Abstract
We have investigated the significance of autophagy in the production of the β-lactam antibiotic penicillin (PEN) by the filamentous fungus Penicillium chrysogenum. In this fungus PEN production is compartmentalized in the cytosol and in peroxisomes. We demonstrate that under PEN-producing conditions significant amounts of cytosolic and peroxisomal proteins are degraded via autophagy. Morphological analysis, based on electron and fluorescence microscopy, revealed that this phenomenon might contribute to progressive deterioration of late subapical cells. We show that deletion of the P. chrysogenum ortholog of Saccharomyces cerevisiae serine-threonine kinase atg1 results in impairment of autophagy. In P. chrysogenum atg1 cells, a distinct delay in cell degeneration is observed relative to wild-type cells. This phenomenon is associated with an increase in the enzyme levels of the PEN biosynthetic pathway and enhanced production levels of this antibacterial compound.
The discovery of penicillins in 1929 by Fleming is probably the most important observation in the history of therapeutic medicine development. Penicillins belong to the group of β-lactam antibiotics and are produced as secondary metabolites by several filamentous fungi (3). For industrial production the filamentous fungus Penicillium chrysogenum is used. The first steps of the penicillin (PEN) biosynthetic pathway take place in the cytosol. The amino acid precursors l-α-aminoadipic acid (L-α-AAA), l-cysteine, and l-valine are condensed into the tripeptide δ-(l-α-aminoadipyl)- l-cysteinyl- d-valine (ACV) by the enzyme ACV synthetase (ACVS). Isopenicillin N synthase (IPNS) catalyzes the oxidative ring closure of the linear ACV tripeptide, which leads to the formation of isopenicillin N (IPN), which has a bicyclic ring structure. The final step of penicillin biosynthesis, in which the hydrophilic l-α-AAA side chain of IPN is exchanged for a hydrophobic acyl group, occurs inside peroxisomes via isopenicillin N acyltransferase (IAT) and phenylacetyl coenzyme A ligase (PCL) (2).
Peroxisomes are single-membrane-bound organelles present in all eukaryotes. These cellular compartments are involved in various metabolic pathways. The importance of peroxisomes for efficient penicillin production in P. chrysogenum has been well documented (23, 24). Muller et al. (24) first suggested a correlation between penicillin production and the volume fraction of peroxisomes. Later, it was shown that the high-producing strain DS17690 has enhanced numbers of these organelles relative to the original NRRL 1951 strain (41). Moreover, induction of peroxisome proliferation via overproduction of Pex11 in P. chrysogenum resulted in enhanced levels of penicillin production in two laboratory strains (14).
A remarkable feature of filamentous fungi is the differentiation of cells along the hyphae. These structures can be divided into actively growing regions (apical cells), metabolically active nongrowing regions (subapical cells), and the oldest part of the hyphae, which are comprised of degenerating, highly vacuolated cells (27). Interestingly, it was suggested that β-lactam production is restricted only to some compartments of the hyphae in P. chrysogenum. Based on a structured kinetic model describing growth, differentiation, and penicillin production in submerged P. chrysogenum fermentations, it was suggested that antibiotic production is related to the amount of the metabolically active subapical regions of the hyphae (27).
Autophagy is a highly conserved mechanism in which organelles and proteins are degraded and recycled in the vacuolar lumen. This mechanism is crucial for maintenance of cellular homeostasis, survival during nutrient starvation, and orchestration of an efficient cellular response to stress (40). Although autophagy is generally considered a prosurvival mechanism, under specific conditions this process can also participate in cell death (32, 34, 46). In filamentous fungi autophagy was shown to be involved in nutrient recycling under starvation conditions and during developmental processes (30).
In this study we examined whether inhibition of autophagy-related processes is associated with a delay in the degeneration of late subapical cells. We demonstrate that autophagic degradation of cellular components occurs mainly in older subapical compartments of the hyphae under PEN production conditions. This phenomenon may contribute to progressive deterioration of these cells. Furthermore, the delayed deterioration observed in an autophagy-deficient P. chrysogenum mutant strain resulted in a significant increase in the amount of PEN produced.
MATERIALS AND METHODS
Strains and cultivation conditions.
