Abstract
To investigate the contribution of LamB in Klebsiella pneumoniae antimicrobial resistance, we determined the MICs of various antibiotics and the frequency of mutation to increased cefoxitin or meropenem resistance of the strains CSUB10S (expressing only OmpK36), CSUB10R (lacking OmpK35 and OmpK36), and their derived isogenic insertion-duplication mutants deficient in LamB. Expression of LamB was indispensable in order for CSUB10S to lose OmpK36 and become resistant to cefoxitin, while in CSUB10R, LamB deficiency promoted increased resistance to carbapenem.
An increasing emergence of multidrug resistance among Klebsiella pneumoniae nosocomial isolates has limited the therapeutic options for the treatment of the intrahospital infections caused by this opportunistic pathogen. A multidrug resistance phenotype results from the progressive accumulation of different mechanisms of resistance in the same microorganism, including high-level production of extended-spectrum β-lactamases (ESBLs) and porin deficiency. K. pneumoniae produces two major porins, OmpK35 (8) and OmpK36 (1). However, most ESBL-expressing K. pneumoniae clinical isolates produce only OmpK36 (11). Clinical and experimental evidences indicate that loss of these major porins in K. pneumoniae strains producing ESBLs causes resistance to cefoxitin, increased resistance to expanded-spectrum cephalosporins and monobactams, and decreased susceptibility to fluoroquinolones (8, 9, 12-14, 15).
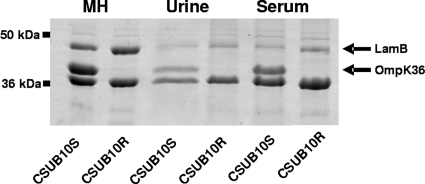
Besides the major porins, K. pneumoniae may express other porins, such as PhoE, OmpK37, and LamB (3, 7, 13). These alternative porins may be crucial for the microorganism in the absence of OmpK35/36, although their contribution to antimicrobial resistance has been poorly investigated. In a previous study, the outer membrane protein (OMP) profile of the susceptible K. pneumoniae clinical isolate CSUB10S (expressing only OmpK36) and that of its resistant clinical derivative CSUB10R (lacking both OmpK35 and OmpK36) were compared (3). It was observed that LamB may be a predominant OMP in the resistant isolate compared to the susceptible one, suggesting that LamB may contribute to resistance in K. pneumoniae. To verify this result, the amounts of LamB present in the outer membrane of CSUB10S and CSUB10R grown in cation-supplemented Mueller-Hinton broth (MH) or in human pools of urine or serum were analyzed by SDS-PAGE as previously described (11). As shown in Fig. 1, the outer membrane of the resistant isolate contained larger amounts of LamB than that of the susceptible one, both in MH and in the physiological fluids. Densitometric analysis of the intensity of the band of LamB normalized for the band of OmpA of three independent gels demonstrated that in MH, CSUB10R produces 1.86-fold ± 0.09-fold more LamB than CSUB10S (P = 0.001, two-tailed t test). Furthermore, reverse transcription-PCR (RT-PCR) quantification of the amount of lamB mRNA in both strains showed that CSUB10R expresses 2.17-fold ± 0.6-fold more lamB than CSUB10S (P = 0.04, two-tailed t test).
FIG. 1.
Representative SDS-PAGE analysis of the OMPs of K. pneumoniae CSUB10S and CSUB10R grown in cation-supplemented MH, urine, or serum. Molecular markers are indicated on the left. Black arrows indicate the positions of LamB and OmpK36.
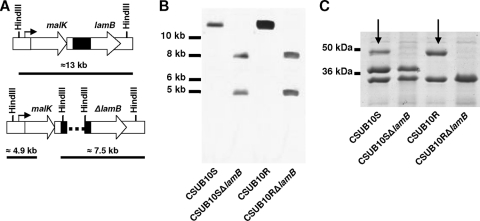
In order to investigate the role of LamB in K. pneumoniae antimicrobial resistance, we used insertion-duplication mutagenesis to construct LamB-deficient knockouts from both CSUB10S and CSUB10R, following the strategy previously described (16) (Fig. 2A). Southern blot analysis of the genomic DNA of both knockouts using a specific probe for lamB confirmed that two incomplete copies of the gene were generated by the integration of the plasmid (Fig. 2B). SDS-PAGE analysis confirmed that the lamB mutation abolished the expression of LamB in both knockouts (Fig. 2C).
