Abstract
Multidrug-resistant Klebsiella pneumoniae strains that produce a serine carbapenemase (KPC) are emerging worldwide, with few therapeutic options that retain consistent susceptibility. The objective of this study was to determine the effect of combination therapy with tigecycline versus tigecycline alone against KPC-producing isolates (KPC isolates). An in vitro pharmacodynamic model was used to simulate adult steady-state epithelial lining fluid concentrations of tigecycline (50 mg every 12 h) given alone and in combination with either meropenem (2 g by 3-hour infusion every 8 h) or rifampin (600 mg every 12 h). Five KPC isolates with various phenotypic profiles were exposed over 48 h. Time-kill curves were constructed, and the areas under the bacterial killing and regrowth curves (AUBCs) were calculated. No regimens tested were able to maintain bactericidal reductions in CFU over 48 h. The AUBCs for tigecycline and meropenem monotherapies at 48 h ranged from 375.37 to 388.11 and from 348.62 to 383.83 (CFU-h/ml), respectively. The combination of tigecycline plus meropenem significantly reduced the AUBCs at 24 and 48 h for isolates with tigecycline MICs of ≤2 μg/ml and meropenem MICs of ≤16 μg/ml (P < 0.001) but added no additional activity when the meropenem MIC was 64 μg/ml (P = 0.5). Rifampin provided no additional reduction in CFU or AUBC over tigecycline alone (P = 0.837). The combination of tigecycline with high-dose, prolonged-infusion meropenem warrants further study as a potential treatment option for these multidrug-resistant organisms.
Carbapenems are often recommended as first-line therapy for serious infections caused by extended-spectrum β-lactamase (ESBL)-producing bacteria (33). Therefore, the development of resistance to carbapenems and the global dissemination of multiple variants of Klebsiella pneumoniae producing carbapenemases (KPC) are alarming. The first KPC (KPC-1) was isolated in North Carolina in 2001 (46). Nine other variants were subsequently identified, with KPC-2 and KPC-3 being the most commonly reported variants in the United States, predominantly among the mid-Atlantic and northeastern states (24, 31). These pathogens have primarily caused nosocomial infections, including pneumonia and bacteremia in critically ill patients. In addition, infection with KPC-producing K. pneumoniae was associated with significantly higher mortality than infection with carbapenem-susceptible K. pneumoniae (32.1% versus 9.9%) in a recent epidemiologic study (21). Further complicating the management of patients with KPC-producing K. pneumoniae infections is the scarcity of currently available treatment options. Most isolates are resistant to all β-lactams, fluoroquinolones, and aminoglycosides while retaining consistent in vitro susceptibility only to tigecycline and colistin (7, 24, 28).
Tigecycline is a glycylcycline antibiotic with broad-spectrum activity against a variety of multidrug-resistant bacteria, including methicillin-resistant Staphylococcus aureus, vancomycin-resistant enterococci, and some Acinetobacter species. Additionally, because tigecycline is unaffected by β-lactamase enzymes, it retains microbiological activity against Gram-negative organisms that produce ESBLs as well as KPC-2 enzymes. All KPC-2-producing isolates (KPC-2 isolates) collected during a study in Brooklyn, NY, were found to be susceptible to tigecycline, with a MIC of 2 μg/ml; the MIC50 and MIC90 were 0.5 μg/ml and 1 μg/ml, respectively (8). The in vitro activity of tigecycline against serine carbapenemase-producing Enterobacteriaceae was also evaluated using isolates collected during the 2000 to 2005 SENTRY Antimicrobial Surveillance Program (10). All of the 104 Enterobacteriaceae isolates that produced a carbapenemase were susceptible to tigecycline, with a MIC50 and MIC90 of 0.5 μg/ml and 1 μg/ml, respectively. Specifically, the MIC50 and MIC90 among the 60 Klebsiella spp. in that study were 1 μg/ml and 2 μg/ml, respectively.
Although in vitro data suggest that tigecycline will be a useful antibiotic in managing infections caused by KPC-producing organisms, clinical and experimental data to support its use are limited. To date, one case report has described the use of tigecycline monotherapy for pneumonia and empyema caused by KPC-producing K. pneumoniae (15). While the patient's pneumonia was treated successfully, the empyema recurred and the tigecycline MIC increased from 0.5 to 2 μg/ml while the patient underwent prolonged tigecycline therapy. In a retrospective review of an outbreak of infections caused by KPC-2-producing K. pneumoniae among 22 patients in Greece, 8 patients with pneumonia were treated with tigecycline in combination with colistin and/or garamycin (27). Clinical cure was achieved in 5 of the 8 cases. Since tigecycline was administered in combination with colistin, which also has in vitro activity against KPC-producing K. pneumoniae, it is difficult to ascertain the effects of tigecycline alone (7, 24, 28). Another case series described the successful use of tigecycline against KPC-producing K. pneumoniae infections in 5 of 7 cases; however, only 3 of these patients had pulmonary tract infections (45).
Experience with other antibiotic therapies for serious KPC infections is also limited. Despite continued susceptibility of some strains, carbapenems have been used with mixed success in a few cases of KPC-producing pulmonary tract infections, often in combination with a variety of other agents, making the additive effects of the carbapenems difficult to establish (6, 45). In particular, continuous- and prolonged-infusion carbapenem regimens are potentially more effective than standard dosing regimens against KPC-producing K. pneumoniae. This is due to their ability to achieve an adequate percentage of the dosing interval in which free drug concentrations exceed the MIC (ƒT>MIC) against some KPC phenotypes (23, 41). Rifampin has been shown to have activity against other multidrug-resistant Gram-negative pathogens, including carbapenem-resistant Acinetobacter species, in a limited number of experimental pneumonia models of infection, both as monotherapy and combined with imipenem (30). Since resistance to rifampin develops rapidly, this drug should be combined with another agent if it is used clinically (35). Similarly, some efficacy was demonstrated with the combination of rifampin and imipenem in a small clinical study (40). Furthermore, one study has also reported in vitro synergy of rifampin and polymyxin B against a KPC-2-producing K. pneumoniae isolate (8).
Given the scarcity of new antibiotics with activity against KPC-producing K. pneumoniae and its inevitable spread worldwide, the purpose of the current study was to assess the effect of a human simulated pulmonary exposure of tigecycline, alone and in combination with either prolonged-infusion meropenem or rifampin, against KPC-producing K. pneumoniae isolates, using an in vitro pharmacodynamic model.
