Abstract
The antibacterial compound tropodithietic acid (TDA) is produced by bacteria of the marine Roseobacter clade and is thought to explain the fish probiotic properties of some roseobacters. The aim of the present study was to determine the antibacterial spectrum of TDA and the likelihood of development of TDA resistance. A bacterial extract containing 95% TDA was effective against a range of human-pathogenic bacteria, including both Gram-negative and Gram-positive bacteria. TDA was bactericidal against Salmonella enterica serovar Typhimurium SL1344 and Staphylococcus aureus NCTC 12493 and killed both growing and nongrowing cells. Several experimental approaches were used to select mutants resistant to TDA or subpopulations of strains with enhanced tolerance to TDA. No approach (single exposures to TDA extract administered via different methods, screening of a transposon library for resistant mutants, or prolonged exposure to incremental concentrations of TDA) resulted in resistant or tolerant strains. After more than 300 generations exposed to sub-MIC and MIC concentrations of a TDA-containing extract, strains tolerant to 2× the MIC of TDA for wild-type strains were selected, but the tolerance disappeared after one passage in medium without TDA extract. S. Typhimurium mutants with nonfunctional efflux pump and porin genes had the same TDA susceptibility as wild-type strains, suggesting that efflux pumps and porins are not involved in innate tolerance to TDA. TDA is a promising broad-spectrum antimicrobial in part due to the fact that enhanced tolerance is difficult to gain and that the TDA-tolerant phenotype appears to confer only low-level resistance and is very unstable.
The Roseobacter clade, which belongs to the alphaproteobacteria, constitutes a major part of oceanic bacterioplankton communities (6, 17, 45), and many Roseobacter clade strains are easily culturable (3, 20, 40). The success of Roseobacter clade bacteria may be explained by several factors, one of which is their production of secondary metabolites, that may give them a competitive advantage over other species (19, 27, 30). This includes the antibacterial compound tropodithietic acid (TDA), which is produced by several Roseobacter species such as Phaeobacter gallaeciensis, Phaeobacter inhibens, and Ruegeria mobilis (3, 5, 40).
P. gallaeciensis and R. mobilis are of great interest in the aquaculture sector for use as probiotics, as they kill or inhibit the growth of several fish-pathogenic bacteria and improve the survival of fish larvae in challenge trials (14, 22, 39). The killing or inhibitory effect is related to TDA production (3, 22, 41). Thus, D'Alvise et al. previously showed that the fish-pathogenic bacterium Vibrio anguillarum strain 90-11-287 was killed only by TDA-producing wild-type Phaeobacter strain 27-4 and not by a mutant deficient in TDA production (13).
The ability of TDA to inhibit several bacteria that are pathogenic to fish and the possible use of TDA-producing bacteria as probiotics in aquaculture raise a number of issues and questions, and some are addressed in the present study. TDA has antibacterial action against bacteria outside the marine or aquatic environment, including human-pathogenic bacteria (3, 26, 48), and could also be of interest as an antibacterial outside the aquaculture sector. However, a widespread use of TDA could be potentially detrimental if resistance and subsequent cross-resistance to other agents develop, as has been the case for the majority of drugs discovered so far (11).
TDA is a disulfide tropolone (8), and it exists in a tautomeric form with thiotropocin (21). Thiotropocin (or tropodithietic acid) was first isolated from a soil bacterium identified as a Pseudomonas species (8, 26), and in addition to Roseobacter clade bacteria, the compound has also been isolated from marine Caulobacter spp. and Pseudovibrio strains (15, 25). It is antibacterial against several bacteria, including Escherichia coli and Pseudomonas aeruginosa (3, 26, 48). Tropolones with side groups other than a disulfide are also known to be antibacterial (32, 33) and display antifungal and antiviral activities (7, 26).
Bacteria have evolved resistance to virtually all antibiotics introduced into clinical practice (11), leading to treatment failures of infections of both animals and humans. The purpose of the present study was to investigate the potential for TDA exposure to select for mutants resistant to TDA.
MATERIALS AND METHODS
Bacterial strains and growth media.
The strains used in this study are listed in Table 1. All strains were grown on LB agar (catalog number CM1021; Oxoid, Basingstoke, United Kingdom) or in LB broth (catalog number CM1018; Oxoid) at 37°C except for Phaeobacter strain 27-4, Phaeobacter strain JBB1001, and V. anguillarum 90-11-287, which were cultured on marine agar (MA) 2216 (Difco, BD, Sparks, MD) or in marine broth (MB) 2216 (Difco) at 25°C. Phaeobacter gallaeciensis DSM 17395 was cultured on MA at 25°C. Unless otherwise stated, all strains were incubated for 1 day except for Phaeobacter 27-4 and Phaeobacter JBB1001, which were incubated for 2 to 3 days. The strains were stored either on Protect beads or in a freeze medium (30.0 g tryptone soya broth [catalog number CM129B; Oxoid], 5.0 g glucose, 20.0 g skim-milk powder, 40.0 g glycerol, 1,000 ml H2O) at −80°C.
TABLE 1.