FIG. 2.
Construction of LamB-deficient mutants from K. pneumoniae strains CSUB10S and CSUB10R. (A) Schematic representation of the insertion-duplication mutagenesis of lamB in K. pneumoniae CSUB10S and CSUB10R. The K. pneumoniae chromosome in the strains CSUB10S and CSUB10R is shown the upper part, and the chromosome of their isogenic lamB mutants is in the lower part. DNA fragment size in the schematic is not to scale. The dashed line in the mutant genome represents the DNA plasmid integrated into the chromosome. The black boxes indicate the probe used in the Southern blot analysis. The arrow indicates the promoter of the operon. The expected sizes of the HindIII fragments that hybridize with the probe described above are indicated in kilobases. (B) Southern blot analysis of K. pneumoniae CSUB10S and CSUB10R and the isogenic lamB-deficient mutant chromosomes digested with HindIII. Molecular size markers (in kilobases) are shown to the left of the blots. (C) SDS-PAGE analysis of the OMPs from K. pneumoniae CSUB10S and CSUB10R and the isogenic lamB-deficient mutants. Molecular markers are indicated on the left. Arrows indicate the position of LamB.
The susceptibilities of the clinical isolates CSUB10S and CSUB10R and their derived isogenic LamB-deficient knockouts, CSUB10SΔlamB and CSUB10RΔlamB, to a number of antibiotics were determined using Etest strips (bioMérieux, Marcy l'Etoile, France), following the manufacturer's instructions (Table 1). The absence of LamB in the susceptible isolate CSUB10S did not caused significant changes in the MICs of the agents tested. Only modest reductions in the MICs of ceftazidime, cefepime, cefotaxime, tobramycin, amikacin, and levofloxacin were observed. This was an expected result given that in the presence of OmpK36, the contribution of LamB to the penetration of these antimicrobial agents is probably negligible. In contrast, LamB deficiency in the OmpK36-deficient resistant isolate CSUB10R slightly increased the MICs of cefepime, piperacillin-tazobactam, cefotaxime, imipenem, meropenem, and ertapenem, suggesting that LamB may contribute to the penetration of these agents in the porin-deficient strains.
TABLE 1.
MICs of CSUB10S, CSUB10R, and their derived LamB-deficient mutants
| Strain | MIC (μg/ml) of druga |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAZ | FEP | PIP | TZP | FOX | CTX | IPM | MEM | ETP | GEN | TOB | AMK | CIP | LVX | TET | |
| CSUB10S | 128 | 2 | >256 | 1.5 | 4 | 8 | 0.19 | 0.064 | 0.094 | 4 | 1 | 1 | 0.25 | 1 | 0.5 |
| CSUB10SΔlamB | 64 | 1 | >256 | 1.5 | 4 | 3 | 0.19 | 0.064 | 0.094 | 4 | 0.75 | 0.75 | 0.25 | 0.5 | 0.5 |
| CSUB10R | >256 | 16 | >256 | 8 | 64 | 64 | 0.5 | 1 | 8 | 4 | 1.5 | 1.5 | 2 | 2 | 1.5 |
| CSUB10RΔlamB | >256 | 32 | >256 | 24 | 64 | 96 | 1.5 | 1.5 | 24 | 4 | 1.5 | 1.5 | 2 | 2 | 1.5 |
CAZ, ceftazidime; FEP, cefepime; PIP, piperacillin; TZP, piperacillin-tazobactam; FOX, cefoxitin; CTX, cefotaxime; IPM, imipenem; MEM, meropenem; ETP, ertapenem; GEN, gentamicin; TOB, tobramycin; AMK, amikacin; CIP, ciprofloxacin; LVX, levofloxacin; TET, tetracycline.
