Abstract
The importance of the mammalian intestinal microbiota to human health has been intensely studied over the past few years. It is now clear that the interactions between human hosts and their associated microbial communities need to be characterized in molecular detail if we are to truly understand human physiology. Additionally, the study of such host-microbe interactions is likely to provide us with new strategies to manipulate these complex systems to maintain or restore homeostasis in order to prevent or cure pathological states. Here, we describe the use of high-throughput metabolomics to shed light on the interactions between the intestinal microbiota and the host. We show that antibiotic treatment disrupts intestinal homeostasis and has a profound impact on the intestinal metabolome, affecting the levels of over 87% of all metabolites detected. Many metabolic pathways that are critical for host physiology were affected, including bile acid, eicosanoid, and steroid hormone synthesis. Dissecting the molecular mechanisms involved in the impact of beneficial microbes on some of these pathways will be instrumental in understanding the interplay between the host and its complex resident microbiota and may aid in the design of new therapeutic strategies that target these interactions.
The human body is colonized by a complex community of microbes termed microbiota or microbiome (9, 17, 18, 24). Virtually every surface of the human body that is exposed to the environment has its own microbial assemblage, with different microbial species and distinct functions associated with them. The intestinal tract is by far the most heavily colonized body site; it has been estimated that the human gut harbors some 1014 microbial cells (14, 22, 32). This microbial consortium is critical for human health and has been implicated in the development of the immune system, energy homeostasis, and protection against pathogens, among other processes (18, 24). Conversely, imbalances in the intestinal microbiota have also been associated with many pathological processes, including inflammatory bowel disease, diabetes, asthma, obesity, autism, and others (18, 24).
The use of antibiotics is known to have significant effects on the intestinal microbiota. The acquisition and spread of antibiotic resistance genes between and within bacterial communities due to antibiotic use have also been studied thoroughly (12). This has caused an increased awareness of the importance of a more responsible use of antibiotics. However, it is only recently that studies began to reveal details of the impact of these drugs on intestinal microbial communities. For instance, it is now well established that antibiotic treatment increases susceptibility to enteric infections (10, 23, 25); some of the members of the microbiota involved in this process are currently under investigation. Although these studies are indispensable to our understanding of the effects of antibiotics in the human body and the importance of the intestinal microbiota for human health, the mechanisms involved in these interactions remain mostly unknown.
Much of what we have learned about the intestinal microbiota comes from high-throughput sequencing studies of the intestinal metagenome, which address only the microbial composition of the samples but do not define the metabolic functions involved (3, 34). Recently, metabolomics has been established as a new technology whose aim is to study the complex lexicon of small molecules present in any given biological sample, or the metabolome (6, 8, 19, 31). In order to expand our understanding of the importance of the intestinal microbiota as well as the disturbances elicited by antibiotic treatment, we have used metabolomics to obtain a snapshot of the chemical composition of the intestinal environment before and after antibiotic treatment. By using Fourier transform ion cyclotron resonance mass spectrometry with direct infusion (DI-FT-ICR-MS) (6), we determined that a single, high dose of the antibiotic streptomycin can have a profound impact on the levels of the majority of the compounds detected. Predictive mapping of these compounds to corresponding metabolic pathways showed that many crucial host metabolic functions are disturbed. Among some of the pathways affected are those involved in sugar, amino acid, fatty acid, bile acid, steroid, and eicosanoid metabolism. Additionally, we show that clinically relevant antibiotic doses can also disrupt host eicosanoid metabolism. Our results show that the microbiota has effects on previously unidentified host functions. Additionally, the critical functions of all pathways affected suggest that the impact of antibiotics on mammalian physiology extends far beyond the development of microbial drug resistance.
MATERIALS AND METHODS
Chemical reagents.
Haloperidol, reserpine, acetonitrile, water, formic acid, ammonium hydroxide, streptomycin, vancomycin, tetracycline, and metronidazole were purchased from Sigma-Aldrich (St. Louis, MO). The ES tuning mix standard solution was purchased from Agilent Technologies (Santa Clara, CA).
Antibiotic treatment.
Age- and gender-matched C57BL/6 mice were used. Mice were obtained from The Jackson Laboratory (Bar Harbor, ME) and our own colony, maintained at The University of British Columbia. Fresh feces were collected and stored at −80°C. Mice were then treated with 20 mg of streptomycin through oral gavage, and fresh feces were collected at several time points after treatment and stored at −80°C until use. For treatments with low doses of antibiotics, drugs were added to drinking water and mice were treated for 2 days, after which feces were collected and immediately used. Antibiotic concentrations were as follows: streptomycin, 450 mg/liter; vancomycin, 50 mg/liter; tetracycline, 250 mg/liter; and metronidazole, 750 mg/liter. All animal experiments were approved by the Animal Care Committee of The University of British Columbia and performed in accordance with institutional guidelines.
Sybr green staining.
Fecal samples were homogenized in sterile Dulbecco's phosphate-buffered saline (PBS; HyClone, Logan, UT) using a Mixer Mill MM 301 apparatus (Retsch, Haan, Germany). Total bacterial counts from fecal samples were performed using Sybr green staining as previously described (15). Fecal samples were diluted 1:10 in PBS, fixed, and stored in 3.7% formalin at 4°C. Two microliters was stained with 0.25 μl of Sybr green (Invitrogen, Carlsbad, CA) and viewed with a fluorescence microscope. Cells from three randomly chosen fields were counted and the numbers were averaged. Counts were corrected on the basis of the volume and dilution used and the known diameter of the microscope field of view to determine the number of cells per gram of feces in each sample.