MATERIALS AND METHODS
Bacterial strains and susceptibility testing.
Five clinical K. pneumoniae isolates, all modified Hodge test positive and blaKPC-2 or blaKPC-3 positive, were kindly provided by Stephen Jenkins, New York Presbyterian/Weill Cornell Medical Center (New York, NY), or obtained from the Tigecycline Evaluation and Surveillance Trial (TEST) surveillance study (34). Tigecycline and meropenem MICs were determined by broth microdilution in triplicate, using fresh broth made within 12 h, in accordance with Clinical and Laboratory Standards Institute (CLSI) recommendations. Rifampin MICs were determined using Etest according to the manufacturer's specifications (AB bioMérieux, Solna, Sweden). Escherichia coli ATCC 35922 and Streptococcus pneumoniae ATCC 49619 were tested as control strains. Phenotypic profiles for included isolates are listed in Table 1.
TABLE 1.
Phenotypic profiles and achieved meropenem exposures for KPC-producing Klebsiella pneumoniae isolates tested in this studyd
| Isolate | Tigecycline MIC (μg/ml) | Meropenem MIC (μg/ml) | Modela | Meropenem mean ƒT>MIC (%)b |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Target | 0 to 8 h | 8 to 16 h | 16 to 24 h | 24 to 32 h | 32 to 40 h | 40 to 48 h | ||||
| 351 | 1 | 8 | MEMc | 69 | 69 | 62 | 50 | NA | NA | NA |
| 1 | 8 | TGC-MEM | 74 | 75 | 79 | 84 | 79 | 73 | 56 | |
| 382 | 1 | 8 | MEM | 69 | 63 | 48 | 0 | 0 | 16 | 16 |
| 1 | 8 | TGC-MEM | 74 | 81 | 82 | 81 | 78 | 79 | 78 | |
| 358 | 2 | 16 | MEM | 47 | 15 | 3 | 0 | 0 | 1 | 0 |
| 2 | 16 | TGC-MEM | 54 | 40 | 37 | 25 | 22 | 29 | 0 | |
| 360 | 2 | 16 | MEMc | 47 | 0 | 0 | 0 | NA | NA | NA |
| 2 | 16 | TGC-MEM | 54 | 11 | 36 | 0 | 0 | 0 | 0 | |
| 362 | 2 | 64 | MEM | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 64 | TGC-MEM | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
MEM, meropenem; TGC-MEM, tigecycline-meropenem.
NA, not available.
Data on the 24-h meropenem monotherapy model were taken from the work of Bulik et al. (9).
In all cases, the rifampin MIC was >32 μg/ml.
Antibiotics.
Analytical-grade tigecycline for injection (material 0108837; potency, 95.7%; lot RB5603; expiration date, September 2009) was provided by Wyeth Pharmaceuticals. Meropenem for intravenous injection (lot TC0076; expiration date, 27 November 2011) and rifampin for intravenous injection (lot 1180473; expiration date, March 2010) were obtained from the pharmacy department at Hartford Hospital.
Simulated drug exposures.
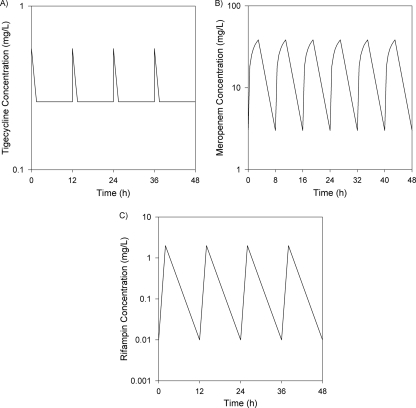
A dosing strategy was developed using mean pharmacokinetic parameters from a previous population pharmacokinetic study of tigecycline to simulate steady-state tigecycline concentrations in epithelial lining fluid (ELF) after 50-mg doses every 12 h, as depicted in Fig. 1A (44). For these calculations, we assumed that the median area under the curve (AUC) for ELF over the dosing interval was 115% of that in serum, as observed in a population pharmacokinetic analysis of ELF data from healthy volunteers (39). Steady-state serum concentrations were calculated using mean pharmacokinetic parameters from this study and a two-compartment model in WinNonLin Professional, version 5.0.1 (Pharsight Corporation, Mountain View, CA). To estimate the pulmonary exposure of tigecycline, and because the pharmacodynamic parameter that drives efficacy for tigecycline is AUC/MIC, serum concentrations were increased proportionally to achieve an AUC that was approximately 115% of that in serum (12, 39). This yielded a 12-h target AUC of approximately 3.1 μg-h/ml. Since only free-fraction tigecycline is assumed to penetrate into ELF, no further adjustments were performed for protein binding.
FIG. 1.
Targeted steady-state concentration-time profiles for tigecycline (50 mg every 12 h) (A), meropenem (2 g every 8 h, administered as a 3-h continuous infusion) (B), and rifampin (600 mg every 12 h) (C).
Since meropenem has been shown to have similar ƒT>MIC values in ELF and serum, a previously developed model simulating free drug concentrations of meropenem in serum following a 2-g dose administered every 8 h as a 3-h continuous infusion was employed (2, 9, 18). This dosing regimen was selected for its high probability of attaining ≥40% ƒT>MIC against isolates with meropenem MICs of up to 16 μg/ml, which is needed to maintain bactericidal activity (23). Briefly, median pharmacokinetic parameters for meropenem were used to simulate steady-state exposure, using WinNonLin (25). All concentrations were then multiplied by 0.97 to adjust for protein binding (3). The simulated peak and trough concentrations for meropenem were 38.4 μg/ml and 3.07 μg/ml, respectively, with a half-life of 1.4 h (Fig. 1B).
The combination of tigecycline and rifampin was then evaluated for additional activity over that of tigecycline alone against select isolates. Rifampin exposures were modeled to simulate mean patient pharmacokinetics in ELF for rifampin given at 600 mg every 12 h. Concentrations were estimated based on a peak serum concentration of 10 μg/ml and a half-life of 3 h (1). Serum concentrations were then reduced using a serum-to-ELF penetration ratio of 21% (1). The resulting estimated ELF concentrations (Fig. 1C) were similar to those described in a recent intrapulmonary pharmacokinetic study of rifampin (13, 22).
In vitro pharmacodynamic model.