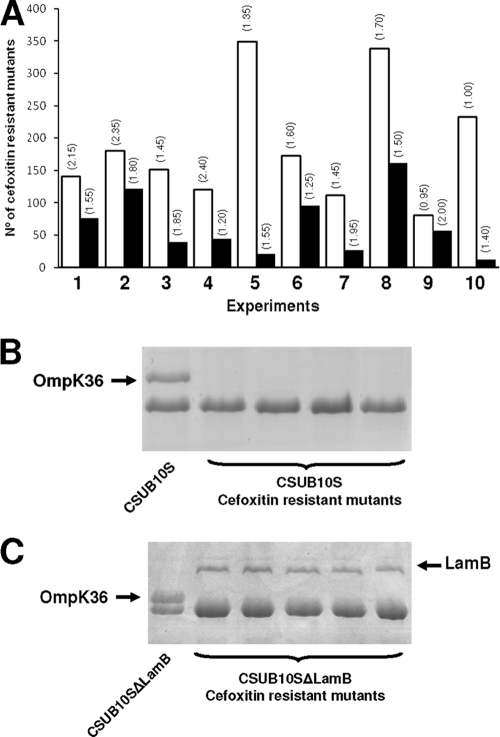
Cefoxitin resistance in CSUB10R is caused by the loss of both OmpK35 and OmpK36 (14). In order to study if the overexpression of LamB compensates the loss of these major porins in the resistant isolate, we determined the frequency of mutation to cefoxitin resistance in CUSB10S and CSUB10SΔlamB. The mutation frequency was examined by determining the proportion of resistant colonies with regard to the total viable cell count as previously described (5). The frequency of mutation to cefoxitin resistance (32 μg/ml) of the parent strain CSUB10S was 10 times higher than that of the LamB-deficient mutant CSUB10SΔlamB (1.86 × 10−8 versus 9.9 × 10−9; P = 0.048) (Fig. 3A). SDS-PAGE analysis of the OMPs of some randomly selected cefoxitin-resistant isolates derived from CSUB10S confirmed the absence of OmpK35/OmpK36 (Fig. 3B). Furthermore, the analysis of the OMPs of some randomly selected cefoxitin-resistant isolates derived from CSUB10SΔlamB revealed that the LamB-deficient phenotype, which is highly stable, reverted to the wild-type phenotype in the presence of cefoxitin (Fig. 3C). This suggests that during the development of resistance to cefoxitin by OmpK35/OmpK36 loss, there are strong selective pressures for the expression of LamB. The mean MICs of the antimicrobial agents tested in Table 1 of 10 randomly selected cefoxitin-resistant mutants derived from either CSUB10S or CSUB10SΔlamB were identical to those reported for CSUB10R in Table 1.
FIG. 3.
Frequency of mutation to cefoxitin resistance of CSUB10S and its derived isogenic LamB-deficient mutant CSUB10SΔlamB. (A) Number of CFU resistant to cefoxitin (32 μg/ml) obtained from CSUB10S (white columns) or CSUB10SΔlamB (black columns) in 10 independent experiments. Numbers in parentheses above the columns indicate the total number of CFU × 109 in each experiment. (B and C) Representative SDS-PAGE analysis of the OMPs of K. pneumoniae CSUB10S (B) or CSUB10SΔlamB (C) and some of their derived cefoxitin-resistant mutants. Black arrows indicate the positions of OmpK36 and LamB.
To investigate if LamB contributes to the penetration of meropenem in CSUB10R, we determined the frequency of mutation to increased meropenem resistance following the protocol described above but using meropenem (2 μg/ml) instead of cefoxitin. The LamB-deficient mutant CSUB10RΔlamB exhibited a higher mutation frequency (1.43 × 10−6) than its parent strain, CSUB10R, for which it was undetectable (<1.0 × 10−11). SDS-PAGE analysis of the OMPs from the meropenem-resistant isolates derived from CSUB10RΔlamB confirmed that the LamB-deficient phenotype was conserved but also revealed the presence of a new protein of about 26 kDa (data not shown).
In this study, we have shown that deficiency of the major porin OmpK36 is associated with the overexpression of LamB. Similar results have been reported for organic solvent-tolerant Escherichia coli mutants (2) and for some Enterobacter aerogenes clinical isolates (10). Despite the fact that this association could be explained in part by coregulation with major porin expression at the level of transcription and/or at the level of translation (4, 6), the contribution of LamB to antimicrobial resistance had not been studied accurately. Our results using isogenic LamB-deficient mutants demonstrate that overexpression of this porin is an essential mechanism to compensate the loss of the major porin OmpK36 and allows K. pneumoniae to show high levels of resistance to various classes of antibiotics. Furthermore, in the porin-deficient background, LamB deficiency promoted decreased susceptibility to carbapenem. Similar results were obtained by Kaczmarek et al. when they studied PhoE (13). In their study, PhoE compensated the loss of the major nonspecific porins, and downregulation of phoE was involved in carbapenem resistance.