Metabolite extraction.
To extract metabolites from feces, acetonitrile was added to samples (approximately 10 to 25 μl of acetonitrile per 1 mg of tissue), which were then homogenized using a tungsten bead and the Mixer Mill. The samples were then cleared by centrifugation and the supernatant was collected and dried at room temperature using a centrifuge equipped with a vacuum pump. All extracts were kept at −80°C until use.
DI-FT-ICR-MS experiments.
For metabolic profiling, the dried extracts from mouse feces were suspended in a 2:3 mixture of water and acetonitrile (10 μl per 1 mg of feces), vortexed, and cleared by centrifugation. Supernatants were collected and used as described below. Extracts were diluted 1:5 with 60% acetonitrile containing either 0.2% formic acid (for positive-ion mode) or 0.5% ammonium hydroxide (for negative-ion mode) and spiked with predefined amounts of the ES tuning mix solution as the internal standard for mass calibration. The solutions were then infused, using a syringe pump (KDS Scientific, Holliston, MA), at a flow rate of 2.5 μl per minute, into a 12-T Apex-Qe hybrid quadrupole-FT-ICR mass spectrometer (Bruker Daltonics, Billerica, MA) equipped with an Apollo II electrospray ionization source, a quadrupole mass filter, and a hexapole collision cell. Data were recorded in positive- and negative-ion modes with broadband detection and an FT acquisition size of 1,024 kilobytes per second within an m/z range of from 150 to 1,100. Under these settings, a mass resolution of ca. 100,000 (full width at half maximum [FWHM]) at m/z 400 and a mass accuracy within 2 ppm or less for all detected components, following internal mass calibration, were observed. Other experimental parameters were as follows: capillary electrospray voltage of 3,600 to 3,750 V, spray shield voltage of 3,300 to 3,450 V, source ion accumulation time of 0.1 s, and collision cell ion accumulation time of 0.2 s. To increase detection sensitivity, survey scan mass spectra in positive- and negative-ion modes were acquired from the accumulation of 200 scans per spectrum, and duplicate acquisitions per sample were carried out to ensure data reproducibility. As an indication of the analytical reproducibility of the methods employed, we report that more than 90% of the peaks detected showed less than 10% variation in intensity between the analytical replicates (data not shown).
DI-FT-ICR-MS data processing.
Raw mass spectrometry data were processed using a custom-developed software package, as described elsewhere (6). First, raw mass spectra acquired from each sample group were batch processed using the instrument vendor's data analysis software, DataAnalysis, but with a home-written VBA script (Visual Basic for Applications) to do automatic internal mass calibration with the reference masses of the spiked calibration standards and a known contaminant, N-butylbenzensulfonamide. Monoisotopic peaks corresponding to the isotopic pattern distributions were then automatically determined, and those with a signal/noise ratio above 3 were picked. Their m/z values were converted to neutral masses by subtracting 1.007276 for positive-ion mode or adding 1.007276 for negative-ion mode. Next, the resulting mass lists from all the mass spectra within each set of untreated or treated groups detected in positive- or negative-ion modes were further processed with another customized software program developed with the LabVIEW graphical programming environment (National Instruments, Austin, TX). Adduct ions were recognized and converted to neutral masses from the mass lists on the basis of the expected mass differences between protonated (M + H)+, (M + Na)+, and/or (M + K)+ ions for positive-ion mode or deprotonated (M − H)− and (M + Cl)− ions for negative-ion mode, within 2 ppm, to yield a list of unique biochemical component masses, together with the sum of their peak intensities. The peak intensities of all the monoisotopic neutral masses were subsequently normalized to the intrasample total ion intensity. Masses observed in at least three of four samples from one of the sample groups (untreated or treated) were aligned and then combined into unique metabolite features from the masses that matched within 2 ppm across all the data. Finally, a two-dimensional data matrix (mass versus relative intensity) was generated for each sample group and saved in a format amenable for further data analysis. Heat maps were then created using the freely available software programs Cluster and Java TreeView (http://rana.lbl.gov/eisensoftware.htm). To identify differences in metabolite composition between untreated and treated samples, we first filtered our list of masses for metabolites that were present in one set of samples (untreated or treated) but not the other. Additionally, we averaged the mass intensities of metabolites in each group and calculated the ratios between averaged intensities of metabolites from untreated and treated samples. To assign possible metabolite identities to the masses present in only one of the sample groups or showing at least a 2-fold change in intensities between the sample groups, the monoisotopic neutral masses of interest were queried against MassTrix (http://masstrix.org), a free-access software designed to incorporate masses into metabolic pathways using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/) (27). Masses were searched against the Mus musculus database within a mass error of 3 ppm.
ELISAs.
Fresh feces were collected and homogenized in PBS, as described for the Sybr green assays. Samples were cleared by centrifugation, and commercially available enzyme-linked immunosorbent assay (ELISA) kits (Cayman Chemical and Assay Designs, Ann Arbor, MI) were used to determine fecal concentrations of leukotriene B4 (LTB4), prostaglandin E2 (PGE2), prostaglandin F2α (PGF2α), and cysteinyl leukotrienes (CysLT).
Statistical analysis.
Data were analyzed by unpaired t tests with 95% confidence intervals using Prism (version 4.0) software (GraphPad Software Inc., San Diego, CA).