A one-compartment in vitro pharmacodynamic model that has been described previously was used for all experiments (9, 17). Each experiment consisted of three independent models (two treatment models and one growth control model) running simultaneously for each isolate and antibiotic regimen. The models were placed in a 37°C circulating water bath for optimal temperature control, and magnetic stir bars were placed in each model to ensure adequate mixing of contents. A starting inoculum of 1 × 106 CFU/ml was prepared from an overnight culture of each test isolate. Each model was filled with cation-adjusted Mueller-Hinton broth (CAMHB) and inoculated 30 min before the start of the experiment to ensure that bacteria were in the logarithmic growth phase prior to antimicrobial exposure.
For experiments modeling tigecycline monotherapy, tigecycline was injected into the model at 0, 12, 24, and 36 h to achieve the desired peak concentration. Fresh CAMHB was pumped continuously into each of the models by use of a peristaltic pump (Masterflex L/S model 7524-40; Cole-Parmer Instrument Company) from 0 to 2 h at a rate that simulated the rapid distribution of tigecycline into tissue. At 2 h, the pump was shut off to maintain a steady trough concentration needed to achieve a 12-h AUC of 3.1 μg-h/ml. This dosing process was then repeated every 12 h over 48 h.
The meropenem monotherapy model was conducted as previously described by our group (9). Briefly, meropenem was injected into the model at 30-min intervals over 3 h while the pump remained off to simulate the rising concentrations of a 3-h infusion. At 3 h, fresh CAMHB was pumped continuously into the model over the next 5 h, at specified rates, to simulate the distribution and terminal half-life of meropenem. The dosing process was repeated every 8 h for 48 h and should have resulted in ƒT>MIC exposures of 69%, 47%, and 0% at the tested MICs of 8, 16, and 64 μg/ml, respectively.
Dosing schemes for both monotherapy models were combined and flow rates adjusted to allow for the modeling of tigecycline and meropenem simultaneously. Tigecycline was injected into the model every 12 h as previously described. CAMHB was infused at a continuous rate over the entire 48 h to simulate the elimination half-life of meropenem. Antibiotic-free CAMHB was infused during the first 1.5 h of each 12-h tigecycline dosing interval to simulate the distribution phase of tigecycline, and then tigecycline-supplemented broth was infused over the remaining 10.5 h of the dosing interval to maintain an adequate tigecycline concentration to achieve the desired 12-h AUC. Meropenem was injected into the model every 30 min over the first 3 h to achieve the desired target concentrations, as described for the monotherapy model; however, volumes were adjusted to account for broth being infused into the model during dosing. The meropenem dosing process was repeated every 8 h, and the tigecycline dosing process and corresponding supplemental broth changes were repeated every 12 h over the 48-h study period. These changes from the monotherapy models resulted in targeted meropenem ƒT>MIC exposures of 74% and 54% for MICs of 8 and 16 μg/ml, respectively.
In order to model exposures of tigecycline and rifampin simultaneously, tigecycline was injected into the model every 12 h as previously described for the monotherapy model, while rifampin-supplemented CAMHB was infused from 0 to 2 h at a rate simulating the distribution phase of tigecycline. Rifampin-supplemented CAMHB was prepared to a concentration needed to achieve the desired rifampin peak at 2 h (4, 47). At 2 h, the rifampin-supplemented CAMHB was stopped and CAMHB supplemented with tigecycline at a concentration equivalent to the tigecycline concentration in the model at 2 h was infused at a rate simulating the elimination half-life of rifampin. This in turn maintained a steady trough concentration of tigecycline needed to achieve the desired target AUC, while allowing for adequate clearance of rifampin from the model.
Samples were obtained from each model at 0, 3, 6, 8, 12, 18, 24, 36, and 48 h and serially diluted in normal saline to assess changes in bacterial density over time. Aliquots of each diluted sample were plated on Trypticase soy agar plates (100-mm diameter) supplemented with 5% sheep blood for quantitative culture. After 18 to 24 h of incubation at 37°C, the change in log CFU/ml over the 48-h interval was calculated, and time-kill curves were constructed by plotting log CFU/ml against time. The areas under the bacterial killing and regrowth curve (AUBCs) over 24 and 48 h were also calculated for each model run, as these are a good measure of overall antibiotic effect over time and have been used in previous experiments to assess antibiotic combinations (16, 20, 43). The lower limit of detection for bacterial density was 101 CFU/ml. Differences in AUBC at 24 and 48 h between all treatment regimens were assessed by repeated-measures analysis of variance (ANOVA) with the Tukey test for multiple comparisons. An a priori P value of <0.05 was considered statistically significant.
Antibiotic concentration determinations.
Samples of CAMHB were taken simultaneously with bacterial density samples and were assayed for tigecycline and meropenem to ensure that targeted exposures were achieved. All samples were immediately frozen and stored at −80°C until analysis by a validated high-performance liquid chromatography (HPLC) method at the Center for Anti-Infective Research and Development, as described previously (19, 26). The tigecycline assay was linear (r = 0.996 to 0.999) over a concentration range of 0.05 to 5 μg/ml. The intraday quality control samples (n = 10) at 0.2 and 4 μg/ml had percent coefficients of variation (%CV) of 5% and 2.09%, respectively. The interday quality control samples (n = 11) at 0.2 and 4 μg/ml had %CV of 4.8% and 3.2%, respectively. The meropenem assay was linear (r = 0.996 to 1.000) over a concentration range of 0.25 to 40 μg/ml. The intraday quality control samples (n = 10) at 0.5 and 30 μg/ml had %CV of 3.32% and 2.41%, respectively. The interday quality control samples (n = 13) at 0.5 and 30 mg/liter had %CV of 5.03% and 5.28%, respectively. Rifampin concentrations were analyzed by the Infectious Disease Pharmacokinetics Laboratory at the University of Florida (Gainesville, FL), using a previously validated HPLC methodology (36).
RESULTS
Pharmacokinetic analysis.
The 12-h tigecycline AUC during all experiments ranged from 3.6 to 5.2 μg-h/ml. Target rifampin concentrations were achieved during all combination therapy experiments with tigecycline. However, the desired ƒT>MIC was not attainable during any of the meropenem monotherapy experiments, likely due to rapid in vitro hydrolysis of the meropenem β-lactam ring by the carbapenemase, as previously observed (9). However, ≥40% ƒT>MIC was achieved during combination experiments over a majority of or for the entire 48-h study period for isolates with a tigecycline MIC of 1 μg/ml and a meropenem MIC of 8 μg/ml (Table 1).