Interestingly, LamB deficiency was associated with the expression of a 26-kDa protein which might compensate the loss of LamB and contribute to carbapenem resistance. Further investigations, currently in progress, are necessary to know the contribution of this novel protein in the porin-deficient isolates resistant to carbapenem.
Acknowledgments
This study was supported by grants from the Ministerio de Ciencia e Innovación (SAF2005-0466 to S.A.) and by the Instituto de Salud Carlos III (Spanish Network for the Research in Infectious Diseases) (REIPI RD06/0008). L.G.-S. is the recipient of predoctoral fellowships from the Ministerio de Ciencia e Innovación.
Footnotes
Published ahead of print on 31 January 2011.
REFERENCES
- 1.Albertí, S., et al. 1995. A porin from Klebsiella pneumoniae: sequence homology, three-dimensional model, and complement binding. Infect. Immun. 63:903-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aono, R., N. Tsukagoshi, and M. Yamamoto. 1998. Involvement of outer membrane protein TolC, a possible member of the mar-sox regulon, in maintenance and improvement of organic solvent tolerance of Escherichia coli K-12. J. Bacteriol. 180:938-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ardanuy, C., et al. 1998. Outer membrane profiles of clonally related Klebsiella pneumoniae isolates from clinical samples and activities of cephalosporins and carbapenems. Antimicrob. Agents Chemother. 42:1636-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Click, E. M., and C. A. Schnaitman. 1989. Export-defective LamB protein is a target for translational control caused by OmpC porin overexpression. J. Bacteriol. 171:616-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crane, G. J., S. M. Thomas, and M. E. Jones. 1996. A modified Luria-Delbruck fluctuation assay for estimating and comparing mutation rates. Mutat. Res. 354:171-182. [DOI] [PubMed] [Google Scholar]
- 6.Diedrich, D. L., and J. A. Fralick. 1982. Relationship between the OmpC and LamB proteins of Escherichia coli and its influence on the protein mass of the outer membrane. J. Bacteriol. 149:156-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doménech-Sánchez, A., S. Hernández-Allés, L. Martínez-Martínez, V. J. Benedí, and S. Albertí. 1999. Identification and characterization of a new porin gene of Klebsiella pneumoniae: its role in beta-lactam antibiotic resistance. J. Bacteriol. 181:2726-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doménech-Sánchez, A., et al. 2003. Role of Klebsiella pneumoniae OmpK35 porin in antimicrobial resistance. Antimicrob. Agents Chemother. 47:3332-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doumith, M., M. J. Ellington, D. M. Livermore, and N. Woodford. 2009. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J. Antimicrob. Chemother. 63:659-667. [DOI] [PubMed] [Google Scholar]
- 10.Gayet, S., R. Chollet, G. Mollet, J. M. Pagès, and J. Chevalier. 2003. Modification of outer membrane protein profile and evidence suggesting an active drug pump in Enterobacter aerogenes clinical strains. Antimicrob. Agents Chemother. 47:1555-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernández-Allés, S., et al. 1999. Porin expression in clinical isolates of Klebsiella pneumoniae. Microbiology 145:673-679. [DOI] [PubMed] [Google Scholar]
- 12.Jacoby, G. A., D. M. Mills, and N. Chow. 2004. Role of beta-lactamases and porins in resistance to ertapenem and other beta-lactams in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48:3202-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaczmarek, F. M., F. Dib-Hajj, W. Shang, and T. D. Gootz. 2006. High-level carbapenem resistance in a Klebsiella pneumoniae clinical isolate is due to the combination of blaACT-1 beta-lactamase production, porin OmpK35/36 insertional inactivation, and down-regulation of the phosphate transport porin PhoE. Antimicrob. Agents Chemother. 50:3396-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martínez-Martínez, L., et al. 1996. In vivo selection of porin-deficient mutants of Klebsiella pneumoniae with increased resistance to cefoxitin and expanded-spectrum cephalosporins. Antimicrob. Agents Chemother. 40:342-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mena, A., et al. 2006. Characterization of a large outbreak by CTX-M-1 producing Klebsiella pneumoniae and mechanisms leading to in vivo carbapenem resistance development. J. Clin. Microbiol. 44:2831-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubirés, X., et al. 1997. A gene (wbbL) from Serratia marcescens N28b (O4) complements the rfb-50 mutation of Escherichia coli K-12 derivatives. J. Bacteriol. 179:7581-7586. [DOI] [PMC free article] [PubMed] [Google Scholar]