s of telithromycin and azithromycin, respectively, and was comparable to that of levofloxacin.
Evaluation of macrolide-resistant genotypes and phenotypes in clinical strains of Gram-positive cocci.
The determination of macrolide-resistant genotypes in Gram-positive cocci was performed by conventional PCR to evaluate the differences in the activities between modithromycin and the other macrolide/ketolide agents tested. Among the 148 clinical strains of S. pneumoniae, the ermB and mefE genes were found in 56.8% and 30.4%, respectively (including 5.4% of the strains carrying both genes), whereas ermTR was not detected in any strain. Among the 94 clinical strains of S. pyogenes, the ermB, ermTR, and mefA genes were found in 4.3%, 2.1%, and 14.9%, respectively. Among the 99 clinical strains of MSSA, the ermA, ermB, and ermC genes were found in 17.1%, 1.0%, and 5.1%, respectively, but in none of the strains carrying the msrA (msrB) gene.
All of these erm-carrying cocci were resistant to erythromycin (MICs, ≥1 μg/ml for streptococci and ≥8 μg/ml for MSSA, according to the CLSI breakpoints). Among the erm-carrying strains, 2 out of 2 ermTR-carrying S. pyogenes strains, 13 out of 17 ermA-carrying MSSA strains, and 5 out of 5 ermC-carrying MSSA strains were identified as being of the inducible resistance phenotype, because they were susceptible to clindamycin and showed a D-shaped inhibition zone in the double-disk diffusion test (Tables 2 to 4 ). All other erm-carrying strains showed resistance to clindamycin. None of the erythromycin-susceptible strains of streptococci and MSSA possessed any of these macrolide resistance genes, but it was interesting that one of the erythromycin-susceptible MSSA strains showed reduced susceptibility to clindamycin, with a MIC of 8 μg/ml.
TABLE 2.
Comparison of susceptibilities of various macrolide-resistant strains of S. pneumoniae
| Test agent | None (n = 27) |
ermB (n = 76) |
mefE (n = 37) |
ermB + mefE (n = 8) |
||||
|---|---|---|---|---|---|---|---|---|
| MIC range (μg/ml) | % I and Ra | MIC range (μg/ml) | % I and R | MIC range (μg/ml) | % I and R | MIC range (μg/ml) | % I and R | |
| Modithromycin | ≤0.063 | NC | ≤0.063-0.5 | NC | ≤0.063-0.5 | NC | 0.125-0.5 | NC |
| Telithromycin | ≤0.063 | 0 | ≤0.063-2 | 1.3 | ≤0.063-0.5 | 0 | ≤0.063-0.5 | 0 |
| Clarithromycin | ≤0.063 | 0 | 1->64 | 100 | 0.25-8 | 91.9 | >64 | 100 |
| Azithromycin | ≤0.063-0.25 | 0 | 4->64 | 100 | 0.25-8 | 86.5 | >64 | 100 |
| Clindamycin | ≤0.063 | 0 | 2->64 | 100 | ≤0.063 | 0 | 64->64 | 100 |
The percentages of intermediate-resistant (I) and resistant (R) strains are shown. The CLSI breakpoint criteria are as follows: telithromycin, 2 and ≥4 μg/ml; clarithromycin, 0.5 and ≥1 μg/ml; azithromycin, 1 and ≥2 μg/ml; clindamycin, 0.5 and ≥1 μg/ml. NC, not calculated (the breakpoints of modithromycin have not been determined).
Comparison of susceptibilities of various macrolide-resistant strains of Gram-positive cocci.
All the erm gene-carrying strains of S. pneumoniae and S. pyogenes showed intermediate resistance or resistance to clarithromycin and azithromycin. However, modithromycin showed MICs of ≤0.5 μg/ml against both erm-carrying streptococci (Tables 2 and 3). Although telithromycin showed potent activity against almost all the ermB-carrying S. pneumoniae strains, one of the strains, which showed susceptibility to modithromycin with a MIC of 0.25 μg/ml, had intermediate resistance to telithromycin, with a MIC of 2 μg/ml. In addition, the ermB-carrying S. pyogenes strains were not susceptible to telithromycin, with MICs of 4 to 8 μg/ml; therefore, the activity of modithromycin was 32-fold or more potent than that of telithromycin against these strains. On the other hand, the activities of modithromycin and telithromycin were little affected by the presence of the Mef efflux pumps, encoded by mefA or mefE, in both streptococci, while it caused intermediate resistance or resistance to clarithromycin and azithromycin. Modithromycin as well as telithromycin and clindamycin had potent activity against MSSA with the inducible resistance phenotype of ermA and ermC, while clarithromycin and azithromycin did not (Table 4). However, MSSA with the constitutive resistance phenotype of ermA and ermB showed high-level resistance to modithromycin and telithromycin as well as other macrolide agents and clindamycin.