DI-FT-ICR-MS data accession number.
The mass spectrometry data have been deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE25687.
RESULTS
Antibiotic treatment causes drastic changes in microbial and chemical compositions of feces.
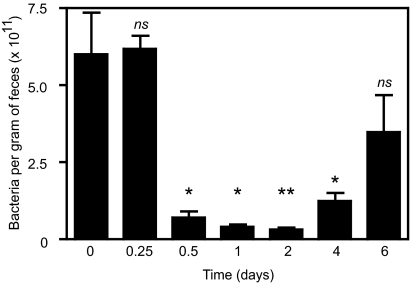
In order to determine the effect of antibiotic treatment on intestinal microbial populations, we determined the number of bacterial cells in feces before and at several time points following streptomycin treatment, until the first signs of recovery appeared. We used this antibiotic because it has previously been shown to affect the structure and function of the intestinal microbiota and increase susceptibility to enteric infection (1, 25). Figure 1 shows that after 6 h of streptomycin treatment the number of bacterial cells in feces remains unaltered. However, 12 h after the initial treatment, we observed a statistically significant (P = 0.0189), 90% reduction in the number of fecal bacteria. At 1 and 2 days posttreatment, the reduction in the number of fecal bacteria was approximately 95%, and these differences were also statistically significant (P = 0.0319 and 0.0013, respectively). At 4 days after treatment, recovery of the intestinal microbiota began, although bacterial numbers were still significantly different from those for the control (P = 0.0306). On day 6, an additional increase in bacterial numbers was observed, and the data at this time point were not significantly different from those for the control samples before antibiotic treatment. These data show that the effect of the antibiotic is most evident from 12 to 48 h after treatment and that this represents an appropriate time frame for the analysis of the intestinal metabolome in order to identify changes caused by disturbances in the microbiota.
FIG. 1.
Dynamics of killing and recovery of intestinal microbial populations upon antibiotic treatment. The numbers of microbial cells present in feces were determined through Sybr green staining before and several time points after mice received 20 mg of streptomycin through oral gavage. The numbers of mice used were 3 (0.25 and 1 day after treatment), 4 (0.5, 4, and 6 days after treatment), and 7 (0 and 2 days after treatment). ns, not significant (P > 0.05); *, P < 0.04; **, P < 0.002.
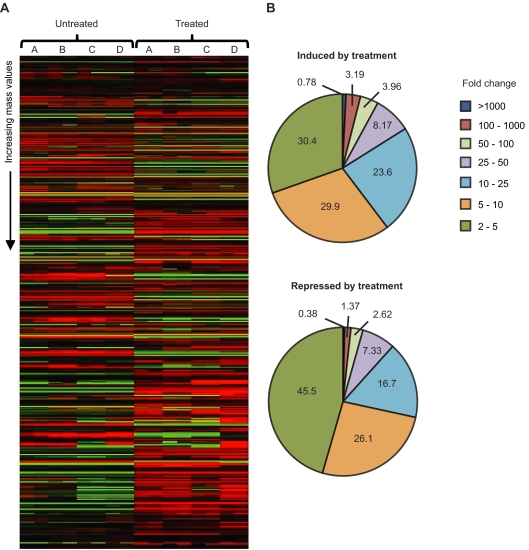
To study the effect of antibiotic treatment in the chemical composition of the intestinal environment, we used DI-FT-ICR-MS to detect and relatively quantify more than 2,000 small-molecule features detected from fecal samples of mice before and after antibiotic treatment. Because the kinetics of metabolite production and degradation can vary widely between compounds, the effect in the number of bacterial cells seen may not be directly correlated with the amounts of the compounds affected by them. Therefore, we chose to analyze samples obtained 24 h after antibiotic treatment to allow some time between the initial killing and recovery of bacterial populations and our sample collection. This increases the chances that a metabolite whose levels are increased in the presence of the intestinal microbiota but that is relatively stable has time to be degraded so that differences in its levels can be detected when the samples are collected. Conversely, our strategy aims at allowing metabolites whose levels are decreased in the presence of the intestinal microbiota to accumulate after antibiotic treatment, but before sample collection, so that changes in their levels caused by treatment can be detected. To study the chemical composition of samples, we prepared acetonitrile extracts from fresh feces from mice before and after antibiotic treatment. DI-FT-ICR-MS analysis of fecal extracts yielded a total of 2,230 different masses (metabolite features) from both untreated and treated samples (Table 1), which were detected from combined positive- and negative-ion modes. To investigate which of these masses were present at different levels in untreated and treated samples, we first selected those that were present only in untreated or treated samples. Additionally, we calculated the average intensities of all masses and compared values from each of the sample groups (untreated and treated). Masses that showed changes of 2-fold or more were combined with those that were observed only in untreated or treated samples for further analyses. Based on this, we found that antibiotic treatment altered the levels of 1,958 of the total 2,230 metabolite features detected (Table 1 and Fig. 2). This represents 87.8% of all masses detected, showing that antibiotic treatment has a profound impact on the biochemical composition of feces. Metabolite levels were affected in various degrees, with changes ranging from 2-fold to over 10,000-fold (Fig. 2).
TABLE 1.
Overview of DI-FT-ICR-MS results and impact of antibiotic treatment on the intestinal metabolomea
| Metabolites | No. of metabolite masses |
|---|---|
| Metabolites detected | |
| Negative ionization | 1,043 |
| Positive ionization | 1,386 |
| Overlap | 199 |
| Total | 2,230 |
| Metabolites changed | |
| Untreated > treated | 793 |
| Treated > untreated | 1,165 |
| Total changed | 1,958 |
The total number of changed metabolites represents 87.8% of all metabolite masses detected.
