Abstract
Among 222 Proteus mirabilis clinical isolates collected from 17 hospitals in Korea in 2008, 28 (12.6%) and 8 (3.6%) isolates exhibited extended-spectrum β-lactamase (ESBL) and AmpC phenotypes, respectively. The most common type of ESBL gene identified by PCR and sequencing experiments was blaCTX-M-14a (n = 12). The blaCTX-M-90 (n = 4), blaCTX-M-15 (n = 3), blaCTX-M-12 (n = 3), blaCTX-M-2 (n = 2), blaCTX-M-14b (n = 1), blaTEM-52 (n = 5), and blaSHV-12 (n = 1) genes were also detected. Eight isolates carried an AmpC β-lactamase gene, such as blaCMY-2 (n = 6) or blaDHA-1 (n = 2). All bla genes encoding CTX-M-1- and CTX-M-9-type enzymes and all blaCMY-2 genes were preceded by ISEcp1-like elements. The blaCTX-M-2 gene found in two isolates was located on a complex class 1 integron. The blaDHA-1 gene was preceded by a transcriptional regulator gene and was followed by phage shock protein genes. The blaCTX-M genes were located on the chromosome in 21 isolates. A plasmid location for the blaCTX-M gene was found in only four isolates: the blaCTX-M-14a gene was located on ∼150-kbp IncA/C plasmids in three isolates and on a ∼50-kbp IncN plasmid in one isolate. The blaTEM-52 gene was located on ∼50-kbp IncN plasmids in all five isolates. The AmpC β-lactamase genes were located on the chromosome in seven of eight isolates; one isolate carried the blaCMY-2 gene on a ∼150-kbp IncA/C plasmid. Our results show that a chromosomal location of CTX-M ESBL and AmpC β-lactamase genes in P. mirabilis is no longer an unusual phenomenon in hospital environments.
Proteus mirabilis is the second most common cause of urinary tract infections and an important cause of nosocomial infections (19). Oxyimino-cephalosporins have been used as the drugs of choice to treat infections caused by ampicillin-resistant P. mirabilis. However, as with other Enterobacteriaceae, P. mirabilis strains exhibiting resistance to expanded-spectrum β-lactam agents have been widely reported in many parts of the world (1, 5, 12, 18). Because P. mirabilis lacks species-specific chromosome-borne β-lactamases, resistance in this microorganism is wholly dependent on the acquisition of plasmid-encoded β-lactamases, such as extended-spectrum β-lactamases (ESBLs), AmpC enzymes, and metallo-β-lactamases (10, 15, 24).
Although uncommon, a chromosomal location for AmpC and CTX-M ESBL genes in P. mirabilis has been reported. Since the first report of a clinical strain of P. mirabilis carrying a chromosomally encoded CMY-2-like enzyme, designated CMY-3 (3), P. mirabilis strains harboring chromosomal CMY-4, CMY-12, CMY-14, CMY-15, or CMY-16 from France, Italy, and Poland have been described (9, 10, 14). Two P. mirabilis strains carrying chromosomally encoded CTX-M ESBL, CTX-M-25 or CTX-M-41 have also been reported (17).
Here we report a high prevalence of clinical P. mirabilis isolates carrying the chromosome-borne blaCTX-M and/or AmpC β-lactamase genes in Korea.
MATERIALS AND METHODS
Bacterial strains.
In a nationwide survey conducted between July and September 2008, a total of 222 consecutive nonduplicate P. mirabilis isolates were collected from 17 hospitals in 11 cities in Korea. The isolates were recovered from urine (n = 138), blood (n = 10), respiratory specimens (n = 18), and pus (n = 56). The isolates were identified using a Vitek GNI card (bioMérieux, Marcy-l'Etoile, France).
Antimicrobial susceptibility testing.
The phenotypic confirmatory test for ESBL and/or AmpC β-lactamase production using boronic acid (BA) as an AmpC β-lactamase inhibitor was performed as described previously (21). MICs of β-lactams in the absence or presence of clavulanic acid (at a fixed concentration of 4 μg/ml; Sigma, St. Louis, MO) were determined by the agar dilution method according to CLSI guidelines (6).
Conjugal transfer experiments.
The agar mating method was used to test the transferability of oxyimino-cephalosporin resistance by using azide-resistant Escherichia coli J53 as a recipient (13). Transconjugants were selected on Mueller-Hinton agar plates supplemented with either 2 μg/ml ceftazidime, 2 μg/ml cefotaxime, or 8 μg/ml cefoxitin and 100 μg/ml sodium azide.