TABLE 3.
Comparison of susceptibilities of various macrolide-resistant strains of S. pyogenes
| Test agent | None (n = 74) |
ermTR (n = 2) |
ermB (n = 4) |
mefA (n = 14) |
||||
|---|---|---|---|---|---|---|---|---|
| MIC range (μg/ml) | % I and Ra | MIC range (μg/ml) | % I and R | MIC range (μg/ml) | % I and R | MIC range (μg/ml) | % I and R | |
| Modithromycin | ≤0.063 | NC | ≤0.063 | NC | ≤0.063-0.25 | NC | ≤0.063-0.25 | NC |
| Telithromycin | ≤0.063 | NC | ≤0.063 | NC | 4-8 | NC | ≤0.063-0.5 | NC |
| Clarithromycin | ≤0.063 | 0 | 0.5-1 | 100 | >64 | 100 | 0.5-8 | 100 |
| Azithromycin | ≤0.063-1.25 | 0 | 4-8 | 100 | >64 | 100 | 2-8 | 100 |
| Clindamycin | ≤0.063 | 0 | ≤0.063 | 0 | 16->64 | 100 | ≤0.063 | 0 |
The percentages of intermediate-resistant (I) and resistant (R) strains are shown. The CLSI breakpoint criteria are as follows: clarithromycin, 0.5 and ≥1 μg/ml; azithromycin, 1 and ≥2 μg/ml; clindamycin, 0.5 and ≥1 μg/ml. NC, not calculated (the breakpoints of modithromycin and telithromycin have not been determined).
TABLE 4.
Comparison of susceptibilities of various macrolide-resistant strains of MSSA
| Test agent | None (n = 76) |
ermA (i)a (n = 13) |
ermA (c)b (n = 4) |
ermB (c) (n = 1) |
ermC (i) (n = 5) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| MIC range (μg/ml) | % I and Rc | MIC range (μg/ml) | % I and R | MIC range (μg/ml) | % I and R | MIC range (μg/ml) | % I and R | MIC range (μg/ml) | % I and R | |
| Modithromycin | ≤0.063-0.125 | NC | ≤0.063-0.125 | NC | 16->64 | NC | >64 | NC | 0.125 | NC |
| Telithromycin | ≤0.063-0.125 | 0 | ≤0.063-0.125 | 0 | >64 | 100 | >64 | 100 | ≤0.063-0.125 | 0 |
| Clarithromycin | ≤0.063-0.25 | 0 | 64->64 | 100 | >64 | 100 | >64 | 100 | >64 | 100 |
| Azithromycin | 0.25-1 | 0 | >64 | 100 | >64 | 100 | >64 | 100 | >64 | 100 |
| Clindamycin | ≤0.063-8 | 1.3 | ≤0.063-0.125 | 0 | >64 | 100 | >64 | 100 | ≤0.063-0.25 | 0 |
i, inducible resistance phenotype.
c, constitutive resistance phenotype.
The percentages of intermediate-resistant (I) and resistant (R) strains are shown. The CLSI breakpoint criteria are as follows: telithromycin, 2 and ≥4 μg/ml; clarithromycin, 4 and ≥8 μg/ml; azithromycin, 4 and ≥8 μg/ml; clindamycin, 1 to 2 and ≥4 μg/ml. NC, not calculated (the breakpoints of modithromycin have not been determined).