P. chrysogenum strains used in this study are listed in Table 1. For biochemical and ultrastructural analyses, P. chrysogenum strains were cultivated on PEN induction medium (9) supplemented with 0.05% phenoxyacetic acid. In order to induce nonselective autophagy, mycelia were grown for 24 h on PEN induction medium and then harvested by centrifugation and resuspended in PEN induction medium without a nitrogen source. For genetic manipulation purposes, P. chrysogenum strains were grown on YGG medium (0.8% KCl, 1.6% glucose, 0.67% yeast nitrogen base [Difco], 0.15% citric acid, 0.6% K2HPO4, 0.2% yeast extract, pH 6.2, supplemented with penicillin and streptomycin [Gibco]). P. chrysogenum mycelia were cultivated at 25°C at 200 rpm in batch cultivation mode.
TABLE 1.
P. chrysogenum strains used in this study
| Strain | Genotype or characteristics | Reference or source |
|---|---|---|
| DS17690 | High-PEN-producing strain, AmdS− | 8 |
| DS54465 | DS17690 with deletion of hdfA gene, AmdS− | 37 |
| DS54465 GFP.SKL | DS54465 with integrated PgpdA-GFP.SKL-TpenDE cassette at niaD locus, AmdS−, chlorate resistant | W. H. Meijer et al., submitted for publication |
| DS17690 GFP | DS17690 with integrated PpcbC-GFP-TpenDE cassette, AmdS+ | This study |
| atg1 | DS54465 with deletion of atg1 gene, phleomycin resistant, AmdS− | This study |
| atg1::GFP | atg1 with PgpdA-GFP-TpenDE cassette integrated at niaD locus, phleomycin resistant, AmdS−, chlorate resistant | This study |
| atg1::GFP.SKL | atg1 with Pgpda-GFP.SKL-TpenDE cassette integrated at niaD locus, phleomycin resistant, AmdS−, chlorate resistant | This study |
| atg1::GFP + atg1 | atg1::GFP with PATG1-ATG1-TATG1 cassette integrated at pyrG locus | This study |
P. chrysogenum niaD-deficient transformants were selected on plates containing 1.25% KClO3 and supplemented with 0.185% adenine as sole nitrogen source. Phleomycin-resistant (Bler) P. chrysogenum strains were selected on Phleo-plates containing 50 μg/ml of phleomycin (Invitrogen) (17). To induce conidiospore formation, R-agar was used (0.52% [vol/vol] glycerol, 0.75% [vol/vol] beet molasses, 0.5% yeast extract, 300 mM NaCl, 0.2 mM MgSO4·7H2O, 0.370 mM KH2PO4, 3.3 μM NH4Fe(SO4)2·12H2O, 0.4 μM CuSO4·5H2O, and 1.45 mM CaSO4·2H2O).
Escherichia coli DH5α and XL1-Blue cells, which were used for cloning purposes, were grown in super optimal broth with catabolite repression (SOC) or lysogeny broth (LB) (33) medium supplemented with the appropriate antibiotics.
Micrococcus luteus ATCC 9341, which was used for the PEN bioassay, was cultivated at 30°C on 2× TY medium (6).
Molecular techniques.
Plasmids and oligonucleotides used in this study are listed in Tables 2 and 3, respectively. Standard DNA manipulations were performed according to established procedures described by Sambrook et al. (33). Some of the cloning procedures were performed using the Multisite Gateway technology (Invitrogen). Transformation of P. chrysogenum protoplasts was performed as described previously (4). Restriction enzymes (Fermentas and Roche), T4 DNA ligase (Fermentas), and other DNA-modifying enzymes were used as recommended by the suppliers. PCRs were carried out with Phusion (Fermentas) or Expand (Roche) high-fidelity polymerase. After cloning, all DNA fragments obtained via PCR were sequenced (ServiceXS). Southern blotting was performed according to the DIG High Prime labeling and detection kit (Roche). For in silico DNA sequence analysis, the Clone Manager 5 software (Scientific and Education Software, Durham, NC) was used.
TABLE 2.