Characterization of genes encoding β-lactamases.
Genes coding for ESBLs and AmpC β-lactamases were detected by PCR amplification as described previously (20, 22). The templates for PCR amplification in clinical isolates were whole-cell lysates, and the PCR products were subjected to direct sequencing. Both strands of the PCR products were sequenced twice with an automatic sequencer (model 3730xl; Applied Biosystems, Weiterstadt, Germany).
Analysis of the genetic environments surrounding the bla genes.
The genetic organization of the bla genes of CTX-M and AmpC enzymes was investigated by PCR mapping and sequencing of the regions surrounding the genes by using the primers listed in Table 1.
TABLE 1.
Sequences of PCR primers used in a genetic environment study
| Name | Target gene | Sequence (5′ to 3′) |
|---|---|---|
| CTX-M1-mF | blaCTX-M-1 cluster | CGC GCT ACA GTA CAG CGA TA |
| CTX-M1-mR | AGC TTA TTC ATC GCC ACG TT | |
| CTX-M2-mF | blaCTX-M-2 cluster | ACAGAGCGAGAGCGATAAGC |
| CTX-M2-mR | TATTGCCCTTAAGCCACGTC | |
| CTX-M9-mF | blaCTX-M-9 cluster | ACG TGG CTC AAA GGC AAT AC |
| CTX-M9-mR | TCA ATT TGT TCA TGG CGG TA | |
| CMY-2bF | blaCMY-2 cluster | GAT TAG GCT GGG AGA TGC TG |
| DHA-fF | blaDHA | TTA CGG TCA TTA CGC CAC TG |
| DHA-mF | ATT TTC AGT GAC CGG CTG TT | |
| DHA-fR | TAA TTC AGC GCA GAT TGT GG | |
| DHA-mR | CAT TAA CGG TGT GAC CAA CG | |
| DHA AmpR-mR | CAG GGT AAA GCG GTG AAC AT | |
| IRL ISEcp1-F | ISEcp1-like | CCT AGA TTC TAC GTC AGT ACT TCA AAA |
| ISEcp1-F | TCT GCT CCT TGA GAA TGC AA | |
| ISEcp1-R | TCG CCC AAA ATG ACT TTA GC | |
| ISEcp1-bmF | TCA AAG AAG CCA AAT ACG ACA | |
| ISCR1-F | ISCR1 | GCG AGT CAA TCG CCC ACT |
| ISCR1-R | CGA CTC TGT GAT GGA TCG AA | |
| ISCR1-mF | GAT GCC GAG AAT ACG TGG TT | |
| ISCR1-pR | CAG ACG CTC GTG ATG ACA AT | |
| IS903-F | IS903 | TGG CCC ACC TAC AAT AAA GC |
| IS903-R | GGC ATA CCT GCT TTC GTC AT | |
| IS903-bF | CTG TTC GGG GGT TCA CTG AC | |
| IS903-mR | CAG ATG CAG CTT ACG CCA TA | |
| pspA-fF | pap | GCA TTG CCC GAT ATC AGA AC |
| pspB-F | TAT TTC TGG CCA TCC CGT TA | |
| pspD-mF | ACG TCC GCT AAA AAT GTT GC | |
| pspB-mR | CGA CTG GAA CGA TTG CTG TA | |
| pspD-bR | TGC TTG CTT TTG TTG TTT CG | |
| IntI1-F | IntI1 | CCA AGC TCT CGG GTA ACA TC |
| IntI1-R | CAT GAA AAC CGC CAC TGC | |
| 5CS-R | CTT AAG GAG GCT ACG GCT TTC | |
| qacEΔ1-F | qacEΔ1-sul1 | GAA AGG CTG GCT TTT TCT TG |
| qacEΔ1-R | GCA GCG ACT TCC ACG ATG | |
| 3CS-mR | TGT GAA AGG CGA GAT CAT CA | |
| sul1-F | TCA CCG AGG ACT CCT TCT TC | |
| sul1-mF | GGG TTT CCG AGA AGG TGA TT | |
| sul1-R | ACG AGA TTG TGC GGT TCT TC | |
| orf1-F | orf1 | GCT GTT CGT CGC TTT CTC TT |
| cat-F | cat | CCA AAA GGA CAC GGA GAT GT |
| cat-fF | AAG CAA GCT CAG GAG GTG AA | |
| aadA1-mF | aadA1 | ACA TCA TTC CGT GGC GTT AT |
| aadA1-fF | AGG TCC GTT TTT GGT CAC AG | |
| aadA1-mR | AGG TTT CAT TTA GCG CCT CA | |
| aadA1-fR | AGC CGA GGG AAG TTA AAA GC | |
| ORF477-F | orf477 | GAG CGA TAT GCA GCA ACA GA |
| ORF477-R | GCA TAC CAG GCC ATA AGC TC | |
| blc-R | blc | GTA GGG GTC CGT GAA AGG AT |
PCR-based replicon typing of plasmids.