Bactericidal activity.
The average bacterial density of the starting inoculum was 6.14 ± 0.11 log10 CFU/ml. Control isolates grew to 8.41 ± 0.12 log10 CFU/ml. Growth control models that ran with each experiment were similar throughout the study, regardless of pump flow rate. Figures 2 through 4 summarize the time-kill curves for all isolates after exposure to human simulated ELF exposures of tigecycline alone, meropenem alone, tigecycline plus meropenem, and tigecycline plus rifampin. Data are plotted as the means for the two models for the treatments and the means for all corresponding growth control isolates.
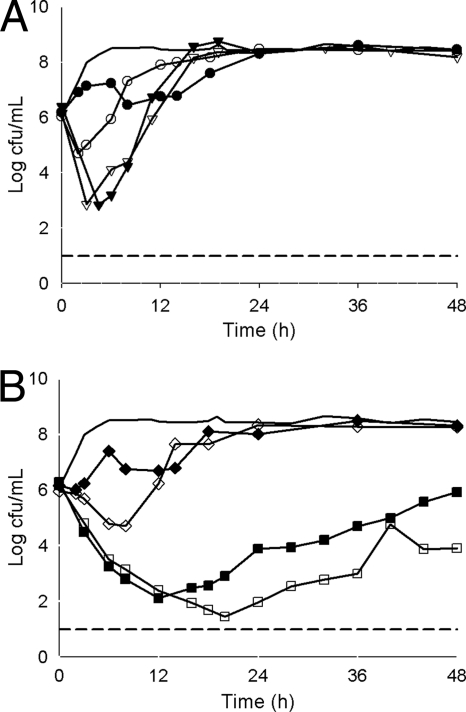
FIG. 2.
Mean bacterial densities over 48 h for KPC-producing Klebsiella pneumoniae isolates with a tigecycline MIC of 1 μg/ml and a meropenem MIC of 8 μg/ml. (A) Monotherapy models. Thick solid line, control; closed circles, tigecycline (isolate 351); open circles, tigecycline (isolate 382); closed triangles, meropenem (isolate 351); open triangles, meropenem (isolate 382). (B) Combination therapy models. Thick solid line, control; closed squares, tigecycline and meropenem (isolate 351); open squares, tigecycline and meropenem (isolate 382); closed diamonds, tigecycline and rifampin (isolate 351); open diamonds, tigecycline and rifampin (isolate 382). The lower limit of detection (dashed line) was set to 101 CFU/ml.
A reduction in log CFU/ml was not observed in any of the tigecycline monotherapy experiments, with the exception of one isolate with a tigecycline MIC of 1 μg/ml (isolate 382) (Fig. 2A). For this isolate, tigecycline achieved an approximately 1-log CFU reduction by 2 h, followed by regrowth. All isolates experienced regrowth approaching the level of the control by approximately 18 h (Fig. 2A, 3A, and 4). The AUBCs for tigecycline monotherapy at 24 h and 48 h ranged from 171.31 to 185.67 and from 375.37 to 388.11 CFU-h/ml, respectively (Table 2).
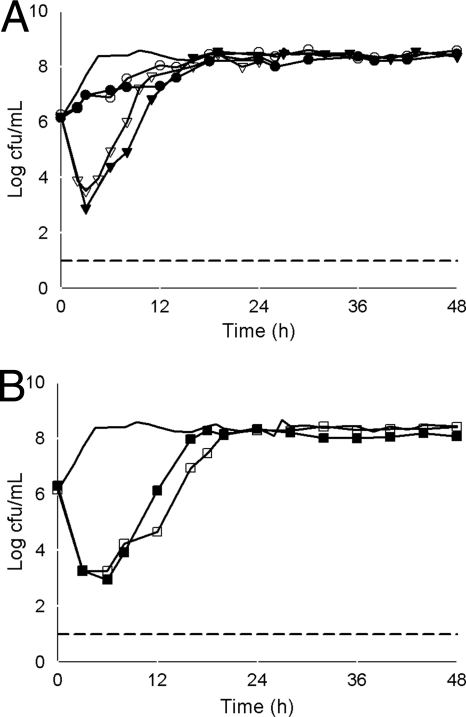
FIG. 3.
Mean bacterial densities over 48 h for KPC-producing Klebsiella pneumoniae isolates with a tigecycline MIC of 2 μg/ml and a meropenem MIC of 16 μg/ml. (A) Monotherapy models. Thick solid line, control; closed circles, tigecycline (isolate 358); open circles, tigecycline (isolate 360); closed triangles, meropenem (isolate 358); open triangles, meropenem (isolate 360). (B) Combination therapy models. Thick solid line, control; closed squares, tigecycline and meropenem (isolate 358); open squares, tigecycline and meropenem (isolate 360). The lower limit of detection (dashed line) was set to 101 CFU/ml.
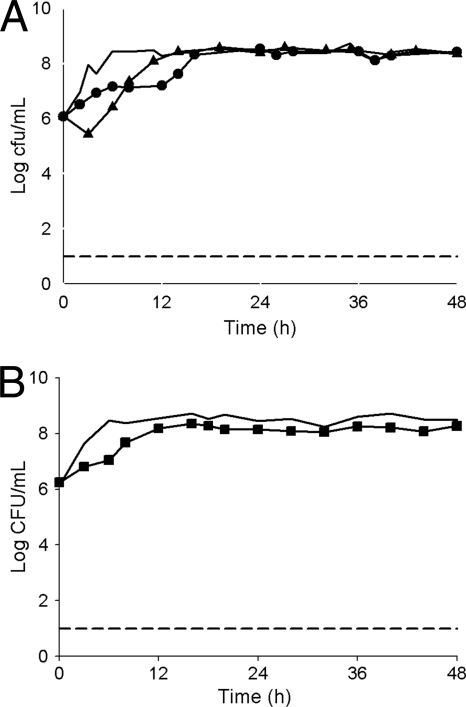
FIG. 4.
Mean bacterial densities over 48 h for KPC-producing Klebsiella pneumoniae isolates with a tigecycline MIC of 2 μg/ml and a meropenem MIC of 64 μg/ml. (A) Monotherapy models. Thick solid line, control; closed circles, tigecycline (isolate 362); closed triangles, meropenem (isolate 362). (B) Combination therapy model. Thick solid line, control; closed squares, tigecycline and meropenem (isolate 362). The lower limit of detection (dashed line) was set to 101 CFU/ml.