FIG. 2.
Antibiotic treatment has a profound impact on the chemical composition of feces. (A) The heat maps show the impact of streptomycin treatment on the levels of metabolites from mouse feces. Data were median centered using cluster analysis, and heat maps were constructed using Java TreeView (http://rana.lbl.gov/eisensoftware.htm). Masses are presented from lowest (top) to highest (bottom). Green, masses with signal intensities higher than the median; red, signal intensities lower than the median; black, missed values or values with no difference from the median signal intensity. Each of the letters above the map (A, B, C, and D) indicates one mouse used. Each letter corresponds to the two columns of data under it in the heat map due to the duplicate data acquisitions performed. (B) Distribution of metabolites affected, based on fold changes. Numbers inside and around the pie charts represent the percentage of the total number of metabolites affected.
Multiple metabolic pathways are affected by antibiotic treatment.
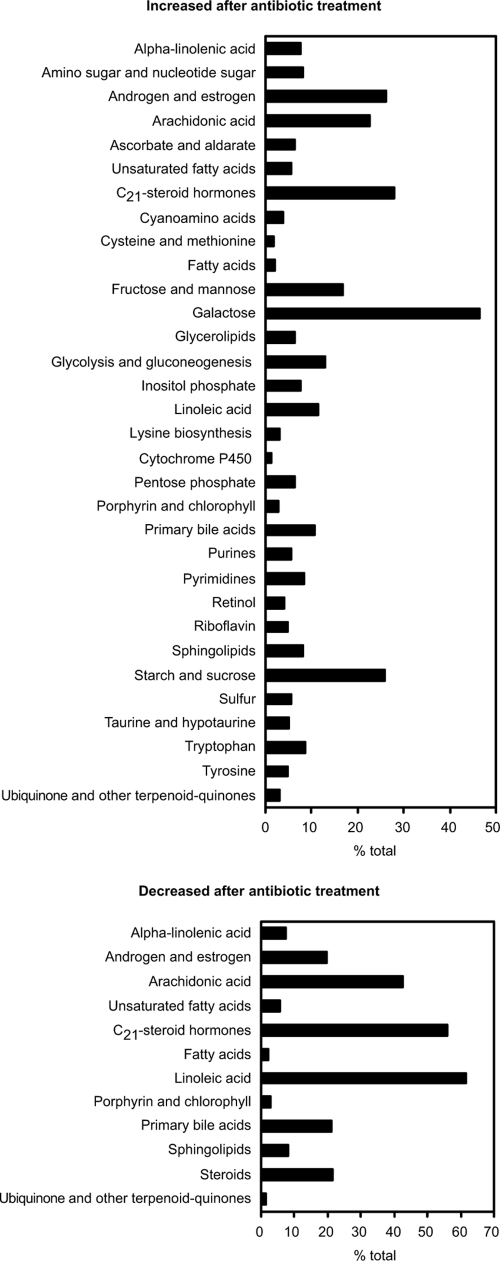
In order to identify the metabolic pathways affected by streptomycin treatment, we interrogated the MassTrix database (http://masstrix.org) using the masses whose levels were affected by the treatment. Figure 3 shows the metabolic pathways affected by antibiotic treatment. The pathways identified involve sugar, amino acid, fatty acid, bile acid, steroid, and eicosanoid metabolism, among others. All of these metabolites were matched within mass errors ranging from −0.4 to +0.65 ppm, based on the comparison of their calculated monoisotopic masses with the FT-MS-measured accurate masses (data not shown). The effect of antibiotic treatment in some of these pathways is detailed below.
FIG. 3.
Multiple metabolic pathways are affected by antibiotic treatment. Masses of interest were searched against the KEGG database using MassTrix (http://masstrix.org). Bars indicate the percentage of metabolites from each KEGG pathway that was affected by treatment.
Bile acid metabolism is highly impacted by antibiotic treatment.
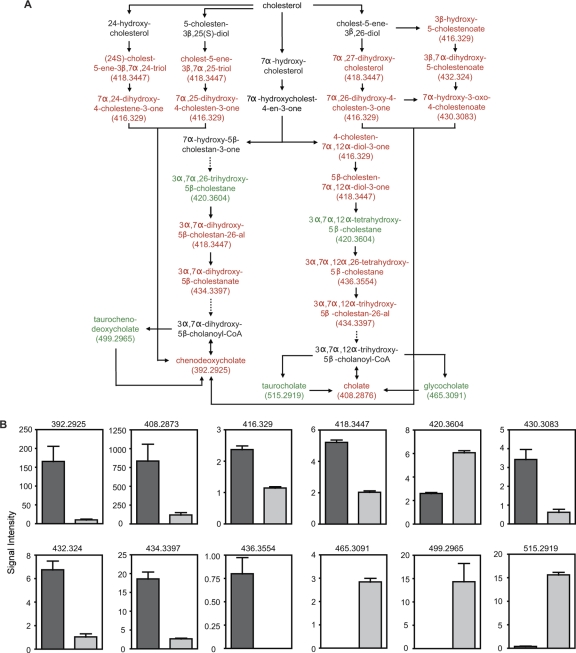
One of the pathways significantly affected by antibiotic treatment involves the synthesis of primary bile acids (Fig. 3 and 4). Of all metabolites in the pathway, 17 potential metabolites showed decreased levels after antibiotic treatment. Conversely, the levels of 5 potential metabolites were increased by antibiotic treatment. Because multiple metabolites in any given pathway may have identical masses, we manually screened the potential metabolites assigned by MassTrix to determine the number of different masses that were affected by antibiotic treatment. This analysis revealed that 8 different masses were decreased upon treatment, whereas 4 masses had their levels increased after antibiotic treatment, confirming the significant effect of antibiotic treatment on bile acid metabolism (Fig. 4).