MICs of tropodithietic acid determined in LB brotha
| Species | Strain(s) | Genotype or description | Reference or source | MIC (mg/liter) |
|---|---|---|---|---|
| Gram negative | ||||
| Salmonella Typhimurium | SL1344 | Wild type | 50 | 625 |
| Salmonella Typhimurium | SL1344 | ompF::aph | 12 | 625 |
| Salmonella Typhimurium | SL1344 | ompC::aph | 12 | 625 |
| Salmonella Typhimurium | ATCC 14028s | Wild type | ATCC | 625 |
| Salmonella Typhimurium | ATCC 14028s | ΔtolC::Cmr | 34 | 625-1,250 |
| Salmonella Typhimurium | ATCC 14028s | ΔacrB::Kmr | 34 | 625-1,250 |
| Salmonella Typhimurium | ATCC 14028s | ΔacrAB::Cmr | 34 | 625-1,250 |
| Salmonella Typhimurium | ATCC 14028s | ΔacrD::Cmr | 34 | 625-1,250 |
| Salmonella Typhimurium | ATCC 14028s | ΔacrEF::Cmr | 34 | 625-1,250 |
| Salmonella Typhimurium | ATCC 14028s | ΔmdtABC::Cmr | 34 | 625-1,250 |
| Salmonella Typhimurium | ATCC 14028s | ΔmdsABC::Cmr | 34 | 625-1,250 |
| Salmonella Typhimurium | ATCC 14028s | ΔermAB::Cmr | 34 | 625 |
| Salmonella Typhimurium | ATCC 14028s | ΔmdfA::Cmr | 34 | 625 |
| Salmonella Typhimurium | ATCC 14028s | ΔmdtK::Cmr | 34 | 625-1,250 |
| Salmonella Typhimurium | ATCC 14028s | ΔmacAB::Cmr | 34 | 625 |
| Salmonella Typhimurium | L3 | Human pretherapy clinical isolate | 38 | 625-1,250 |
| Salmonella Typhimurium | L10 | Human posttherapy clinical isolate; acrAB+++ | 38 | 625-1,250 |
| Enterobacter cloacae | NCTC 10005 | NCTC | 625-1,250 | |
| Serratia marcescens | NCTC 2847 | NCTC | 625-1,250 | |
| Klebsiella pneumoniae | NCTC 10896 | NCTC | 625-1,250 | |
| Klebsiella pneumoniae | NCTC 9633 | NCTC | 625 | |
| Escherichia coli | NCTC 10538, K-12 | NCTC | 155-625 | |
| Morganella morganii | NCTC 235 | NCTC | 155 | |
| Pseudomonas aeruginosa | NCTC 10662 | NCTC | 625 | |
| Vibrio anguillarum | 90-11-287 (from rainbow trout) | 46 | 40-80 | |
| Phaeobacter sp. | 27-4 (from Spanish turbot unit) | Wild type | 22 | 310 |
| Phaeobacter sp. | JBB1001 | tdaB::EZ-Tn5; Kan | 16 | 155 |
| P. gallaeciensis | DSM 17395 | DSMZ | ||
| Gram positive | ||||
| Staphylococcus aureus | NCTC 8532 (MSSA) | NCTC | 155 | |
| Staphylococcus aureus | NCTC 12493 (MRSA) | NCTC | 155 | |
| Enterococcus faecalis | NCTC 775 | NCTC | 80 | |
| Enterococcus faecalis | NCTC 7171 | NCTC | 80 | |
| Listeria monocytogenes | NC12427 | NCTC | 80 |
MICs were determined in two independent experiments, and if all experiments gave the same value, this is stated in the table. For some strains, MICs varied by a factor of 2 between experiments, as indicated. TDA was extracted to 95% purity from Phaeobacter strain 27-4. ATCC, American Type Culture Collection; NCTC, National Collection of Type Cultures (HPA, Colindale, London, United Kingdom); DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH; Cm, chloramphenicol; MSSA, methicillin-sensitive Staphylococcus aureus; MRSA, methicillin-resistant Staphylococcus aureus; Kan, kanamycin.
Purification of TDA extract.
Phaeobacter 27-4 was grown in 1.5 liters of MB for 4 to 5 days at 25°C under stagnant conditions. The culture was extracted twice with an equal volume of ethyl acetate containing 1% formic acid and shaken for 10 min in a separation funnel. After phase separation the upper phase was filtered through anhydrous Na2SO4 (∼100 g per liter ethyl acetate) and evaporated in vacuo. The extract from 1.5 liters of culture was then redissolved in 10 ml 15% acetonitrile and loaded onto a 1-g Strata-XC column (Phenomenex, Torrance, CA), which was washed with 20 ml 15% acetonitrile and eluted with 10 ml 70% acetonitrile, which was evaporated using a stream of N2.
The purity of TDA was verified by liquid chromatography-UV-high resolution-mass spectrometry (LC-UV-HR-MS) analysis as described previously (5) as well as ultra-high-performance liquid chromatography (UHPLC)-UV-Vis analysis, which was performed with a rapid-separation liquid chromatography (RSLC) Ultimate 3000 system (Dionex, Sunnyvale, CA) using a 150-mm- by 2-mm-internal-diameter (i.d.), 2.6-μm Kinetex C18 column (Phenomenex), running at 800 μl/min at 60°C using a binary linear solvent system of water (solvent A) and acetonitrile (solvent B) (both buffered with 50 μl/liter trifluoroacetic acid). The gradient program was as follows: t of 0 at 15% solvent B, t of 0.5 min at 25% solvent B, t of 6 min at 65% solvent B, and t of 7 min at 100% solvent B, keeping this for 1 min and then reverting to 15% solvent B in 1 min.