TABLE 2.
Mean AUBCs after 24 and 48 h of exposure to tested regimens for each KPC-producing Klebsiella pneumoniae isolate tested
| Isolate | Time (h) | Mean AUBC (CFU-h/ml) ± SDe |
|||
|---|---|---|---|---|---|
| Tigecycline | Meropenem | Tigecycline + meropenem | Tigecycline + rifampin | ||
| 351 | 24 | 171.31 ± 1.32 | 148.27 ± 11.38 | 85.59 ± 10.73a | 171.46 ± 1.35b |
| 48 | 375.37 ± 2.46 | 348.62 ± 11.27 | 234.62 ± 32.61a | 365.65 ± 6.06b | |
| 382 | 24 | 173.99 ± 0.76 | 149.91 ± 2.11 | 72.91 ± 2.92a | 157.22 ± 1.23b |
| 48 | 376.49 ± 4.77 | 351.49 ± 1.42 | 153.11 ± 8.19a | 356.33 ± 0.84b | |
| 358 | 24 | 179.86 ± 2.52 | 157.22 ± 0.20 | 144.45 ± 0.13c | ND |
| 48 | 377.98 ± 3.38 | 359.79 ± 0.34 | 339.10 ± 0.64c | ND | |
| 360 | 24 | 185.67 ± 3.68 | 162.70 ± 0.53 | 135.31 ± 4.97c | ND |
| 48 | 388.11 ± 4.57 | 359.32 ± 2.91 | 335.41 ± 6.36c | ND | |
| 362 | 24 | 180.77 ± 0.26 | 181.08 ± 2.75 | 218.41 ± 0.43d | ND |
| 48 | 381.37 ± 0.09 | 383.83 ± 3.26 | 381.17 ± 0.87d | ND | |
Data are for a combined total of 4 observations each at 24 and 48 h for the two isolates with a tigecycline MIC of 1 μg/ml and a meropenem MIC of 8 μg/ml. Data are significantly different from those for tigecycline or meropenem alone by repeated-measures ANOVA and the Tukey test for multiple comparisons (P < 0.001).
Data are for a combined total of 4 observations each at 24 and 48 h for two isolates with a tigecycline MIC of 1 μg/ml and a rifampin MIC of >32 μg/ml. Data are not significantly different from those for tigecycline alone by repeated-measures ANOVA and the Tukey test for multiple comparisons (P = 0.416 at 24 h and P = 0.837 at 48 h).
Data are for a combined total of 4 observations each at 24 and 48 h for two isolates with a tigecycline MIC of 2 μg/ml and a meropenem MIC of 16 μg/ml. Data are significantly different from those for tigecycline or meropenem alone by repeated-measures ANOVA and the Tukey test for multiple comparisons (P < 0.001 for comparison with tigecycline at 24 and 48 h, and P = 0.004 and 0.001 for comparisons with meropenem at 24 and 48 h, respectively).
Data are for a total of 2 observations each at 24 and 48 h for two isolates with a tigecycline MIC of 2 μg/ml and a meropenem MIC of 64 μg/ml. Data are not significantly different from those for tigecycline or meropenem alone by repeated-measures ANOVA and the Tukey test for multiple comparisons (P = 0.5).
ND, not done; SD, standard deviation.
Meropenem monotherapy produced a ≥3-log CFU/ml reduction for isolates with meropenem MICs of 8 and 16 μg/ml at 3 h, followed by regrowth (Fig. 2A and 3A). A 1-log CFU/ml reduction was observed for the isolate with a meropenem MIC of 64 μg/ml, before regrowth to the control level by 12 h (Fig. 4). The AUBCs for meropenem monotherapy at 24 and 48 h ranged from 148.27 to 181.08 and from 348.62 to 383.83 CFU-h/ml, respectively (Table 2).
Colonies began to regrow or regrew to control levels by 48 h in all combination experiments (Fig. 2B and 3B). The combination of tigecycline and meropenem, however, produced a significant reduction in AUBC at both 24 h and 48 h compared with either tigecycline or meropenem alone for isolates with a tigecycline MIC of 1 μg/ml and a meropenem MIC of 8 μg/ml (P < 0.001) and for those with tigecycline and meropenem MICs of 2 μg/ml and 16 μg/ml, respectively (P ≤ 0.004), but not for the isolate with a tigecycline MIC of 2 μg/ml and a meropenem MIC of 64 μg/ml (P = 0.05). In contrast, combining rifampin with tigecycline had no effect on bacterial killing and produced an insignificant (P = 0.416 and 0.837 at 24 and 48 h, respectively) decrease in AUBC compared with tigecycline monotherapy (Table 2). Since no effect was observed with the addition of rifampin against isolates with the lowest tigecycline MIC, further experiments utilizing the combination of tigecycline and rifampin against isolates with higher tigecycline MICs were not conducted.
DISCUSSION
KPC-producing K. pneumoniae is often resistant to many antibiotics, retaining consistent susceptibility to polymyxin B or colistin and tigecycline only. As a result, there are few options available to treat serious infections, such as nosocomial pneumonia, caused by this organism. In vitro data from two large surveillance studies reported tigecycline susceptibility rates of 93.7% and 100% for ESBL-producing and KPC-producing K. pneumoniae isolates, respectively (5, 10); however, few clinical or experimental data are available to support the utility of tigecycline as monotherapy or in combination with other agents. The aim of the current study was to provide further insight into the antimicrobial action of tigecycline, with and without meropenem or rifampin, against KPC-producing K. pneumoniae, using an in vitro pharmacodynamic model.