DISCUSSION
During the last few decades, the incidence of clinical strains of macrolide-resistant Gram-positive cocci has been increasing worldwide. The prevalence rate of resistant organisms varies considerably among countries, presumably because the use of these antimicrobial agents also varies among countries (3, 4, 5, 6). In the PROTEKT (Prospective Resistant Organism Tracking and Epidemiology for the Ketolide Telithromycin) study, clinical strains of S. pneumoniae resistant to erythromycin accounted for 24.2% of S. pneumoniae strains in the United Kingdom, 34.7% in the United States, 53.1% in France, 81.9% in Japan, and 97.6% in Taiwan (11). In the present study, approximately 80% of S. pneumoniae strains collected from a Japanese medical facility had the macrolide resistance genes of ermB and/or mefE and showed resistance to macrolide agents, such as clarithromycin and azithromycin. The frequency of the resistant strains was similar to the results of a previous surveillance study in Japan (14). All of the macrolide-resistant streptococci in this study had one or more genes that have been reported to cause resistance to macrolide agents, so there seemed to be no resistant strains with unknown mechanisms of resistance. The predominant resistance genotype in S. pneumoniae strains was ermB rather than mefE in this study. It has also been reported that ermB is the most predominant resistance genotype among clinical strains in Japan and most European countries (12, 14). We did not observe the msr gene, which is plasmid mediated, in any of the MSSA strains tested in this study. This observation was consistent with the previous report on msr being prevalent in coagulase-negative staphylococci, such as Staphylococcus epidermidis and Staphylococcus haemolyticus (17, 18, 21). One erythromycin-susceptible MSSA strain showed resistance to clindamycin, with an MIC of 8 μg/ml. Because no cross-resistance between clindamycin and the macrolide agents tested was observed, clindamycin resistance mechanisms such as those involving the mph and lnu genes, encoding a lincosamide inactivator, may exist in this MSSA isolate (17).
To overcome the macrolide resistance mechanisms, including target site modification and active efflux, the ketolide antimicrobial agent telithromycin was developed (20, 28). Telithromycin has unique structural features, such as an additional aromatic side chain, which enhances the affinity of binding to the ribosomal domain II as well as domain V, and the C-3 ketone, due to removal of the cladinose sugar responsible for the inducibility of macrolide resistance and the possibility of macrolide recognition by the mef efflux pump. A novel bicyclolide, modithromycin, which has a unique 6,11-bridged ether ring and aromatic side chain, in addition to the C-3 ketone, was also designed to improve the antibacterial activity against macrolide-resistant organisms (28). Our results revealed that modithromycin has excellent antibacterial activity against all clinical strains of S. pneumoniae, including macrolide-resistant ermB- and/or mefE-carrying strains. In addition, modithromycin was also active against one ermB-carrying strain which showed intermediate resistance to telithromycin, with an MIC of 2 μg/ml. The telithromycin resistance mechanism of this strain has not been revealed yet; however, it has been reported that specific mutations of 23S rRNA, ribosomal protein, or the ermB upstream region cause intermediate or high-level resistance to telithromycin in S. pneumoniae (1, 17, 28). Modithromycin also showed activity against both ermB- and ermTR-carrying S. pyogenes strains. In particular, modithromycin was 32- to >64-fold more active than telithromycin against ermB-carrying strains. Douthwaite et al. have reported that the telithromycin resistance in S. pyogenes correlates with the degree of rRNA dimethylation by Erm (9). These findings demonstrated that the bicyclolide nucleus and aromatic side chain of modithromycin would contribute to the increased affinity for the dimethylated ribosome of streptococci, including telithromycin-resistant strains. Unlike the Erm phenotype in streptococci, except for ErmTR, the expression of the Erm phenotype can be determined to be inducible or constitutive in S. aureus by the inducibility test using erythromycin and clindamycin (17, 27). Against the ermA-, ermB-, or ermC-carrying MSSA strains, clarithromycin and azithromycin were inactive regardless of the inducible or constitutive resistance phenotypes. Modithromycin, as well as telithromycin and clindamycin, was active against ermA- and ermC-carrying strains with the inducible resistance phenotype but was not active against ermA- and ermB-carrying strains with the constitutive resistance phenotype. Although it is not fully understood why the activity of modithromycin against ermB-carrying strains differed between streptococci and MSSA, we speculate that possible reasons may be differences in the degree of rRNA dimethylation or in the ribosomal region of modithromycin binding.