Plasmid replicon typing of the isolates carrying genes encoding CTX-M ESBLs and/or AmpC β-lactamases was performed using the PCR-base method with 18 pairs of primers described by Carattoli et al. (4).
Plasmid analysis.
Plasmid DNA was purified from clinical P. mirabilis isolates using the NucleoBond Xtra Midi kit (Macherey-Nagel GmbH, Düren, Germany). Purified plasmids were digested with the restriction endonucleases HindIII, NdeI, and SpeI, and their restriction patterns were compared with one another by agarose gel electrophoresis.
PFGE and Southern blotting.
Plugs containing whole genomic DNA of the P. mirabilis isolates were digested with SfiI, I-CeuI, and S1 nuclease separately. DNA fragments were separated by pulsed-field gel electrophoresis (PFGE) using a CHEF-DRII device (Bio-Rad, Hercules, CA). The PFGE conditions of SfiI macrorestriction analysis were 6 V/cm for 20 h, with pulse times ranging from 0.5 to 60 s, at a temperature of 14°C. Pulse times for I-CeuI and S1 nuclease restriction analysis were 9 to 90 s. DNA treated with I-CeuI or S1 nuclease was blotted onto a nylon membrane (Zeta-Probe blotting membranes; Bio-Rad) and was hybridized with probes specific for genes encoding AmpC β-lactamases or CTX-M ESBLs, various specific replicons, and 16S rRNA genes. The probes were obtained by PCR experiments as described above. Probe labeling, hybridization, and detection were performed with the DIG DNA labeling and detection kit (Roche Diagnostics, Indianapolis, IN) according to the manufacturer's protocols.
Nucleotide sequence accession number.
The nucleotide sequence data for the blaCTX-M-90 gene are available in the GenBank nucleotide sequence database under accession no. GU065288.
RESULTS AND DISCUSSION
Prevalence and types of ESBLs and AmpCs.
The phenotypic confirmatory test detected 28 (12.6%) ESBL producers and 8 (3.6%) AmpC producers among 222 P. mirabilis isolates (data not shown). Three isolates gave positive results for both an ESBL and an AmpC enzyme simultaneously. P. mirabilis isolates exhibiting the ESBL and AmpC phenotypes were recovered from 12 and 7 hospitals, respectively.
The most common type of class A ESBL gene identified using PCR and sequencing experiments was blaCTX-M-14a (n = 12). The blaCTX-M-90 (n = 4), blaCTX-M-15 (n = 3), blaCTX-M-12 (n = 3), blaCTX-M-2 (n = 2), blaCTX-M-14b (n = 1), blaTEM-52 (n = 5), and blaSHV-12 (n = 1) genes were also detected. Three isolates each carried two ESBL genes (Table 2). No bla genes of the PER, VE-, GES, or TLA type, or of the CTX-M-8 cluster, were detected in this survey.
TABLE 2.