To our knowledge, this is the first in vitro pharmacodynamic assessment of tigecycline monotherapy against the KPC phenotype. Although tigecycline exposures alone resulted in regrowth of the five KPC isolates studied, tigecycline did delay regrowth by approximately 18 h. The lack of effect in this in vitro pharmacodynamic model was disconcerting given the fact that tigecycline has been used clinically against KPC-producing K. pneumoniae, with success, in some reports (27, 45). While in vitro modeling attempts to simulate the effect of the antibiotic at achievable concentrations at the site of infection and may be preferable to checkerboard or time-kill studies that analyze only single drug concentrations, it still cannot mimic the contributions of the host immune system, nor can we simulate all scenarios of the extensive variability in ELF penetration portrayed by tigecycline (39). We speculate that these differences likely account for the discrepancy between our observations with tigecycline and what has been demonstrated in case reports. Clearly, the immune system plays a significant role in modeling the effects of tigecycline, as demonstrated by an in vivo murine thigh model used to characterize the pharmacodynamic profile of tigecycline. In that study, the cumulative 50% and 80% effective pharmacodynamic indices (EI50 and EI80, expressed as the AUC for the free, unbound fraction of the drug [ƒAUC]/MIC) were reduced from 2.01 and 3.27, respectively, in a neutropenic model to 1.59 and 1.70, respectively, in an immunocompetent model against a K. pneumoniae isolate that produced an ESBL (32). Of note, a single KPC isolate was also evaluated in the experiment, but only with the immunocompromised model. Despite a lower MIC (0.5 μg/ml versus 2 μg/ml), the KPC isolate displayed a larger EI50 (4.53) and EI80 (7.48) for tigecycline than those for the previously described ESBL-producing isolate.
Tigecycline portrayed significant variability in ELF penetration with healthy volunteer data. We selected the median penetration of 115% for these in vitro studies but acknowledge that exposures for half of the volunteers studied could have been as much as 500% higher. Additionally, the presence of infection could potentially affect antibiotic penetration into ELF; however, no studies have been conducted to date to evaluate this phenomenon for these antimicrobials in humans. One study conducted in a murine pneumonia model did observe that tigecycline penetration into ELF increased in the face of infection (14). Collectively, this means that the simulated exposure in this study may be a conservative estimate of AUC exposure achieved in patients with pneumonia, which may account for the discordance between our observations and reports of clinical success with tigecycline.
Tigecycline pharmacodynamics have been evaluated in numerous in vitro experiments with variable success, but primarily against Acinetobacter baumannii or non-KPC-producing Enterobacteriaceae (11, 38, 42). Importantly, these studies were conducted using time-kill or checkerboard experiments, with tigecycline concentrations ranging from 0.5 to 4 times the MICs of the tested isolates. A single in vitro time-kill experiment simulated a tigecycline concentration of 2 μg/ml against 16 blaKPC-2-carrying K. pneumoniae isolates with tigecycline MICs ranging from 0.25 to 2 μg/ml (8). In that study, a mean reduction in log CFU/ml of 0.4 ± 1.6 was observed at 4 h. Tigecycline generally produced a bacteriostatic effect over 24 h (mean change of −0.06 ± 1.9 log CFU/ml) but was bactericidal against 2 of the 16 isolates. However, this experiment simulated a higher concentration of tigecycline than those used in our model, and that concentration was maintained over the entire course of the experiment. Clearly, this is not the concentration-time profile exhibited by tigecycline administered clinically at a dose of 50 mg every 12 h, even in the ELF (12, 39, 44). To achieve an fAUC exposure in ELF of 3.1 μg-h/ml, our model simulated a steady-state tigecycline peak concentration of 0.57 μg/ml, followed rapidly by an exponential decline in the concentration to approximately 0.21 μg/ml, which was then maintained for the 12-h dosing interval (Fig. 1A). Given these achievable exposures and the results from the previous time-kill experiment with tigecycline, it was not surprising that tigecycline did not maintain a 48-h bactericidal or even bacteriostatic effect against these KPC isolates.
Our observations with meropenem monotherapy against these KPC isolates were not unlike our previous experiences with a similar model (9). It is very difficult to maintain meropenem concentrations in the in vitro pharmacodynamic model due to rapid hydrolysis of the compound by the carbapenemase released into the broth. However, even with ƒT>MIC exposures of >40% (Table 1), bactericidal activity could not be maintained with this high-dose, prolonged-infusion meropenem regimen. The combination of tigecycline plus high-dose meropenem did achieve a statistically significant reduction in the AUBC against isolates with meropenem MICs of ≤16 μg/ml, as well as improving the meropenem exposures achieved in the model. Nevertheless, the synergistic combination was still not able to maintain bactericidal activity against any of the isolates, and regrowth occurred over the 48-h experiment. The primary aim of this study was to determine if efficacy could even be demonstrated with these combinations. Further work is needed to determine how resistance may have contributed to the regrowth observed. The presence of heteroresistant subpopulations that are able to grow at much higher concentrations of meropenem has been described previously and likely contributed to the eventual regrowth observed within the model (37). This lack of killing was ultimately why the AUBC was selected as a measure of the additive or synergistic effect of the antimicrobial combinations. Despite regrowth, combination therapy clearly affected both the extent of killing and the rate of regrowth of selected organisms. These differences would not be observed by simply comparing bacterial densities at 48 h.
Previous in vitro experiments assessing combination therapy by using time-kill and checkerboard analyses have observed synergistic, enhanced, or additive effects of tigecycline plus imipenem, tigecycline plus amikacin, and tigecycline plus polymyxin B against multidrug-resistant A. baumannii, although with many of these experiments, regrowth by 24 or 48 h was still observed (29, 38, 42). In contrast, an additive effect with the combination of rifampin plus tigecycline was not observed, which was also reported in one other time-kill study of A. baumannii (40). However, rifampin has been demonstrated to have synergistic effects with other antimicrobial agents against a multitude of multidrug-resistant Gram-negative pathogens, including KPC-producing K. pneumoniae (8, 30, 40).
In this in vitro pharmacodynamic model simulating exposures likely to occur in the ELF of patients with pneumonia, tigecycline had little activity against KPC-producing K. pneumoniae isolates when simulated alone or in combination with rifampin. In contrast, a statistically significant synergistic antimicrobial effect was noted when tigecycline and meropenem were simulated together for KPC isolates with MICs of ≤2 and ≤16 μg/ml, respectively. Although none of the studied regimens maintained a bactericidal effect over the full 48-h study period, the combination of tigecycline plus high-dose meropenem deserves additional attention in pulmonary infection experiments that incorporate the presence of the immune system.
Acknowledgments
This study was sponsored by Wyeth, which was acquired by Pfizer Inc. (New York, NY) in October 2009.
We acknowledge Stephen Jenkins (New York Presbyterian/Weill Cornell Medical Center, New York, NY) and Meredith Hackel (International Health Management Associates, Schaumburg, IL) for providing KPC-producing K. pneumoniae isolates, Charles Peloquin (University of Florida School of Pharmacy, Gainesville, FL) for conducting rifampin HPLC analysis, and Christina Sutherland for conducting tigecycline and meropenem HPLC analyses. We also thank Hank Christensen for assistance with all in vitro model experiments.