Purification and NMR validation of TDA.
Phaeobacter gallaeciensis strain DSM 17395 produces TDA at a much higher yield than does strain 27-4, and we purified TDA from 6 liters of culture grown for 3 days. Cells were removed by centrifugation at 10,000 × g. The broth was acidified with 1% formic acid, loaded onto a 20-g C18 column on an Isolera Flash chromatography system (Biotage, Uppsala, Sweden), and subsequently eluted with a water-methanol (containing 1% formic acid) gradient. The TDA-containing fraction was purified on a 10-g Biotage diol silica column eluted from heptanes over dichloromethane, ethyl acetate, and methanol. The TDA-containing fraction was fine-polished by preparative high-performance liquid chromatography (HPLC) on 25-cm, 5-μm Luna preparative columns using an acidic water-acetonitrile gradient. Analysis of the 1H nuclear magnetic resonance (NMR) spectrum in d6-benzene on a Bruker 500-MHz instrument showed data identical to the data obtained previously by Liang (29) with d6-benzene. Integrals showed a purity of >95% (see Fig. S1 in the supplemental material).
Antimicrobial activity of the TDA-containing extract.
The MICs of TDA against a range of bacteria were determined by the broth microdilution method (1, 2). The TDA extract (see above) was diluted in 10% ethanol (2,530 mg/liter), and 2-fold dilutions were prepared in microtiter plates. After inoculation with approximately 105 CFU/ml, the human-pathogenic bacteria were incubated at 37°C for 1 day, and the marine bacteria were incubated at 25°C for 2 days (Table 1). Growth was determined visually. All MIC determinations were repeated two times in independent experiments. To determine if porins or efflux pumps were involved in innate tolerance to TDA, MIC values of TDA for mutants of Salmonella enterica serovar Typhimurium (here referred to as S. Typhimurium) with disruptions in the relevant genes (Table 1) were determined as described above. The MIC of TDA extract in agar was also determined for S. Typhimurium SL1344, E. coli NCTC 10538, P. aeruginosa NCTC 10662, and Staphylococcus aureus NCTC 12493. Twofold dilutions of TDA extract stock solution were incorporated into LB agar (1, 2). Cultures were diluted, and 104 CFU was spotted onto the agar. Results (growth or no growth) were read after 1 day of incubation at 37°C.
Effect of TDA extract upon bacterial growth.
To determine the killing kinetics of TDA, S. Typhimurium SL1344 and S. aureus NCTC 12493 were grown in LB broth at 37°C for 1 day (300 rpm) and inoculated to 106 CFU/ml in either LB broth, phosphate-buffered saline (PBS), or LB broth or PBS containing TDA extract at concentrations equaling the MIC, one-half the MIC, or one-quarter the MIC. The strains were incubated at 37°C (300 rpm), and numbers of bacteria were determined by plate counts at appropriate time points. The experiment was carried out in duplicate.
Selection of TDA-resistant mutants.
Four different experimental approaches were used to select TDA-resistant mutants. All methods included a single exposure to TDA extract administered via different methods.
Exposure to 2× the MIC of TDA on agar.
Cultures of S. Typhimurium SL1344, E. coli NCTC 10538, P. aeruginosa NCTC 10662, and S. aureus NCTC 12493 grown overnight were diluted or concentrated to give a range of inocula of 105 to 109 CFU/ml (37). One hundred microliters of each suspension was spread onto LB agar containing 2 × the MIC of TDA extract for each strain. Plates were checked every day for 3 days for the appearance of TDA-resistant colonies. Colonies that arose were retained, and MIC values of the TDA extract were determined.
Exposure to the MIC or 2× the MIC of TDA in broth.
Cultures of all strains (apart from the Salmonella mutants with gene disruptions) were inoculated (at 107 and 108 CFU/ml) into liquid medium containing TDA extract at either the MIC or 2× the MIC in microtiter trays. After incubation for 1 day (human pathogens) or 2 days (marine strains), a drop of liquid culture was plated onto LB or MA plates, and any colonies arising were retained. The MIC of TDA extract for these colonies was determined as described above.
TDA diffusion mutant selection.
Cultures of S. Typhimurium SL1344, P. aeruginosa NCTC 10662, E. coli NCTC 10538, S. aureus NCTC 12493, and V. anguillarum 90-11-287 were diluted in PBS to approximately 105, 106, 107, and 108 CFU/ml and were plated onto agar plates. Paper disks or filter paper in different patterns were placed onto the bacterial lawns, and TDA extract stock solution was pipetted onto the disks or paper. Wells were also drilled into the agar, and 70 μl of TDA extract stock solution was added. Plates were incubated for 1 day (human pathogens) or 2 days (V. anguillarum 90-11-287) and observed for the appearance of colonies within the clearing zone formed by the diffusion of TDA extract into the agar. Colonies were isolated and retained, and the MIC of TDA extract was determined.
TDA-resistant Tn5 mutants.
A random transposon insertion library in S. Typhimurium SL1344 based on the Tn5 and Mu transposons (9) was screened for mutants with increased TDA tolerance. Approximately 104 insertion mutants were screened for the ability to grow in TDA extract (2× the MIC) and kanamycin (25 mg/liter). Kanamycin was added to retain the transposon insertion in the bacteria. Cultures grown without TDA extract served as a control. After incubation at 37°C for 1 day, growth was observed visually, and cultures were plated onto LB agar plates.