FIG. 4.
Bile acid metabolism is disturbed by antibiotic treatment. (A) Schematic of the bile acid synthetic pathway. Red, metabolites decreased by antibiotic treatment; green, metabolites increased upon treatment; black, metabolites not detected or unchanged. Masses (Da) for metabolites affected are shown in parentheses. Solid arrows, direct steps; dashed arrows, multiple steps that are not shown. (B) Levels of masses affected by antibiotic treatment. Masses (Da) are shown at the top of each graph. The y axis indicates mass signal intensity. Dark gray bars, levels before treatment; light gray bars, levels after antibiotic treatment. Four mice (n = 4) were used, and averages with standard errors of the means are shown. Because masses 392.2925, 408.2876, 430.3083, 432.324, and 499.2965 were detected in both positive and negative ionization modes, the intensities of these masses in both ionization modes were combined before analysis (n = 8). All differences were statistically significant (P < 0.05). CoA, coenzyme A.
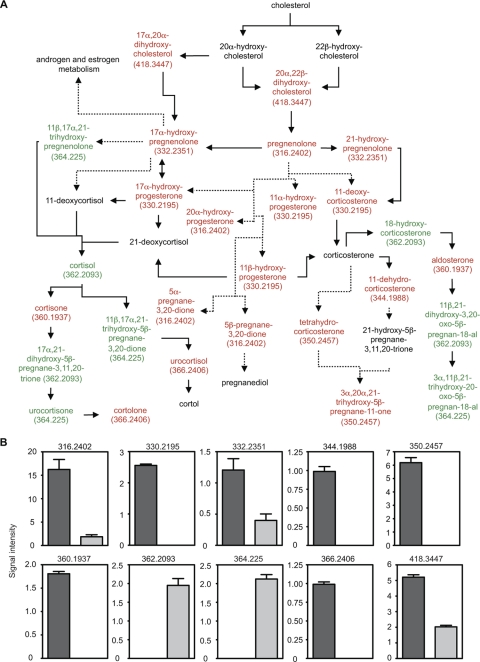
Antibiotic treatment disrupts steroid hormone homeostasis.
Of the many metabolic pathways affected by antibiotic treatment, the metabolism of steroid hormones was the most profoundly impacted (Fig. 3 and 5). The levels of 8 potential metabolites predicted to be part of the C21-steroid hormone metabolic pathway were increased in feces after antibiotic treatment. Additionally, the levels of 19 potential metabolites were decreased by the treatment (Fig. 5). Further analyses revealed that metabolites of 2 different masses were increased upon treatment, whereas metabolites of 8 masses were decreased (Fig. 5), corroborating the predictive analysis that suggested that the steroid pathway is significantly affected by antibiotic treatment.
FIG. 5.
Antibiotic treatment disrupts steroid hormone metabolism. (A) Schematic of the steroid hormone metabolic pathway. Red, metabolites decreased by antibiotic treatment; green, metabolites increased upon treatment; black, metabolites not detected or unchanged. Masses (Da) for metabolites affected are shown in parentheses. Solid arrows, direct steps; dashed arrows, multiple steps that are not shown. (B) Levels of masses affected by antibiotic treatment. Masses (Da) are shown at the top of each graph. The y axis indicates mass signal intensity. Dark gray bars, levels before treatment; light gray bars, levels after antibiotic treatment. Four mice (n = 4) were used, and averages with standard errors of the means are shown. All differences were statistically significant (P < 0.025).
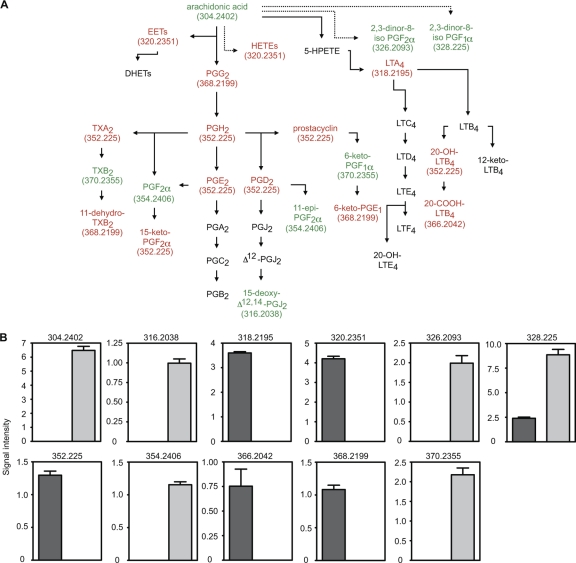
Eicosanoid hormone levels are affected by antibiotic treatment.