Characteristics of P. mirabilis isolates producing ESBLs and/or AmpC β-lactamases
| Isolate | Hospital | Warda | Specimen | β-Lactamase |
MIC (μg/ml)b |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| ESBL | AmpC | CTX | CTX-CA | CAZ | CAZ-CA | FOX | ||||
| SSM6 | Seoul-1 | ER | Urine | TEM-52 | 8 | <0.06 | 4 | <0.06 | 2 | |
| SSM10 | Seoul-1 | GW | Pus | CTX-M-90, TEM-52 | 32 | 0.12 | 8 | 0.12 | 2 | |
| SSM12 | Seoul-1 | ER | Urine | CTX-M-14, SHV-12 | DHA-1 | 32 | 0.25 | 16 | 0.12 | 16 |
| trcSSM12 | CTX-M-14 | 16 | <0.06 | 0.25 | <0.06 | 2 | ||||
| KNS26 | Seoul-2 | GW | Urine | CTX-M-12 | 16 | <0.06 | 2 | <0.06 | 4 | |
| KNS34 | Seoul-2 | GW | Urine | CMY-2 | 8 | 8 | 8 | 4 | 32 | |
| KDS43 | Seoul-3 | GW | Pus | CMY-2 | 16 | 16 | 16 | 16 | 64 | |
| YS126 | Seoul-4 | GW | Pus | CTX-M-12 | 32 | 0.12 | 0.5 | 0.12 | 4 | |
| YS132 | Seoul-4 | GW | Sputum | CTX-M-90, TEM-52 | 64 | 0.12 | 4 | 0.5 | 2 | |
| trcYS132 | TEM-52 | 32 | <0.06 | 16 | 0.25 | 2 | ||||
| YS143 | Seoul-4 | GW | Pus | CTX-M-14 | 32 | <0.06 | 0.25 | <0.06 | 4 | |
| JYS190 | Seoul-5 | GW | Pus | CTX-M-14 | 32 | 0.12 | 0.25 | <0.06 | 2 | |
| JYS195 | Seoul-5 | GW | Pus | CTX-M-14 | 16 | <0.06 | 0.25 | <0.06 | 2 | |
| JYS201 | Seoul-5 | ER | Pus | CTX-M-14 | 16 | <0.06 | 0.25 | <0.06 | 4 | |
| JHS209 | Seoul-6 | GW | Pus | CTX-M-12 | 16 | 0.12 | 2 | 0.25 | 2 | |
| JHS214 | Seoul-6 | GW | Urine | CTX-M-15 | 32 | <0.06 | 8 | <0.06 | 4 | |
| JHS215 | Seoul-6 | ICU | Pus | CTX-M-14 | CMY-2 | 32 | 16 | 4 | 4 | 32 |
| JHS218 | Seoul-6 | ICU | Urine | CTX-M-14 | CMY-2 | 16 | 16 | 4 | 4 | 32 |
| trcJHS218 | CMY-2 | 16 | 8 | 16 | 16 | 64 | ||||
| JHS222 | Seoul-6 | GW | Urine | CTX-M-14 | 32 | <0.06 | 0.25 | <0.06 | 2 | |
| MG63 | Goyang | ER | Blood | CTX-M-14 | 32 | <0.06 | 0.25 | <0.06 | 2 | |
| trcMG63 | CTX-M-14 | 16 | <0.06 | 0.12 | <0.06 | 2 | ||||
| MG67 | Goyang | ICU | Blood | CMY-2 | 8 | 8 | 8 | 4 | 32 | |
| MG73 | Goyang | GW | Pus | CTX-M-15 | >128 | <0.06 | 8 | <0.06 | 2 | |
| MG74 | Goyang | GW | Urine | CTX-M-15 | 128 | 0.12 | 4 | 0.12 | 2 | |
| MG77 | Goyang | ER | Urine | CTX-M-14 | 32 | <0.06 | 0.25 | <0.06 | 8 | |
| BDC115 | Sungnam | GW | Pus | CTX-M-14 | 32 | 0.12 | 0.5 | 0.12 | 4 | |
| WJC161 | Wonju | GW | Sputum | CTX-M-2 | 64 | 0.25 | 2 | 0.12 | 8 | |
| WJC164 | Wonju | GW | Urine | CTX-M-2 | 64 | 0.25 | 1 | 0.12 | 8 | |
| WJC168 | Wonju | GW | Urine | DHA-1 | 1 | 2 | 1 | 2 | 64 | |
| GM58 | Daegu | ICU | Sputum | CMY-2 | 4 | 8 | 2 | 2 | 32 | |
| GS56 | Jinju | GW | Sputum | CTX-M-14 | 32 | 0.12 | 2 | 0.12 | 4 | |
| PS79 | Busan-1 | ER | Blood | TEM-52 | 16 | <0.06 | 32 | <0.06 | 2 | |
| IJP86 | Busan-2 | GW | Pus | CTX-M-14 | 32 | <0.06 | 0.12 | <0.06 | 4 | |
| trcIJP86 | CTX-M-14 | 16 | <0.06 | 0.12 | <0.06 | 2 | ||||
| IJP88 | Busan-2 | GW | Urine | TEM-52 | 32 | <0.06 | >128 | <0.06 | 4 | |
| IJP92 | Busan-2 | ER | Urine | CTX-M-90 | 32 | 0.12 | 0.25 | 0.12 | 4 | |
| JJH178 | Jeju | GW | Urine | CTX-M-90 | 128 | 0.25 | 0.25 | <0.06 | 4 | |
ER, emergency room; GW, general ward; ICU, intensive-care unit.