Selection of TDA-tolerant strains.
Three approaches were used to select subpopulations of strains with elevated tolerance to TDA extract.
Exposure to one-half the MIC to 2× the MIC of TDA.
Cultures (Table 1 strains apart from the Salmonella mutants with gene disruptions) were inoculated (105 CFU/ml) into medium containing one-half the MIC of TDA extract, and after incubation overnight, 20 μl of the cultures was inoculated into fresh broth (200 μl) containing TDA extract at the MIC and incubated again, and on the following day, 20 μl of culture was inoculated into fresh broth containing 2× the MIC of TDA extract. Experiments were conducted in triplicates.
Exposure to one-half the MIC of TDA.
S. Typhimurium SL1344, P. aeruginosa NCTC 10662, E. coli NCTC 10538, S. aureus NCTC 12493, V. anguillarum 90-11-287, and Phaeobacter 27-4 were reinoculated (30 μl) six times in medium (2,970 μl) containing one-half the MIC of TDA extract. These transfers equaled 42 to 48 generations. Human-pathogenic bacteria were incubated for 1 day at 37°C, whereas marine bacteria were incubated for 2 days at 25°C.
Exposure to 1/16 the MIC to the MIC of TDA.
S. Typhimurium SL1344 was reinoculated (10 μl) five times into 990 μl LB broth and then 40 times into gradually increasing concentrations of TDA extract (from 1/16 the MIC to the MIC), which equals 315 to 360 generations in total. The experiment was carried out with triplicate lineages. A control in duplicate lineages was also reinoculated (10 μl into 990 μl LB broth) 45 times (315 to 360 generations) but without TDA extract. All cultures were inoculated once per day and incubated at 37°C. At the end of all three adaptive approaches, the MICs of TDA extract were determined both for cultures exposed to TDA extract and for control cultures.
RESULTS
Antimicrobial activity of TDA extract.
The TDA extract from Phaeobacter 27-4 inhibited the growth of both Gram-negative (625 to 1,250 mg/liter) and Gram-positive (80 to 155 mg/liter) bacteria in broth (Table 1). The solvent of TDA (10% ethanol) was not inhibitory at the concentrations used. TDA is degraded at high temperatures (5), so as a control, the values were also determined for TDA extract that had been incubated at 37°C or 25°C prior to inoculation. This gave the same MIC values as inoculation straightaway (data not shown), and hence, the degradation of TDA extract did not take place during our assay. None of the porin or efflux pump mutants of S. Typhimurium had major altered tolerance to TDA extract compared with their parental strains.
Kintaka et al. previously reported MIC values of thiotropocin for human-pathogenic bacteria that were lower than those found in the present work (26). Kintaka et al. purified TDA from 1,200 liters of a Pseudomonas culture and did not specify the method used for MIC determinations. We did, however, suspect that the Phaeobacter 27-4 TDA extract, despite being 95% pure, contained pigment residues that interfered with mass determinations. We were able to purify 2 mg of pure TDA (as determined by NMR) from a Phaeobacter gallaeciensis strain that produces much more TDA than does 27-4 (see Fig. S1 in the supplemental material). MIC values for S. Typhimurium SL1344, E. coli NCTC 10535, P. aeruginosa NCTC 10662, S. aureus NCTC 8532, and V. anguillarum 90-11-287 were 3 to 25 mg/liter when using pure TDA, which is in the same range as those reported previously for thiotropocin (26), which is considered a tautomer of TDA (21).
Effect of TDA extract on bacterial growth.
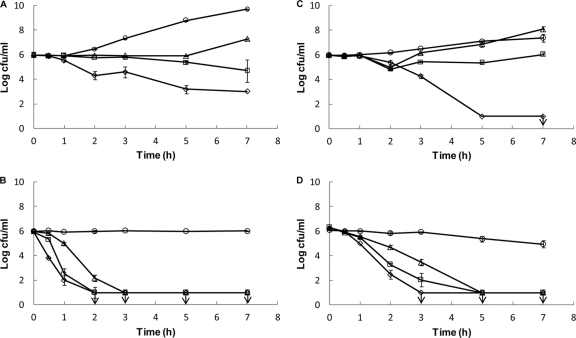
TDA extract is bactericidal, as both S. Typhimurium SL1344 and S. aureus NCTC 12493 were killed at the MIC value in both LB broth (Fig. 1A and C) and PBS (Fig. 1B and D). The reduction in cell numbers was greater for S. aureus (≥5-log reduction) than with the Gram-negative bacterium (3-log reduction) in LB broth. At one-half the MIC and one-quarter the MIC, killing of both S. Typhimurium SL1344 and S. aureus NCTC 12493 in PBS was seen. A dose-response kinetic relationship was observed for both the growth media and the buffer. As the concentration of TDA extract increased, the bacteria were killed faster or the growth of the bacteria was inhibited longer.
FIG. 1.
Killing kinetics of tropodithietic acid (TDA) against Salmonella enterica serovar Typhimurium strain SL1344 (A and B) and Staphylococcus aureus NCTC 12493 (C and D) in LB medium (A and C) and in phosphate-buffered saline (B and D). The following concentrations of TDA were used: the MIC (⋄), one-half the MIC (□), one-quarter the MIC (▵), and a growth control containing no TDA (○). Counts are means of duplicates, and error bars represent standard deviations. Vertical arrows indicate detection limits. TDA was extracted to 95% purity from Phaeobacter strain 27-4.