D.P.N. has received research grants from and is a member of the advisory board and speakers’ bureau for Pfizer Inc. J.L.K. has received research grants from Pfizer Inc.
Footnotes
Published ahead of print on 31 January 2011.
REFERENCES
- 1.Acocella, G. 1978. Clinical pharmacokinetics of rifampicin. Clin. Pharmacokinet. 3:108-127. [DOI] [PubMed] [Google Scholar]
- 2.Allegranzi, B., et al. 2000. Concentrations of single-dose meropenem (1g iv) in bronchoalveolar lavage and epithelial lining fluid. J. Antimicrob. Chemother. 46:319-322. [DOI] [PubMed] [Google Scholar]
- 3.AstraZeneca. 2009. Merrem I.V. (meropenem for injection) product information. AstraZeneca, Wilmington, DE.
- 4.Blaser, J. 1985. In-vitro model for simultaneous simulation of the serum kinetics of two drugs with different half-lives. J. Antimicrob. Chemother. 15(Suppl. A):125-130. [DOI] [PubMed] [Google Scholar]
- 5.Bouchillon, S. K., et al. 2005. In vitro activity of tigecycline against 3989 gram-negative and gram-positive clinical isolates from the United States tigecycline evaluation and surveillance trial (TEST program: 2004). Diagn. Microbiol. Infect. Dis. 52:173-179. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, P. A., et al. 2004. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 β-lactamases in New York City. Clin. Infect. Dis. 39:55-60. [DOI] [PubMed] [Google Scholar]
- 7.Bratu, S., et al. 2005. Rapid spread of carbapenem-resistant Klebsiella pneumoniae in New York City. Arch. Intern. Med. 165:1430-1435. [DOI] [PubMed] [Google Scholar]
- 8.Bratu, S., et al. 2005. Carbapenemase-producing Klebsiella pneumoniae in Brooklyn, NY: molecular epidemiology and in vitro activity of polymyxin B and other agents. J. Antimicrob. Chemother. 56:128-132. [DOI] [PubMed] [Google Scholar]
- 9.Bulik, C. C., et al. 2010. Comparative activity of human simulated, high-dose, prolonged infusion of meropenem against Klebsiella producing the KPC carbapenamase versus Pseudomonas aeruginosa in an in vitro pharmacodynamic model. Antimicrob. Agents Chemother. 54:804-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castanheira, M., H. S. Sader, L. M. Deshpande, T. R. Fritsche, and R. N. Jones. 2008. Antimicrobial activities of tigecycline and other broad-spectrum antimicrobials tested against serine carbapenemase- and metallo-β-lactamase-producing Enterobacteriaceae: report from the SENTRY antimicrobial surveillance program. Antimicrob. Agents Chemother. 52:570-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cha, R. 2008. In vitro activity of cefepime, imipenem, tigecycline, and gentamicin, alone and in combination, against extended-spectrum beta-lactamase-producing Klebsiella pneumoniae and Escherichia coli. Pharmacotherapy 28:295-300. [DOI] [PubMed] [Google Scholar]
- 12.Conte, J. E., J. A. Golden, M. G. Kelly, and E. Zurlinden. 2005. Steady-state serum and intrapulmonary pharmacokinetics and pharmacodynamics of tigecycline. Int. J. Antimicrob. Agents 25:523-529. [DOI] [PubMed] [Google Scholar]
- 13.Conte, J. E., J. A. Golden, J. E. Kipps, E. T. Lin, and E. Zurlinden. 2004. Effect of sex and AIDS status on the plasma and intrapulmonary pharmacokinetics of rifampicin. Clin. Pharmacokinet. 43:395-404. [DOI] [PubMed] [Google Scholar]
- 14.Crandon, J. L., A. Kim, and D. P. Nicolau. 2009. Comparison of tigecycline penetration into the epithelial lining fluid of infected and uninfected murine lungs. J. Antimicrob. Chemother. 64:837-839. [DOI] [PubMed] [Google Scholar]
- 15.Daly, M. W., D. J. Riddle, N. A. Ledeboer, W. M. Dunne, and D. J. Ritchie. 2007. Tigecycline for the treatment of pneumonia and empyema caused by carbapenemase-producing Klebsiella pneumoniae. Pharmacotherapy 27:1052-1057. [DOI] [PubMed] [Google Scholar]
- 16.den Hollander, J. G., A. M. Horrevorts, M. L. van Goor, H. A. Verbrugh, and J. W. Mouton. 1997. Synergism between tobramycin and ceftazidime against a resistant Pseudomonas aeruginosa strain, tested in an in vitro pharmacokinetic model. Antimicrob. Agents Chemother. 41:95-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeRyke, C. A., X. Du, and D. P. Nicolau. 2006. Evaluation of bacterial kill when modeling the bronchopulmonary pharmacokinetic profile of moxifloxacin and levofloxacin against parC-containing isolates of Streptococcus pneumoniae. J. Antimicrob. Chemother. 58:601-609. [DOI] [PubMed] [Google Scholar]
- 18.Drusano, G. L., et al. 2004. Pharmacokinetics and penetration of meropenem into epithelial lining fluid in patients with ventilator-associated pneumonia, abstr. A-15. Abstr. 44th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC. [DOI] [PMC free article] [PubMed]
- 19.Elkhaïli, H., et al. 1996. High-performance liquid chromatographic assay for meropenem in serum. J. Chromatogr. B Biomed. Appl. 686:19-26. [DOI] [PubMed] [Google Scholar]
- 20.Firsov, A. A., S. N. Vostrov, A. A. Shevchenko, and G. Cornaglia. 1997. Parameters of bacterial killing and regrowth kinetics and antimicrobial effect examined in terms of area under the concentration-time curve relationships: action of ciprofloxacin against Escherichia coli in an in vitro dynamic model. Antimicrob. Agents Chemother. 41:1281-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gasink, L. B., P. H. Edelstein, E. Lautenbach, M. Synnestvedt, and N. O. Fishman. 2009. Risk factors and clinical impact of Klebsiella pneumoniae carbapenemase-producing K. pneumoniae. Infect. Control Hosp. Epidemiol. 30:1180-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goutelle, S., et al. 2009. Population modeling and Monte Carlo simulation study of the pharmacokinetics and anti-tuberculosis pharmacodynamics of rifampin in lungs. Antimicrob. Agents Chemother. 53:2974-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuti, J. L., P. K. Dandekar, C. H. Nightingale, and D. P. Nicolau. 2003. Use of a Monte Carlo simulation to design an optimized pharmacodynamic dosing strategy for meropenem. J. Clin. Pharmacol. 43:1116-1123. [DOI] [PubMed] [Google Scholar]