CTX, cefotaxime; CA, clavulanic acid; CAZ, ceftazidime; FOX, cefoxitin.
Compared with that found by a survey carried out in 2004 (18), the prevalence of ESBL-producing P. mirabilis has increased from 9.7% to 12.6% in Korea. In 2004, 6.7% (9/134) of P. mirabilis clinical isolates were reported to produce CTX-M ESBLs, while in 2008, the prevalence of these enzymes showed a notable increase, reaching 11.3% (25/222). These results suggest that the significant increase in the incidence of ESBL-producing P. mirabilis has been due to the dissemination of strains producing CTX-M enzymes.
All eight isolates exhibiting the AmpC phenotype carried an AmpC β-lactamase gene, such as blaCMY-2 (n = 6) or blaDHA-1 (n = 2). Moreover, bla genes encoding ESBLs and AmpC β-lactamases were detected simultaneously in three isolates. The phenotypic characteristics of ESBL- and/or AmpC-producing isolates and their transconjugants are shown in Table 2.
Analysis of the genetic contexts of the bla genes.
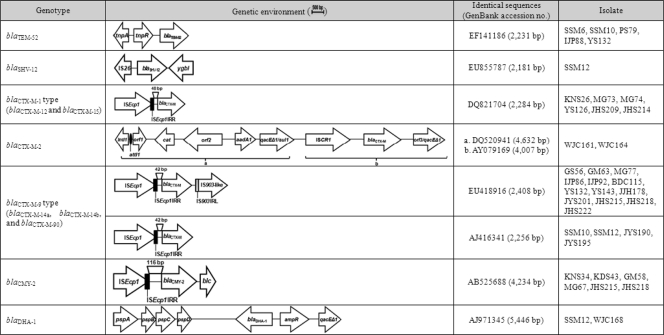
All bla genes encoding CTX-M-1- and CTX-M-9-type enzymes were preceded by ISEcp1-like elements (Fig. 1). The ISEcp1-like elements were observed 48 bp upstream of the open reading frames (ORFs) encoding CTX-M-1-type enzymes or 42 bp upstream of the ORFs encoding CTX-M-9-type enzymes, as reported previously (11). The IS903-like element was detected downstream of the blaCTX-M-9-type genes in 13 isolates (Fig. 1).
FIG. 1.
Schematic representation of the genetic environments surrounding the ESBL and AmpC genes in Proteus mirabilis clinical isolates. IRR, right inverted repeat sequences; IRL, left inverted repeat sequences.
The blaCTX-M14b gene was first detected in E. coli strains isolated in Spain, and it exhibits three silent mutations compared with the blaCTX-M-14a gene (16). Navarro et al. (16) reported that the blaCTX-M-14a gene was associated with ISEcp1 and IS903 located on an IncK plasmid, whereas the blaCTX-M-14b gene was associated with In60 located on an IncHI2 plasmid, like blaCTX-M-9. In this study, one isolate (JY195) was found to contain a blaCTX-M-14b gene, but it was linked to ISEcp1, like the blaCTX-M-14a gene, and was located on the chromosome (see below).
The blaCTX-M-2 gene in two isolates was located on a complex class 1 integron. The 4,632-bp region from the 5′ conserved segment (5′-CS) to the first copy of 3′-CS (3′-CS1) showed 100% homology with a class 1 integron previously characterized from a P. mirabilis strain (12). The 4,007-bp region from ISCR1 to the second copy of 3′-CS (3′-CS2) also showed 100% homology to that of an unusual class 1 integron, In35 (2) (Fig. 1).
The blaTEM-52 gene in all five isolates was preceded by resolvase (tnpR) and transposase (tnpA) genes identical to those of transposon Tn3 (7). The blaSHV-12 gene in isolate SSM12 was preceded by IS26 and was followed by a transcriptional regulator gene (ygbI), showing the same arrangement as that found on plasmid pE19 from an Enterobacter cloacae strain (GenBank accession no. GU205813) (Fig. 1).
The blaCMY-2 gene in all six isolates was also preceded by ISEcp1-like elements. The ISEcp1-like elements were observed 116 bp upstream of the ORF encoding the CMY-2 enzyme. The blaDHA-1 gene in two isolates was preceded by a transcriptional regulator gene (ampR) and followed by phage shock protein genes (pspA, pspB, pspC, and pspD), as reported previously (23) (Fig. 1).
Locations of the bla genes.