Selection of TDA-resistant mutants.
None of the experimental approaches used in this study resulted in stable TDA-resistant mutants. Although S. Typhimurium SL1344, E. coli NCTC 10538, P. aeruginosa NCTC 10662, and S. aureus NCTC 12493 all gave rise to colonies on agar containing 2× the MIC of TDA extract, the MIC of TDA was the same as that prior to TDA extract exposure after one inoculation in fresh medium. Independent of the inoculation level (107 or 108 CFU/ml), none of the strains survived exposure to 2× the MIC of TDA extract in broth (except S. aureus NCTC 8532). When inoculating in broth containing the MIC of TDA extract, 10 of 18 strains survived (S. Typhimurium SL1344, ATCC 14028s, and L3; Enterobacter cloacae NCTC 10005; Serratia marcescens NCTC 2847; P. aeruginosa NCTC 10662; Klebsiella pneumoniae NCTC 10896; Morganella morganii NCTC 235; S. aureus NCTC 8532; Enterococcus faecalis NCTC 775 and NCTC 7171; and Listeria monocytogenes NC12427). However, after one passage in fresh medium, no altered MIC values were observed compared to the original strains. In experiments with the diffusion of TDA extract into agar, no colonies appeared inside the clearing zones for any of the five strains tested by any of the three diffusion methods used. A transposon insertion library in S. Typhimurium SL1344 was exposed to 2× the MIC in liquid medium and plated onto agar plates. However, no colonies with decreased susceptibility to TDA extract appeared.
Selection of TDA-tolerant strains.
Two of the three adaptation experiments resulted in “variants” that tolerated slightly elevated TDA extract concentrations. However, the phenotype was not stable after passage in medium not containing TDA. In the first adaptation experiment, concentrations of TDA extract were increased from one-half the MIC to the MIC and to 2× the MIC, and 9 of the 18 strains (S. Typhimurium L10, E. cloacae NCTC 10005, S. marcescens NCTC 2847, P. aeruginosa NCTC 10662, K. pneumoniae NCTC 10896 and NCTC 9633, E. coli NCTC 10538, V. anguillarum 90-11-287, and L. monocytogenes NC12427) were able to survive and grow at 2× the MIC of TDA extract. However, when retested, none of these nine strains had decreased susceptibility to TDA extract. In the second adaptation experiment, where S. Typhimurium SL1344, P. aeruginosa NCTC 10662, E. coli NCTC 10538, S. aureus NCTC 12493, V. anguillarum 90-11-287, and Phaeobacter 27-4 were exposed to six reinoculations at one-half the MIC of TDA extract, the MICs of TDA extract required to inhibit the final populations were 2-fold higher than those for parental strains. However, after one passage in fresh medium, none of the strains had altered susceptibility to TDA extract. In the last experiment, S. Typhimurium SL1344 was exposed to gradually increasing concentrations of TDA extract for a prolonged period (40 transfers). The MIC of TDA extract was 2-fold higher for the final cultures arising in the experiment (all three lineages). However, after one inoculation in fresh medium, the MIC again reverted to the original wild-type level.
DISCUSSION
It was previously shown that TDA-producing Roseobacter species have the ability to inhibit or kill other marine bacteria (3, 13, 22, 41) and that purified TDA alone has a growth-inhibitory effect on marine bacteria (3). This study shows that TDA is a broad-spectrum antibacterial compound effective against all the human-pathogenic strains tested, including methicillin-sensitive Staphylococcus aureus and an S. Typhimurium strain harboring acrB, which encodes a multidrug efflux pump. The tautomeric form of TDA, thiotropocin, produced by Caulobacter and Pseudomonas strains also inhibited the growth of all tested bacteria (26, 48). The TDA-containing extract from strain 27-4 did contain pigment impurities, and we suspected that these impurities caused mass interference, resulting in high MIC values. Indeed, by testing the MIC of a pure preparation, we found levels (3 to 25 mg/liter) comparable to those reported previously for thiotropocin (26).
Neither the porin mutants nor efflux pump mutants of S. Typhimurium had elevated levels of tolerance to TDA extract compared to their parental strains. This indicates that perhaps TDA is neither transported into the bacterial cell through porins nor exported out again through the major efflux pumps. Keeping in mind that TDA works very well on nongrowing cells, these results could collectively indicate that TDA may not have to enter the bacterial cell in order to exert its effect. Thereby, the cell envelope could be the bacterial target for TDA. This hypothesis is supported by the results of an unpublished biosensor method using gene reporter fusions in Salmonella. This method can identify if ribosome inhibition, the cell wall, DNA, fatty acid biosynthesis, or membrane damage is the target for novel compounds. The reporters are calibrated using known antibiotics, and results are scored between 0 and 1, where 1 indicates a high confidence for a particular target. The TDA-containing extract gave scores of 3.08 × 10−5, 1, 0, 0, and 1.08 × 10−41 for the five targets indicating cell envelope damage (D. Bumann, personal communication).