One of the metabolic pathways extensively affected by antibiotic treatment was that of arachidonic acid, the main precursor of eicosanoid hormones (Fig. 3 and 6). Treatment caused an increase in the levels of 8 potential metabolites. Additionally, antibiotic treatment caused a decrease in the levels of 14 potential metabolites (Fig. 6). As with the other pathways analyzed, we also looked for the individual masses affected, and this analysis revealed that 6 different masses were increased after antibiotic treatment, whereas 5 masses were decreased (Fig. 6), confirming the impact of antibiotic treatment on the eicosanoid synthesis pathway.
FIG. 6.
Antibiotic treatment has a profound impact on eicosanoid hormone metabolism. (A) Schematic of the eicosanoid hormone metabolic pathway. Red indicates metabolites decreased by antibiotic treatment whereas green indicates metabolites increased upon treatment. Black indicates metabolites not detected or unchanged. Masses (Da) for metabolites affected are shown in parentheses. Solid arrows indicate direct steps and dashed arrows indicate multiple steps that are not shown. (B) Levels of masses affected by antibiotic treatment. Masses (Da) are shown at the top of each graph. The y axis indicates mass signal intensity. Dark gray bars, levels before treatment; light gray bars, levels after antibiotic treatment. Four mice (n = 4) were used, and averages with standard errors of the means are shown. Mass 326.2093 was detected in both positive- and negative-ion modes, and therefore, its intensity values from both ionization modes were combined before analysis (n = 8). All differences were statistically significant (P ≤ 0.006). PG, prostaglandin; LT, leukotriene; TX, thromboxane; EET, epoxyeicosatrienoic acid; HETE, hydroxyeicosatetraenoic acid; HPETE, hydroperoxyeicosatetraenoic acid; DHET, dihydroxyeicosatrienoic acid.
Clinically relevant doses of multiple antibiotics disrupt eicosanoid metabolism.
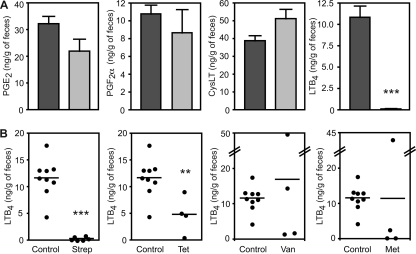
The results described above showed that a single, high-dose treatment with streptomycin greatly disturbed the levels of eicosanoids and other metabolites in the gastrointestinal tract of mice. In order to investigate whether this phenomenon was a consequence of the high dose of antibiotic used and if it was specific to streptomycin, we decided to test if clinically relevant doses of antibiotics could also affect eicosanoid metabolism. To do so, we first screened the effect of high-dose streptomycin treatment on the concentrations of multiple eicosanoids through ELISAs, with a view of establishing a screening assay that could be used to test lower streptomycin concentrations and other drugs. To this end, we tested the effect of high-dose streptomycin treatment on the fecal concentrations of PGE2, PGF2α, CysLT, and LTB4. As can be seen from Fig. 7, fecal levels of PGE2, PGF2α, and CysLT were mostly unaffected by streptomycin treatment. Although these data may seem to contradict our metabolomics results, cross-reactivity of ELISA antibodies with other molecules in our samples may have masked potential differences in the concentrations of these molecules. Also, because FT-ICR-MS assigns metabolite identity on the basis of mass alone, it is possible that these particular hormones were not affected by antibiotic treatment and that other compounds with identical masses may have caused these metabolites to “light up” during our database searches. Nevertheless, when we compared the fecal concentrations of LTB4 in control mice and mice that had been treated with streptomycin, we found that treatment with this antibiotic caused an 88-fold reduction in LTB4 levels (Fig. 7). Therefore, we used the concentration of LTB4 in feces as an indicator of eicosanoid metabolism disturbance to test the effects of multiple antibiotics. To do so, we treated mice with streptomycin (450 mg/liter), vancomycin (50 mg/liter), metronidazole (750 mg/liter), or tetracycline (250 mg/liter) in drinking water for 2 days and measured the levels of LTB4 in feces. We chose these doses of antibiotics because they are known to have very little impact on the total number of microbes in the gastrointestinal tract but significantly alter the species composition of the intestinal microbiota (25; unpublished observations). Figure 7 shows that the low-dose streptomycin treatment employed was sufficient to cause a drastic reduction (34-fold) in the fecal levels of LTB4. Tetracycline treatment also caused a significant reduction in LTB4 levels (2.4-fold, P = 0.0086). Metronidazole and vancomycin, on the other hand, did not significantly affect LTB4 levels. However, it is important to note that treatments with these two antibiotics caused major changes in the levels of LTB4 of individual mice, with some showing levels much higher than those obtained with untreated controls and others showing drastic reductions in the levels of this eicosanoid. Although the wide range of results obtained with these two drugs resulted in average LTB4 concentrations that are similar to those of untreated controls, our results suggest that these two antibiotics can also disturb eicosanoid homeostasis.
FIG. 7.
Clinically relevant doses of antibiotics affect eicosanoid metabolism. (A) Fecal levels of multiple eicosanoids were measured through ELISAs before and after high-dose streptomycin treatment. Dark gray bars, levels before treatment; light gray bars, levels after antibiotic treatment. Averages with standard errors of the means are shown. The numbers of mice used were as follows: CysLT before treatment, 11; CysLT after treatment, 6; PGE2 before and after treatment, 4 each; PGF2α before and after treatment, 6 each; LTB4 before and after treatment, 6 each. (B) Fecal levels of LTB4 were measured in groups of untreated mice and mice treated with clinically relevant doses of streptomycin (Strep), metronidazole (Met), vancomycin (Van), and tetracycline (Tet). Each dot represents one mouse, and bars indicate the averages of the results. **, P < 0.009; ***, P < 0.0001.