- 24.Landman, D., et al. 2007. Evolution of antimicrobial resistance among Pseudomonas aeruginosa, Acinetobacter baumannii and Klebsiella pneumoniae in Brooklyn, NY. J. Antimicrob. Chemother. 60:78-82. [DOI] [PubMed] [Google Scholar]
- 25.Li, C., J. L. Kuti, C. H. Nightingale, and D. P. Nicolau. 2006. Population pharmacokinetic analysis and dosing regimen optimization of meropenem in adult patients. J. Clin. Pharmacol. 46:1171-1178. [DOI] [PubMed] [Google Scholar]
- 26.Li, C., C. A. Sutherland, C. H. Nightingale, and D. P. Nicolau. 2004. Quantitation of tigecycline, a novel glycylcycline [corrected], by liquid chromatography. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 811:225-229. [DOI] [PubMed] [Google Scholar]
- 27.Maltezou, H. C., et al. 2009. Outbreak of infections due to KPC-2 producing Klebsiella pneumoniae in a hospital in Crete (Greece). J. Infect. 58:213-219. [DOI] [PubMed] [Google Scholar]
- 28.Marchaim, D., S. Navon-Venzia, M. J. Schwaber, and Y. Carmeli. 2008. Isolation of imipenem-resistant Enterobacter species; emergence of KPC-carbapenemase, molecular characterization, epidemiology and outcomes. Antimicrob. Agents Chemother. 52:1413-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moland, E. S., et al. 2008. In vitro activity of tigecycline against multidrug-resistant Acinetobacter baumannii and selection of tigecycline-amikacin synergy. Antimicrob. Agents Chemother. 52:2940-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montero, A., et al. 2004. Antibiotic combinations for serious infections caused by carbapenem-resistant Acinetobacter baumannii in a mouse pneumonia model. J. Antimicrob. Chemother. 56:1085-1091. [DOI] [PubMed] [Google Scholar]
- 31.Munoz-Price, L. S., and J. P. Quinn. 2009. The spread of Klebsiella pneumoniae carbapenemases: a tale of strains, plasmids, and transposons. Clin. Infect. Dis. 49:1739-1741. [DOI] [PubMed] [Google Scholar]
- 32.Nicasio, A. M., J. L. Crandon, and D. P. Nicolau. 2009. In vivo pharmacodynamic profile of tigecycline against phenotypically diverse Escherichia coli and Klebsiella pneumoniae isolates. Antimicrob. Agents Chemother. 53:2756-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nordmann, P., G. G. Cuzon, and T. Naas. 2009. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 9:228-236. [DOI] [PubMed] [Google Scholar]
- 34.Norskov-Lauritsen, N., H. Marchandin, and M. J. Dowzicky. 2009. Antimicrobial susceptibility of tigecycline and comparators against bacterial isolates collected as part of the TEST study in Europe (2004-2007). Int. J. Antimicrob. Agents 34:121-130. [DOI] [PubMed] [Google Scholar]
- 35.Pachon-Ibanez, M. E., F. Fernandez-Cuenca, F. Docobo-Perez, J. Pachon, and A. Pascual. 2006. Prevention of rifampicin resistance in Acinetobacter baumannii in an experimental pneumonia model, using rifampicin associated with imipenem or sulbactam. J. Antimicrob. Chemother. 58:689-692. [DOI] [PubMed] [Google Scholar]
- 36.Peloquin, C. A., R. Namdar, M. D. Singleton, and D. E. Nix. 1999. Pharmacokinetics of rifampin under fasting conditions, with food, and with antacids. Chest 115:12-18. [DOI] [PubMed] [Google Scholar]
- 37.Pournaras, S., et al. 2010. Characteristics of meropenem heteroresistance in Klebsiella pneumoniae carbapenemase (KPC)-producing clinical isolates of K. pneumoniae. J. Clin. Microbiol. 48:2601-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prinipe, L., S. D'Arezzo, A. Capone, N. Petrosillo, and P. Visca. 2009. In vitro activity of tigecycline in combination with various antimicrobials against multidrug resistant Acinetobacter baumannii. Ann. Clin. Microbiol. Antimicrob. 8:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rubino, C. M., et al. 2007. Evaluation of tigecycline penetration into colon wall tissue and epithelial lining fluid using a population pharmacokinetic model and Monte Carlo simulation. Antimicrob. Agents Chemother. 51:4085-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saballs, M., et al. 2006. Rifampicin/imipenem combination in the treatment of carbapenem-resistant Acinetobacter baumannii infections. J. Antimicrob. Chemother. 58:697-700. [DOI] [PubMed] [Google Scholar]
- 41.Sakka, S. G., et al. 2007. Population pharmacokinetics and pharmacodynamics of continuous versus short-term infusion of imipenem-cilastatin in critically ill patients in a randomized, controlled trial. Antimicrob. Agents Chemother. 51:3304-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheetz, M. H., et al. 2007. In vitro activities of various antimicrobials alone and in combination with tigecycline against carbapenem-intermediate or -resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:1621-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tisdale, J. E., M. T. Pasko, and J. M. Mylotte. 1989. Antipseudomonal activity of simulated infusions of gentamicin alone or with piperacillin assessed by serum bactericidal rate and area under the killing curve. Antimicrob. Agents Chemother. 33:1500-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Wart, S. A., et al. 2006. Population pharmacokinetics of tigecycline in patients with complicated intra-abdominal or skin and skin structure infections. Antimicrob. Agents Chemother. 50:3701-3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weisenberg, S. A., D. J. Morgan, R. Espinal-Witter, and D. H. Larone. 2009. Clinical outcomes of patients with Klebsiella pneumoniae carbapenemase-producing K. pneumoniae after treatment with imipenem or meropenem. Diagn. Microbiol. Infect. Dis. 64:233-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yigit, H., et al. 2001. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zinner, S. H., J. Blaser, B. B. Stone, and M. C. Groner. 1985. Use of an in-vitro kinetic model to study antibiotic combinations. J. Antimicrob. Chemother. 15(Suppl. A):221-226. [DOI] [PubMed] [Google Scholar]