V. anguillarum 90-11-287 was very sensitive to TDA. The TDA producer Phaeobacter strain 27-4 was isolated specifically due to its antagonistic effect on this strain (22). The Gram-negative TDA producer strain itself is also inhibited by a low TDA concentration and did not display the expected resistance to TDA in our assay. It is possible that these data were obtained because the conditions used for the MIC value determinations with only 2 days of incubation did not support the production of TDA by Phaeobacter 27-4. TDA is produced in the late exponential phase and requires a large surface/volume ratio (4, 5). We hypothesize that only when TDA is produced does the producer organism express the mechanism(s) by which it becomes resistant to its own drug.
The TDA extract was bactericidal against both S. Typhimurium SL1344 and S. aureus 12493, and the MIC value for each strain was also lethal in rich growth medium (LB broth). This indicates that the MIC and the minimum bactericidal concentration (MBC) are the same for TDA, as was reported previously for some host defense peptides (18). Bacteria treated with TDA extract were unable to multiply but remained viable for 1 h, after which the bacteria died. The same phenomenon was described previously for the host defense peptide plectasin (44). When carrying out killing kinetics with a nutrient-poor medium (PBS), where essentially no cell division occurs, all tested concentrations of TDA extract killed the bacteria rapidly. These results suggest that TDA is active against both growing and nongrowing cells. This was reported previously for glycan-targeting antibiotics like moenomycin and teicoplanin (10, 49).
Several different methods for selecting TDA-resistant mutants or strains with tolerance to TDA extract were explored in this study. Most of the methods reported in the literature for such a purpose were used. We did not use the gradient gel method (10), as this technique requires large volumes of the compound of interest, and our available, pure supply of TDA was limited.
None of the methods of single exposure to TDA extract resulted in mutants resistant to TDA. This could indicate that more than one mutation is needed in order to gain resistance. If this is the case, TDA may resemble “dirty drugs,” such as biocides or antimicrobial peptides, that have multiple targets in bacteria and thus require mutations in multiple loci in order to evolve tolerance (36, 42). Another possibility is that TDA targets functions (genes) which are essential for bacterial survival and growth. A slow adaptation to gradually increasing concentrations of the drug has proven to be a successful way of obtaining stable strains with enhanced tolerance to an antimicrobial peptide and to biocides (24, 31, 35, 47), but a similar approach was not successful in this study, as S. Typhimurium SL1344 immediately lost the gained tolerance. Previous studies showed a gradual deadaptation of the decreased sensitivity to different biocides (23, 31, 43, 47); however, Sallum and Chen reported previously that tolerance to the antimicrobial peptide cecropin B was abolished after one passage in fresh medium without the peptide (43), as found in the present study with TDA and TDA extract.
Tolerance to a drug, apart from “fixed” mutations, can also occur as a response to environmental stimuli leading to a phenotypic response (28). It is doubtful that true adaptation to TDA was seen in this study, as the tolerance was only 2-fold higher than that of nonadapted cells, whereas in the literature for quaternary ammonium compounds and the antimicrobial peptide cecropin B, it was reported to be many folds higher (23, 31, 43). The increased tolerance to TDA seen in this study could be the result of a phenotypic switch that is rapidly reversed after exposure to TDA.
Based on the results obtained in this study, we conclude that TDA has a wide spectrum of activity against pathogenic bacteria. We also conclude that it is difficult to create stable strains with tolerance to an extract of TDA from a TDA-producing bacterium. We suggest that the compound more resembles antimicrobial peptides and biocides with regard to the mechanism of action than antibiotics or that TDA targets, e.g., genes essential for growth. As only a slight increase in TDA tolerance was observed and any TDA tolerance phenotype was unstable, TDA is an interesting compound for the potential control of bacterial diseases in aquaculture as well as in other settings.
Supplementary Material
Acknowledgments
The project was financed by the Danish Research Council for Technology and Production (project 09-061086). We acknowledge financial support from the Otto Mønsteds Fond, Oticon Fonden, and Knud Højgaards Fond to Cisse Hedegaard Porsby for her visit to the University of Birmingham.
We thank Dirk Bumann, University of Basel; Ingrid Mecklenbräuker, University of Freiburg; and Christian Schleberger, University of Freiburg, for testing TDA in the biosensor assay. We thank Richard Kerry Phipps, Department of Systems Biology, Center for Microbial Biotechnology, Technical University of Denmark, for purifying TDA from Phaeobacter gallaeciensis DSM 17395.