DISCUSSION
It has been increasingly appreciated that global approaches to the study of host-microbe interactions are necessary if we are to truly understand the molecular intricacies of these relationships. To this end, many “-omics” technologies have been applied with considerable success (33). For example, DNA microarrays have been used to study the interactions between many pathogens and their hosts (11, 16, 20, 29). One caveat of most microarray studies of host-microbe interactions performed to date is the fact that they focus on interactions between single pathogens and cultured host cells. This is not representative of the in vivo interactions between humans and their microbes; the human body is colonized by a complex mixture of thousands of bacterial strains that act in synergy or antagonism to exert their effects on each other as well as host cells. These microbes are, in most cases, harmless, and the interactions of humans with beneficial microbes are by far more common than those with pathogenic organisms. Additionally, global studies of host-microbe interactions have been mostly limited to transcriptome analyses. Although these can be highly informative, transcript levels cannot always be correlated with metabolic activity. Also, transcriptome studies of multispecies microbial communities represent a technical challenge due to cross hybridization of nucleic acids from different species. An alternative to transcriptome studies is the analysis of proteins levels, which is a better predictor of metabolic activity, although it does not necessarily reflect the levels of metabolic end products due to posttranslational regulatory events. Recently, metabolomic approaches to study host-microbe interactions have been developed as an alternative that avoids some of the issues with other global approaches (5). As the name implies, metabolomics aims at the detection and relative quantification of as many metabolites as possible in complex biological samples. This allows the prediction of metabolic activity with higher confidence, since the targets of such analyses are the end products of metabolic pathways.
We have used DI-FT-ICR-MS to detect and relatively quantify over 2,000 small-molecule metabolite features from feces of mice before and after antibiotic treatment as a way to assess the contribution of the intestinal microbiota to host physiology and the chemical ecology of the gastrointestinal tract. Strikingly, we found that antibiotic treatment elicited significant changes in the levels of almost 88% of all the metabolite features detected. Predictive mapping of the metabolic functions involved showed that many critical host metabolic pathways were impacted by antibiotic treatment, suggesting that the intestinal microbiota is involved in controlling such functions. Among the pathways affected were sugar, nucleotide, and fatty acid metabolism as well as bile acid, eicosanoid, and steroid hormone synthesis, among others. Although we suggest that the effect of antibiotic treatment on these pathways is due to disruption of the intestinal microbial communities, it is also possible that the antibiotics themselves have direct effects on host cells and that such interactions are the true culprits of the metabolic changes observed. Although we cannot rule out either hypothesis, our results clearly indicate that antibiotic usage can have profound, previously unappreciated effects on mammalian physiology, making it clear that the detrimental effects of indiscriminate use of antibiotics extend beyond the development of microbial drug resistance.
The link between bile acid metabolism and the intestinal microbiota has been known for several years (13, 26). Intestinal commensals are known to metabolize primary bile acids produced in the liver, generating secondary bile acids. In our studies, we found that the levels of primary bile acids were significantly affected by antibiotic treatment, as 22 out of the 47 annotated metabolites were potentially affected by treatment. It is important to emphasize that the metabolites searched do not include secondary bile acids. Therefore, our results differ from the established link between the intestinal microbiota and bile acid synthesis. Our data suggest that the microbiota is involved not only in the chemical modification of primary bile acids but also in the control of their production or degradation by the host.
Another host function that was significantly affected by antibiotic treatment was hormone synthesis. Two major classes of hormones, the steroids and eicosanoids, were affected, suggesting that the intestinal microbiota is involved in the control of such pathways. Hormones have crucial functions in mammals. They are involved in both the maintenance of homeostasis and the responses to insult (2, 4, 28, 30). Steroids and eicosanoids are important inflammatory mediators and have been implicated in immunological responses to infection. It is well established that the intestinal microbiota is critical for the defense against invading pathogens. The fact that antibiotic treatment affects pathways involved in the response to infection suggests that the microbiota may be involved in maintaining an alert state through the control of these potent inflammatory mediators and that this phenomenon might be involved in resistance to infectious processes. It has been known for some time that the intestinal microbiota is involved in the long-term development of gut immune responses (7, 21, 24). Our results suggest that the microbiota may have a more immediate role in controlling the immune system through the modulation of intestinal hormone levels. Studies on the mechanisms of such a phenomenon will be fundamental in understanding the interplay between host, commensals, and pathogens.
Acknowledgments
We thank the scientists who reviewed the manuscript for their constructive criticism.
This work was funded by grants from the Canadian Institutes of Health Research and the Crohn's and Colitis Foundation of Canada, as well as platform funding from Genome Canada and Genome British Columbia. L.C.M.A. is supported by postdoctoral fellowships from the Department of Foreign Affairs and International Trade Canada and the Canadian Institutes of Health Research. R.B.R.F. is funded by a postdoctoral fellowship from the Canadian Institutes of Health Research. B.B.F. is an HHMI International Research Scholar and The University of British Columbia Peter Wall Distinguished Professor.
Footnotes
Published ahead of print on 31 January 2011.