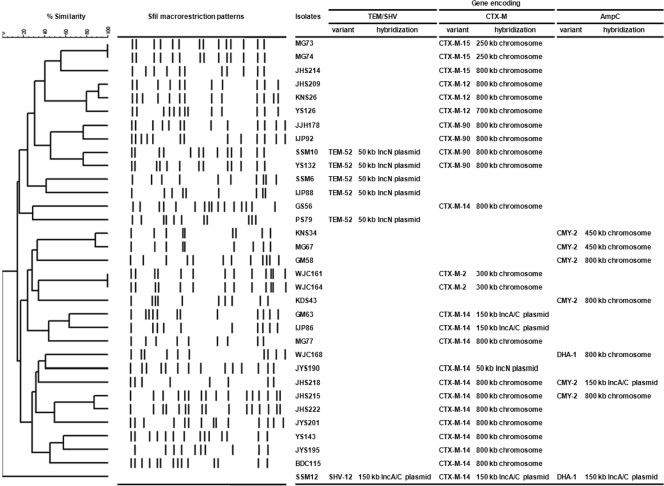
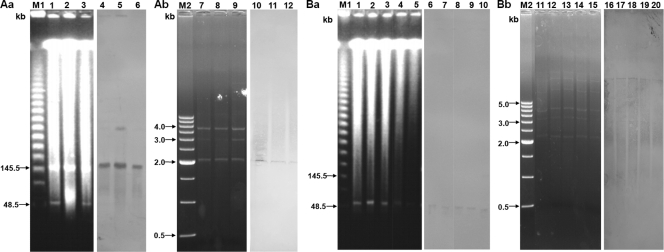
Despite repeated attempts, transconjugants were obtained from only five isolates, with transfer of blaCTX-M-14 in three cases and of blaTEM-52 and blaDHA-1 in the remaining cases (Table 2). The probe specific for the blaCTX-M-14a gene hybridized with a ∼50-kbp IncN plasmid and ∼150-kbp IncA/C plasmids in one and three isolates, respectively (Fig. 2). IncA/C plasmids carrying the blaCTX-M-14a gene from three isolates exhibited restriction patterns identical to one another when digested with endonucleases of HindIII, NdeI, and SpeI (Fig. 3 Ab). The probe specific for the blaTEM-52 gene hybridized with ∼150-kbp IncA/C plasmids in all five isolates. The IncN plasmids also showed restriction patterns identical to one another (Fig. 3Bb). The probe specific for the blaCMY-2 gene hybridized with a ∼50-kbp IncN plasmid in only one isolate. However, probes specific for the CTX-M and AmpC genes did not bind to any plasmids in the remaining isolates. Probes specific for CTX-M genes hybridized strongly with I-CeuI macrorestriction fragments (250 to >800 kbp) from 21 CTX-M ESBL-producing P. mirabilis isolates (2 isolates with CTX-M-2, 3 with CTX-M-12, 9 with CTX-M-14, 3 with CTX-M-15, and 4 with CTX-M-90). Furthermore, probes specific for the blaDHA-1 and blaCMY-2 genes also hybridized with I-CeuI macrorestriction fragments (450 to >800 kbp) from two and five isolates, respectively. Finally, the hybridization of macrorestriction fragments with the probe specific for 16S rRNA genes confirmed the chromosomal location of the bla genes encoding CTX-M ESBLs and AmpC enzymes (data not shown).
FIG. 2.
Dendrogram based on SfiI macrorestriction patterns of P. mirabilis isolates producing ESBLs and/or AmpC β-lactamases, indicating the similarity (expressed as a percentage) of PFGE profiles. Isolate SSM12 repeatedly failed to yield SfiI macrorestriction fragments. Three CTX-M-12-producing isolates showed >80% similarity, despite being recovered from three different hospitals, while other types of ESBL- or AmpC-producing isolates showed lower percentages of similarity. The locations of the bla genes of TEM-, SHV-, and CTX-M-type ESBLs and AmpC enzymes are given on the right.
FIG. 3.