Footnotes
Published ahead of print on 24 January 2011.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Andrews, J. M. 2001. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 48:5-16. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, J. M. 2006. Determination of minimum inhibitory concentrations. British Society for Antimicrobial Chemotherapy, Birmingham, United Kingdom. http://www.bsac.org.uk/Resources/BSAC/Chapter_2_Determination_of_MICs_2006.pdf. [DOI] [PubMed]
- 3.Brinkhoff, T., et al. 2004. Antibiotic production by a Roseobacter clade-affiliated species from the German Wadden Sea and its antagonistic effects on indigenous isolates. Appl. Environ. Microbiol. 70:2560-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruhn, J. B., L. Gram, and R. Belas. 2007. Production of antibacterial compounds and biofilm formation by Roseobacter species are influenced by culture conditions. Appl. Environ. Microbiol. 73:442-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruhn, J. B., et al. 2005. Ecology, inhibitory activity, and morphogenesis of a marine antagonistic bacterium belonging to the Roseobacter clade. Appl. Environ. Microbiol. 71:7263-7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchan, A., J. M. Gonzalez, and M. A. Moran. 2005. Overview of the marine Roseobacter lineage. Appl. Environ. Microbiol. 71:5665-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Budihas, S. R., et al. 2005. Selective inhibition of HIV-1 reverse transcriptase-associated ribonuclease H activity by hydroxylated tropolones. Nucleic Acids Res. 33:1249-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cane, D. E., Z. Wu, and J. E. Vanepp. 1992. Thiotropocin biosynthesis—shikimate origin of a sulfur-containing tropolone derivative. J. Am. Chem. Soc. 114:8479-8483. [Google Scholar]
- 9.Chaudhuri, R. R., et al. 2009. Comprehensive identification of Salmonella enterica serovar Typhimurium genes required for infection of BALB/c mice. PLoS Pathog. 5:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chmara, H., S. Ripa, F. Mignini, and E. Borowski. 1991. Bacteriolytic effect of teicoplanin. J. Gen. Microbiol. 137:913-919. [DOI] [PubMed] [Google Scholar]
- 11.Clatworthy, A. E., E. Pierson, and D. T. Hung. 2007. Targeting virulence: a new paradigm for antimicrobial therapy. Nat. Chem. Biol. 3:541-548. [DOI] [PubMed] [Google Scholar]
- 12.Coldham, N. G., M. Webber, M. J. Woodward, and L. J. V. Piddock. 2010. A 96-well plate fluorescence assay for assessment of cellular permeability and active efflux in Salmonella enterica serovar Typhimurium and Escherichia coli. J. Antimicrob. Chemother. 65:1655-1663. [DOI] [PubMed] [Google Scholar]
- 13.D'Alvise, P. W., J. Melchiorsen, C. H. Porsby, K. F. Nielsen, and L. Gram. 2010. Inactivation of Vibrio anguillarum by attached and planktonic Roseobacter cells. Appl. Environ. Microbiol. 76:2366-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fjellheim, A. J., G. Klinkenberg, J. Skjermo, I. M. Aasen, and O. Vadstein. 2010. Selection of candidate probionts by two different screening strategies from Atlantic cod (Gadus morhua L.) larvae. Vet. Microbiol. 144:153-159. [DOI] [PubMed] [Google Scholar]
- 15.Geng, H., and R. Belas. 2010. Expression of tropodithietic acid biosynthesis is controlled by a novel autoinducer. J. Bacteriol. 192:4377-4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geng, H. F., J. B. Bruhn, K. F. Nielsen, L. Gram, and R. Belas. 2008. Genetic dissection of tropodithietic acid biosynthesis by marine roseobacters. Appl. Environ. Microbiol. 74:1535-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.González, J. M., et al. 2000. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 66:4237-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gottlieb, C. T., et al. 2008. Antimicrobial peptides effectively kill a broad spectrum of Listeria monocytogenes and Staphylococcus aureus strains independently of origin, sub-type, or virulence factor expression. BMC Microbiol. 8:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gram, L., H. P. Grossart, A. Schlingloff, and T. Kiørboe. 2002. Possible quorum sensing in marine snow bacteria: production of acylated homoserine lactones by Roseobacter strains isolated from marine snow. Appl. Environ. Microbiol. 68:4111-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gram, L., J. Melchiorsen, and J. B. Bruhn. 2010. Antibacterial activity of marine culturable bacteria collected from a global sampling of ocean surface waters and surface swabs of marine organisms. Mar. Biotechnol. 12:439-451. [DOI] [PubMed] [Google Scholar]
- 21.Greer, E. M., D. Aebisher, A. Greer, and R. Bentley. 2008. Computational studies of the tropone natural products, thiotropocin, tropodithietic acid, and troposulfenin. Significance of thiocarbonyl-enol tautomerism. J. Org. Chem. 73:280-283. [DOI] [PubMed] [Google Scholar]
- 22.Hjelm, M., et al. 2004. Selection and identification of autochthonous potential probiotic bacteria from turbot larvae (Scophthalmus maximus) rearing units. Syst. Appl. Microbiol. 27:360-371. [DOI] [PubMed] [Google Scholar]
- 23.Jones, M. V., T. M. Herd, and H. J. Christie. 1989. Resistance of Pseudomonas aeruginosa to amphoteric and quaternary ammonium biocides. Microbios 58:49-61. [PubMed] [Google Scholar]
- 24.Karatzas, K. A. G., et al. 2007. Prolonged treatment of Salmonella enterica serovar Typhimurium with commercial disinfectants selects for multiple antibiotic resistance, increased efflux and reduced invasiveness. J. Antimicrob. Chemother. 60:947-955. [DOI] [PubMed] [Google Scholar]
- 25.Kawano, Y., et al. 1998. Biological activity of thiotropocin produced by marine bacterium, Caulobacter sp. PK654. J. Mar. Biotechnol. 6:49-52. [Google Scholar]
- 26.Kintaka, K., H. Ono, S. Tsubotani, S. Harada, and H. Okazaki. 1984. Thiotropocin, a new sulfur-containing 7-membered-ring antibiotic produced by a Pseudomonas sp. J. Antibiot. 37:1294-1300. [DOI] [PubMed] [Google Scholar]
- 27.Lafay, B., et al. 1995. Roseobacter algicola sp. nov., a new marine bacterium isolated from the phycosphere of the toxin-producing dinoflagellate Prorocentrum lima. Int. J. Syst. Bacteriol. 45:290-296. [DOI] [PubMed] [Google Scholar]
- 28.Lewis, K. 2007. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5:48-56. [DOI] [PubMed] [Google Scholar]
- 29.Liang, L. 2003. Investigation of secondary metabolites of North Sea bacteria: fermentation, isolation and structure elucidation and bioactivity. Ph.D. thesis. University of Göttingen, Göttingen, Germany.