REFERENCES
- 1.Barthel, M., et al. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 71:2839-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calder, P. C. 2009. Polyunsaturated fatty acids and inflammatory processes: new twists in an old tale. Biochimie 91:791-795. [DOI] [PubMed] [Google Scholar]
- 3.Frank, D. N., and N. R. Pace. 2008. Gastrointestinal microbiology enters the metagenomics era. Curr. Opin. Gastroenterol. 24:4-10. [DOI] [PubMed] [Google Scholar]
- 4.Funk, C. D. 2001. Prostaglandins and leukotrienes: advances in eicosanoid biology. Science 294:1871-1875. [DOI] [PubMed] [Google Scholar]
- 5.Han, J., L. C. Antunes, B. B. Finlay, and C. H. Borchers. 2010. Metabolomics: towards understanding host-microbe interactions. Future Microbiol. 5:153-161. [DOI] [PubMed] [Google Scholar]
- 6.Han, J., et al. 2008. Towards high-throughput metabolomics using ultrahigh-field Fourier transform ion cyclotron resonance mass spectrometry. Metabolomics 4:128-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hooper, L. V., and A. J. Macpherson. 2010. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 10:159-169. [DOI] [PubMed] [Google Scholar]
- 8.Jansson, J., et al. 2009. Metabolomics reveals metabolic biomarkers of Crohn's disease. PLoS One 4:e6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kunz, C., S. Kuntz, and S. Rudloff. 2009. Intestinal flora. Adv. Exp. Med. Biol. 639:67-79. [DOI] [PubMed] [Google Scholar]
- 10.Lawley, T. D., et al. 2009. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect. Immun. 77:3661-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leroy, Q., and D. Raoult. 2010. Review of microarray studies for host-intracellular pathogen interactions. J. Microbiol. Methods 81:81-95. [DOI] [PubMed] [Google Scholar]
- 12.Levy, S. B., and B. Marshall. 2004. Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10:S122-S129. [DOI] [PubMed] [Google Scholar]
- 13.Lewis, R., and S. Gorbach. 1972. Modification of bile acids by intestinal bacteria. Arch. Intern. Med. 130:545-549. [PubMed] [Google Scholar]
- 14.Ley, R. E., D. A. Peterson, and J. I. Gordon. 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837-848. [DOI] [PubMed] [Google Scholar]
- 15.Lupp, C., et al. 2007. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2:119-129. [DOI] [PubMed] [Google Scholar]
- 16.Mahapatra, S., P. Ayoubi, and E. I. Shaw. 2010. Coxiella burnetii Nine Mile II proteins modulate gene expression of monocytic host cells during infection. BMC Microbiol. 10:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morelli, L. 2008. Postnatal development of intestinal microflora as influenced by infant nutrition. J. Nutr. 138:1791S-1795S. [DOI] [PubMed] [Google Scholar]
- 18.Neish, A. S. 2009. Microbes in gastrointestinal health and disease. Gastroenterology 136:65-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olszewski, K. L., et al. 2009. Host-parasite interactions revealed by Plasmodium falciparum metabolomics. Cell Host Microbe 5:191-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenberger, C. M., M. G. Scott, M. R. Gold, R. E. Hancock, and B. B. Finlay. 2000. Salmonella typhimurium infection and lipopolysaccharide stimulation induce similar changes in macrophage gene expression. J. Immunol. 164:5894-5904. [DOI] [PubMed] [Google Scholar]
- 21.Round, J. L., and S. K. Mazmanian. 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9:313-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savage, D. C. 1977. Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 31:107-133. [DOI] [PubMed] [Google Scholar]
- 23.Sekirov, I., and B. B. Finlay. 2009. The role of the intestinal microbiota in enteric infection. J. Physiol. 587:4159-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sekirov, I., S. L. Russell, L. C. Antunes, and B. B. Finlay. 2010. Gut microbiota in health and disease. Physiol. Rev. 90:859-904. [DOI] [PubMed] [Google Scholar]
- 25.Sekirov, I., et al. 2008. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 76:4726-4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimada, K., K. S. Bricknell, and S. M. Finegold. 1969. Deconjugation of bile acids by intestinal bacteria: review of literature and additional studies. J. Infect. Dis. 119:73-81. [DOI] [PubMed] [Google Scholar]
- 27.Suhre, K., and P. Schmitt-Kopplin. 2008. MassTRIX: mass translator into pathways. Nucleic Acids Res. 36:W481-W484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tait, A. S., C. L. Butts, and E. M. Sternberg. 2008. The role of glucocorticoids and progestins in inflammatory, autoimmune, and infectious disease. J. Leukoc. Biol. 84:924-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson, L. J., et al. 2009. Transcriptional response in the peripheral blood of patients infected with Salmonella enterica serovar Typhi. Proc. Natl. Acad. Sci. U. S. A. 106:22433-22438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vinson, G. P. 2009. The adrenal cortex and life. Mol. Cell. Endocrinol. 300:2-6. [DOI] [PubMed] [Google Scholar]
- 31.Waldram, A., et al. 2009. Top-down systems biology modeling of host metabotype-microbiome associations in obese rodents. J. Proteome Res. 8:2361-2375. [DOI] [PubMed] [Google Scholar]
- 32.Whitman, W. B., D. C. Coleman, and W. J. Wiebe. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U. S. A. 95:6578-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, W., F. Li, and L. Nie. 2010. Integrating multiple ‘omics’ analysis for microbial biology: application and methodologies. Microbiology 156:287-301. [DOI] [PubMed] [Google Scholar]
- 34.Zoetendal, E. G., M. Rajilic-Stojanovic, and W. M. de Vos. 2008. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 57:1605-1615. [DOI] [PubMed] [Google Scholar]