(Aa) Banding patterns of S1 nuclease-digested linearized plasmids from three P. mirabilis clinical isolates carrying the blaCTX-M-14a gene (lanes 1 to 3) and Southern blot analysis of plasmid DNA with a probe specific to the gene (lanes 4 to 6). (Ab) Restriction patterns of ∼150-kbp plasmids carrying the blaCTX-M-14a gene digested with the HindIII, NdeI, or SpeI restriction endonuclease (lanes 7 to 9) and Southern blot analysis of plasmid DNA with a probe specific to the gene (lanes 10 to 12). (Ba) Banding patterns of S1 nuclease-digested linearized plasmids from five P. mirabilis clinical isolates carrying the blaTEM-52 gene (lanes 1 to 5) and Southern blot analysis of plasmid DNA with a probe specific to the gene (lanes 6 to 10). (Bb) Restriction patterns of ∼50-kbp plasmids carrying the blaTEM-52 gene (lanes 11 to 15) and Southern blot analysis of plasmid DNA with a probe specific to the gene (lanes 16 to 20). Lanes M1, Lambda ladder (Promega Co., Madison, WI); lanes M2, 500-bp DNA ladder (Takara Inc., Tokyo, Japan).
Strain typing.
Of 13 CTX-M-14-producing isolates, 1 (SSM12) repeatedly failed to yield SfiI macrorestriction fragments. Among 12 CTX-M-14-producing isolates, only 2 (JHS215 and JHS222), which were recovered from the same hospital, were clonally related to one another, with 80% similarity; others were not clonally related (Fig. 2).
Our results revealed that the spread of the blaCTX-M-14a and blaTEM-52 genes among P. mirabilis might be driven by horizontal transfer of IncA/C and IncN plasmids, respectively. Notably, however, most of the CTX-M ESBL and AmpC β-lactamase genes were found on the chromosome and not on plasmids. A chromosomal location for CTX-M ESBL and AmpC β-lactamase genes in Enterobacteriaceae other than P. mirabilis has rarely been reported, though Coque et al. (8) recently described eight E. coli clinical isolates carrying the chromosome-borne blaCTX-M-15 gene. In contrast to other species, P. mirabilis containing CTX-M ESBL and/or AmpC β-lactamase genes on the chromosomes has been reported uncommonly elsewhere. It is noteworthy that the chromosomal location of CTX-M ESBL and AmpC β-lactamase genes in P. mirabilis is no longer an unusual phenomenon in the hospital environment. Chromosomally located genes are surrounded by a genetic environment similar to that of plasmids, and this supports the idea that the CTX-M ESBL and AmpC β-lactamase genes are integrated into the chromosome via transposable elements in the P. mirabilis species.
In conclusion, the present data suggested that the incidence of ESBL-producing P. mirabilis has increased due to the dissemination of bla genes encoding CTX-M enzymes in Korea. The most common types of ESBL and AmpC β-lactamase in P. mirabilis were CTX-M-14 and CMY-2, respectively. Interestingly, these enzymes were chromosomally located in most P. mirabilis isolates. The mechanism and significance of this phenomenon need to be elucidated.
Acknowledgments
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MEST) (2009-0071195) and by a research grant from the Korea Center for Disease Control (2009-E00520-00).
Footnotes
Published ahead of print on 31 January 2011.
REFERENCES
- 1.Aragón, L. M., et al. 2008. Increase in β-lactam-resistant Proteus mirabilis strains due to CTX-M- and CMY-type as well as new VEB- and inhibitor-resistant TEM-type β-lactamases. J. Antimicrob. Chemother. 61:1029-1032. [DOI] [PubMed] [Google Scholar]
- 2.Arduino, S. M., et al. 2002. blaCTX-M-2 is located in an unusual class 1 integron (In35) which includes orf513. Antimicrob. Agents Chemother. 46:2303-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bret, L., et al. 1998. Chromosomally encoded AmpC-type β-lactamase in a clinical isolate of Proteus mirabilis. Antimicrob. Agents Chemother. 42:1110-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carattoli, A., et al. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219-228. [DOI] [PubMed] [Google Scholar]
- 5.Chanal, C., et al. 2000. Prevalence of β-lactamases among 1,072 clinical isolates of Proteus mirabilis: a 2-year survey in a French hospital. Antimicrob. Agents Chemother. 44:1930-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2009. Performance standards for antimicrobial susceptibility testing; seventeenth informational supplement. M7-A8. CLSI, Wayne, PA.