- 30.Martens, T., et al. 2007. Bacteria of the Roseobacter clade show potential for secondary metabolite production. Microb. Ecol. 54:31-42. [DOI] [PubMed] [Google Scholar]
- 31.Méchin, L., F. Dubois-Brissonnet, B. Heyd, and J. Y. Leveau. 1999. Adaptation of Pseudomonas aeruginosa ATCC 15442 to didecyldimethylammonium bromide induces changes in membrane fatty acid composition and in resistance of cells. J. Appl. Microbiol. 86:859-866. [DOI] [PubMed] [Google Scholar]
- 32.Morita, Y., et al. 2004. Biological activity of alpha-thujaplicin, the isomer of hinokitiol. Biol. Pharm. Bull. 27:899-902. [DOI] [PubMed] [Google Scholar]
- 33.Morita, Y., et al. 2002. Biological activity of 4-acetyltropolone, the minor component of Thujopsis dolabrata SIeb. et Zucc. hondai Mak. Biol. Pharm. Bull. 25:981-985. [DOI] [PubMed] [Google Scholar]
- 34.Nishino, K., T. Latifi, and E. A. Groisman. 2006. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 59:126-141. [DOI] [PubMed] [Google Scholar]
- 35.Perron, G. G., M. Zasloff, and G. Bell. 2006. Experimental evolution of resistance to an antimicrobial peptide. Proc. Biol. Sci. 273:251-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peschel, A., and H. G. Sahl. 2006. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 4:529-536. [DOI] [PubMed] [Google Scholar]
- 37.Piddock, L. J. V., and H. L. Turner. 1992. Activity of meropenem against imipenem-resistant bacteria and selection in vitro of carbapenem-resistant Enterobacteriaceae. Eur. J. Clin. Microbiol. 11:1186-1191. [DOI] [PubMed] [Google Scholar]
- 38.Piddock, L. J. V., D. G. White, K. Gensberg, L. Pumbwe, and D. J. Griggs. 2000. Evidence for an efflux pump mediating multiple antibiotic resistance in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 44:3118-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Planas, M., et al. 2006. Probiotic effect in vivo of Roseobacter strain 27-4 against Vibrio (Listonella) anguillarum infections in turbot (Scophthalmus maximus L.) larvae. Aquaculture 255:323-333. [Google Scholar]
- 40.Porsby, C. H., K. F. Nielsen, and L. Gram. 2008. Phaeobacter and Ruegeria species of the Roseobacter clade colonize separate niches in a Danish turbot (Scophthalmus maximus)-rearing farm and antagonize Vibrio anguillarum under different growth conditions. Appl. Environ. Microbiol. 74:7356-7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prado, S., J. Montes, J. L. Romalde, and J. L. Barja. 2009. Inhibitory activity of Phaeobacter strains against aquaculture pathogenic bacteria. Int. Microbiol. 12:107-114. [PubMed] [Google Scholar]
- 42.Russell, A. D. 2003. Biocide use and antibiotic resistance: the relevance of laboratory findings to clinical and environmental situations. Lancet Infect. Dis. 3:794-803. [DOI] [PubMed] [Google Scholar]
- 43.Sallum, U. W., and T. T. Chen. 2008. Inducible resistance of fish bacterial pathogens to the antimicrobial peptide cecropin B. Antimicrob. Agents Chemother. 52:3006-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider, T., et al. 2010. Plectasin, a fungal defensin, targets the bacterial cell wall precursor lipid II. Science 328:1168-1172. [DOI] [PubMed] [Google Scholar]
- 45.Selje, N., M. Simon, and T. Brinkhoff. 2004. A newly discovered Roseobacter cluster in temperate and polar oceans. Nature 427:445-448. [DOI] [PubMed] [Google Scholar]
- 46.Skov, M. N., K. Pedersen, and J. L. Larsen. 1995. Comparison of pulsed-field gel electrophoresis, ribotyping, and plasmid profiling for typing of Vibrio anguillarum serovar 01. Appl. Environ. Microbiol. 61:1540-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suller, M. T. E., and A. D. Russell. 1999. Antibiotic and biocide resistance in methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus. J. Hosp. Infect. 43:281-291. [DOI] [PubMed] [Google Scholar]
- 48.Tsubotani, S., Y. Wada, K. Kamiya, H. Okazaki, and S. Harada. 1984. Structure of thiotropocin, a new sulfur-containing 7-membered antibiotic. Tetrahedron Lett. 25:419-422. [Google Scholar]
- 49.Vanheijenoort, Y., M. Leduc, H. Singer, and J. Vanheijenoort. 1987. Effects of moenomycin on Escherichia coli. J. Gen. Microbiol. 133:667-674. [DOI] [PubMed] [Google Scholar]
- 50.Wray, C., and W. J. Sojka. 1978. Experimental Salmonella typhimurium infection in calves. Res. Vet. Sci. 25:139-143. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.