Our results also revealed the potent antibacterial activity of modithromycin against Gram-negative respiratory pathogens, such as M. catarrhalis, H. influenzae, and Legionella species. In particular, the anti-Legionella activity of modithromycin was more potent than the activities of the other macrolide and ketolide agents tested and comparable to that of levofloxacin. Legionella is a facultative intracellular pathogen that causes life-threatening pneumonia. Therefore, efficient penetration of the antimicrobial agents into cells at the site of infection is required for therapeutic efficacy against this pathogen. We have already evaluated the in vivo efficacy of modithromycin in a murine model of L. pneumophila pneumonia and found it to be excellent (25), indicating that modithromycin seemed to have preferable pharmacokinetic features for efficacy, in addition to its potent in vitro activity.
In summary, the novel bicyclolide modithromycin was shown to possess a broad spectrum of in vitro activity against both Gram-positive and -negative pathogens that cause respiratory tract infections. In particular, modithromycin was shown to have advantages over currently available macrolide and ketolide agents because it has potent activity against the clinically important macrolide-resistant Gram-positive cocci that have spread worldwide and Gram-negative intracellular bacteria of the genus Legionella.
Acknowledgments
We thank H. Murakami, K. Sugihara, R. Shibuya, A. Ohno, S. Miyazaki, H. Maki, M. Tsuji, H. Miwa, and Y. Yamano for experimental technical support and/or helpful discussion.
T. Sato is an employee of Shionogi & Co., Ltd., but does not have stock or options. We have no other possible conflicts of interest to report.
This work was supported by Shionogi & Co., Ltd.
Footnotes
Published ahead of print on 10 January 2011.
REFERENCES
- 1.Al-Lahham, A., P. C. Appelbaum, M. van der Linden, and R. R. Reinert. 2006. Telithromycin-nonsusceptible clinical isolates of Streptococcus pneumoniae from Europe. Antimicrob. Agents Chemother. 50:3897-3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkinson, M., M. Yanney, T. Stephenson, and A. Smyth. 2007. Effective treatment strategies for paediatric community-acquired pneumonia. Expert Opin. Pharmacother. 8:1091-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baquero, F., G. Baquero-Artigao, R. Cantón, and C. García-Rey. 2002. Antibiotic consumption and resistance selection in Streptococcus pneumoniae. J. Antimicrob. Chemother. 50(Suppl. 2):S27-S37. [DOI] [PubMed] [Google Scholar]
- 4.Bergman, M., et al. 2006. Macrolide and azithromycin use are linked to increased macrolide resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 50:3646-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Čižman, M., M. Pokorn, K. Seme, A. Oražem, and M. Paragi. 2001. The relationship between trends in macrolide use and resistance to macrolides of common respiratory pathogens. J. Antimicrob. Chemother. 47:475-477. [DOI] [PubMed] [Google Scholar]
- 6.Čižman, M., M. Pokorn, K. Seme, M. Paragi, and A. Oražem. 1999. Influence of increased macrolide consumption on macrolide resistance of common respiratory pathogens. Eur. J. Clin. Microbiol. Infect. Dis. 18:522-524. [DOI] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 7th ed. Approved standard M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 8.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing, 16th informational supplement M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA.