- 7.Cloeckaert, A., et al. 2007. Dissemination of an extended-spectrum β-lactamase blaTEM-52 gene carrying IncI1 plasmid in various Salmonella enterica serovars isolated from poultry and humans in Belgium and France between 2001 and 2005. Antimicrob. Agents Chemother. 51:1872-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coque, T. M., et al. 2008. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum β-lactamase CTX-M-15. Emerg. Infect. Dis. 14:195-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Andrea, M. M., et al. 2006. CMY-16, a novel acquired AmpC-type β-lactamase of the CMY/LAT lineage in multifocal monophyletic isolates of Proteus mirabilis from Northern Italy. Antimicrob. Agents Chemother. 50:618-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Decré, D., et al. 2002. Characterization of CMY-type β-lactamases in clinical strains of Proteus mirabilis and Klebsiella pneumoniae isolated in four hospitals in the Paris area. J. Antimicrob. Chemother. 50:681-688. [DOI] [PubMed] [Google Scholar]
- 11.Eckert, C., V. Gautier, and G. Arlet. 2006. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J. Antimicrob. Chemother. 57:14-23. [DOI] [PubMed] [Google Scholar]
- 12.Gionechetti, F., et al. 2008. Characterization of antimicrobial resistance and class 1 integrons in Enterobacteriaceae isolated from Mediterranean herring gulls (Larus cachinnans). Microb. Drug Resist. 14:93-99. [DOI] [PubMed] [Google Scholar]
- 13.Jacoby, G. A., and P. Han. 1996. Detection of extended-spectrum β-lactamases in clinical isolates of Klebsiella penumoniae and Escherichia coli. J. Clin. Microbiol. 34:908-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Literacka, E., et al. 2004. Four variants of the Citrobacter freundii AmpC-type cephalosporinases, including novel enzymes CMY-14 and CMY-15, in a Proteus mirabilis clone widespread in Poland. Antimicrob. Agents Chemother. 48:4136-4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luzzaro, F., et al. 2009. Spread of multidrug-resistant Proteus mirabilis isolates producing an AmpC-type β-lactamase: epidemiology and clinical management. Int. J. Antimicrob. Agents 33:328-333. [DOI] [PubMed] [Google Scholar]
- 16.Navarro, F., et al. 2007. Evidence for convergent evolution of CTX-M-14 ESBL in Escherichia coli and its prevalence. FEMS Microbiol. Lett. 273:120-123. [DOI] [PubMed] [Google Scholar]
- 17.Navon-Venezia, S., I. Chmelnitsky, A. Leavitt, and Y. Carmeli. 2008. Dissemination of the CTX-M-25 family β-lactamases among Klebsiella pneumoniae, Escherichia coli and Enterobacter cloacae and identification of the novel enzyme CTX-M-41 in Proteus mirabilis in Israel. J. Antimicrob. Chemother. 62:289-295. [DOI] [PubMed] [Google Scholar]
- 18.Park, Y. J., et al. 2006. Occurrence of extended-spectrum β-lactamases and plasmid-mediated AmpC β-lactamases among Korean isolates of Proteus mirabilis. J. Antimicrob. Chemother. 57:156-158. [DOI] [PubMed] [Google Scholar]
- 19.Rózalski, A., Z. Sidorezyk, and K. Kotelko. 1997. Potential virulence factors of Proteus bacilli. Microbiol. Mol. Biol. Rev. 61:65-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryoo, N. H., et al. 2005. Dissemination of SHV-12 and CTX-M-type extended-spectrum β-lactamases among clinical isolates of Escherichia coli and Klebsiella pneumoniae and emergence of GES-3 in Korea. J. Antimicrob. Chemother. 56:698-702. [DOI] [PubMed] [Google Scholar]
- 21.Song, W., et al. 2007. Use of boronic acid disk methods to detect the combined expression of plasmid-mediated AmpC β-lactamases and extended-spectrum β-lactamases in clinical isolates of Klebsiella spp., Salmonella spp., and Proteus mirabilis. Diagn. Microbiol. Infect. Dis. 57:315-318. [DOI] [PubMed] [Google Scholar]
- 22.Song, W., et al. 2006. Increasing trend in the prevalence of plasmid-mediated AmpC β-lactamases in Enterobacteriaceae lacking chromosomal ampC gene at a Korean university hospital from 2002 to 2004. Diagn. Microbiol. Infect. Dis. 55:219-224. [DOI] [PubMed] [Google Scholar]
- 23.Verdet, C., et al. 2006. Emergence of DHA-1-producing Klebsiella spp. in the Parisian region: genetic organization of the ampC and ampR genes originating from Morganella morganii. Antimicrob. Agents Chemother. 50:607-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vourli, S., et al. 2006. Emergence of Proteus mirabilis carrying the blaVIM-1 metallo-β-lactamase gene. Clin. Microbiol. Infect. 12:691-694. [DOI] [PubMed] [Google Scholar]