- 9.Douthwaite, S., J. Jalava, and L. Jakobsen. 2005. Ketolide resistance in Streptococcus pyogenes correlates with the degree of rRNA dimethylation by Erm. Mol. Microbiol. 58:613-622. [DOI] [PubMed] [Google Scholar]
- 10.Farrell, D. J., et al. 2005. Emergence and spread of Streptococcus pneumoniae with erm(B) and mef(A) resistance. Emerg. Infect. Dis. 11:851-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farrell, D. J., C. Couturier, and W. Hryniewicz. 2008. Distribution and antibacterial susceptibility of macrolide resistance genotypes in Streptococcus pneumoniae: PROTEKT Year 5 (2003-2004). Int. J. Antimicrob. Agents 31:245-249. [DOI] [PubMed] [Google Scholar]
- 12.Felmingham, D., R. Cantón, and S. G. Jenkins. 2007. Regional trends in β-lactam, macrolide, fluoroquinolone and telithromycin resistance among Streptococcus pneumoniae isolates 2001-2004. J. Infect. 55:111-118. [DOI] [PubMed] [Google Scholar]
- 13.Green, N. M., et al. 2005. Genetic diversity among type emm28 group A Streptococcus strains causing invasive infections and pharyngitis. J. Clin. Microbiol. 43:4083-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inoue, M., et al. 2008. Antimicrobial susceptibility of respiratory tract pathogens in Japan during PROTEKT years 1-5 (1999-2004). Microb. Drug Resist. 14:109-117. [DOI] [PubMed] [Google Scholar]
- 15.Kataja, J., P. Huovinen, M. Skurnik, the Finnish Study Group for Antimicrobial Resistance, and H. Seppälä. 1999. Erythromycin resistance genes in group A streptococci in Finland. Antimicrob. Agents Chemother. 43:48-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klugman, K. P. 2007. Clinical impact of antibiotic resistance in respiratory tract infections. Int. J. Antimicrob. Agents 29(Suppl. 1):S6-S10. [DOI] [PubMed] [Google Scholar]
- 17.Leclercq, R. 2002. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 34:482-492. [DOI] [PubMed] [Google Scholar]
- 18.Lina, G., et al. 1999. Distribution of genes encoding resistance to macrolides, lincosamides and streptogramins among staphylococci. Antimicrob. Agents Chemother. 43:1062-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Low, D. E. 2008. Treatment of community-acquired pneumonia. CMAJ 179:1245-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma, Z., et al. 2001. Novel erythromycin derivatives with aryl groups tethered to the C-6 position are potent protein synthesis inhibitors and active against multidrug-resistant respiratory pathogens. J. Med. Chem. 44:4137-4156. [DOI] [PubMed] [Google Scholar]
- 21.Martineau, F., et al. 2000. Multiplex PCR assays for the detection of clinically relevant antibiotic resistance genes in staphylococci isolated from patients infected after cardiac surgery. J. Antimicrob. Chemother. 46:527-534. [DOI] [PubMed] [Google Scholar]
- 22.Miyazaki, S., et al. 2001. In vitro and in vivo antibacterial activities of L-084, a novel oral carbapenem, against causative organisms of respiratory tract infections. Antimicrob. Agents Chemother. 45:203-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nawaz, M. S., S. A. Khan, A. A. Khan, F. M. Khambaty, and C. E. Cerniglia. 2000. Comparative molecular analysis of erythromycin-resistance determinants in staphylococcal isolates of poultry and human origin. Mol. Cell. Probes 14:311-319. [DOI] [PubMed] [Google Scholar]
- 24.Reynolds, E. D., and J. H. Cove. 2005. Resistance to telithromycin is conferred by msr(A), msrC and msr(D) in Staphylococcus aureus. J. Antimicrob. Chemother. 56:1179-1182. [DOI] [PubMed] [Google Scholar]
- 25.Sato, T., K. Tateda, S. Kimura, Y. Ishii, and K. Yamaguchi. 2011. In vitro intracellular activity and in vivo efficacy of modithromycin, a novel bicyclolide, against Legionella pneumophila. Antimicrob. Agents Chemother. 55:1594-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stout, J. E., K. Sens, S. Mietzner, A. Obman, and V. L. Yu. 2005. Comparative activity of quinolones, macrolides and ketolides against Legionella species using in vitro broth dilution and intracellular susceptibility testing. Int. J. Antimicrob. Agents 25:302-307. [DOI] [PubMed] [Google Scholar]
- 27.Syrogiannopoulos, G. A., et al. 2003. Antimicrobial susceptibility and macrolide resistance inducibility of Streptococcus pneumoniae carrying erm(A), erm(B), or mef(A). Antimicrob. Agents Chemother. 47:2699-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Bambeke, F., J. M. Harms, Y. Van Laethem, and P. M. Tulkens. 2008. Ketolides: pharmacological profile and rational positioning in the treatment of respiratory tract infections. Expert Opin. Pharmacother. 9:267-283. [DOI] [PubMed] [Google Scholar]
- 29.Wolter, N., A. M. Smith, D. E. Low, and K. P. Klugman. 2007. High-level telithromycin resistance in a clinical isolate of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 51:1092-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]