Plasmids used in this study
| Plasmid | Characteristicsa | Reference or source |
|---|---|---|
| pDONRP4-P1R | Multisite Gateway vector, Kanr Cmr | Invitrogen |
| pDONR221 | Multisite Gateway vector, Kanr Cmr | Invitrogen |
| pDONRP2R-P3 | Multisite Gateway vector, Kanr Cmr | Invitrogen |
| pDESTR4-R3 | Multisite Gateway vector, Ampr Cmr | Invitrogen |
| pDESTR4-R3/AMDS | pDESTR4-R3 with PgpdA-amdS cassette, Ampr | 13 |
| pDELatg1 | pDESTR4-R3/AMDS with atg1 deletion cassette, Ampr | This study |
| pENTR41-Patg1 | pDONRP4-P1R with 5′-flanking region of atg1, Kanr | This study |
| pENTR23-Tatg1 | pDONRP2R-P3 with 3′-flanking region of atg1, Kanr | This study |
| pENTR221phleo | pDONR221 vector with PpcbC-phleomycin cassette, Kanr | 16 |
| pMDB005 | Plasmid containing PgpdA-eGFP-TpenDE cassette, Ampr | This study |
| pMDB006 | pDONR221 with PgpdA-eGFP-TpenDE cassette, Kanr | This study |
| pENTR41-5′niaD | pDONRP4-P1R with 5′-flanking region of niaD, Kanr | Meijer et al., submitted |
| pENTR23-3′niaD | pDONRP2R-P3 with 3′-flanking region of niaD, Kanr | Meijer et al., submitted |
| pMDB007 | pDESTR4-R3 with ΔniaD::PgpdA-eGFP -TpenDE cassette, Ampr | This study |
| pGBRH2-eGFP | pGBRH2 plasmid containing PpcbC-eGFP-TpenDE cassette, Ampr | 13 |
| pNiGaNi | Plasmid containing PgpdA-amdS expression cassette flanked by niaD sequences, Ampr | Lab collection |
| pBBK-007 | Plasmid containing PgpdA-DsRed.SKL-TpenDE cassette, Ampr | Meijer et al., submitted |
| pEXP-5′niaD-PgpdA-GFP.SKL-TpenDE-3′niaD | pDESTR4-R3 containing ΔniaD-PgpdA-GFP.SKL-TpenDE cassette, Ampr | Meijer et al., submitted |
| pENTR23-ATG1 | pDONR P2R-P3 vector containing PATG1-ATG1-TATG1 cassette, Kanr | This study |
| pENTR221/AMDS | pDONR 221 vector with PgpdA-amdS cassette, Kanr | Gift from J. G. Nijland |
| LMOPE41pyrG | pDONR P4-P1R with pyrG gene, Kanr | 26 |
| pCOMatg1 | Plasmid containing PATG1-ATG1-TATG1 complementation cassette, pyrG gene, and amdS marker, Ampr | This study |
Kan, kanamycin; Cm, chloramphenicol; Amp, ampicillin.
TABLE 3.
Oligonucleotides used in this study
| Oligonucleotide | Sequencea (5′-3′) |
|---|---|
| PdelATG1_Fow1 | 5′-GGGG ACA ACT TTG TAT AGA AAA GTT GGG AGA TTC CGT CAC GAG ATG-3′ |
| PdelATG1_Rev1 | 5′-GGGG ACT GCT TTT TTG TAC AAA CTT G GC CTG TTC CGT CTC TGG TAA-3′ |
| TdelATG1_Fow1 | 5′-GGGG ACA GCT TTC TTG TAC AAA GTG GTC CGG AAG AAG GTA GCT GTT-3′ |
| TdelATG1_Rev1 | 5′-GGGG ACA ACT TTG TAT AAT AAA GTT G CA TGC CAA TTC CAC GCT GAT-3′ |
| MDB008 | 5′-GGGG ACA AGT TTG TAC AAA AAA GCA GGC TGC TCT GTA CAG TGA CC GGT GAC TC-3′ |
| LMOp012 | 5′-GGGG AC CAC TTT GTA CAA GAA AGC TGG GTT CCCC TGA AAG AGT TGA TAT TGA AGG-3′ |
| FWRprobe | 5′-GCTTCCGTCACCGTACAGTT-3′ |
| REVprobe | 5′-CTACCTCAGCAGCAGCAATG-3′ |
| Phleo.rev | 5′-AACGGCACTGGTCAACTTGG-3′ |
| 5′ atgknout.for | 5′-GCTTCCGTCACCGTACAGTT-3′ |
| ATG1comfor | 5′ GGGG ACA GCT TTC TTG TAC AAA GTG GCC GAC TTT GCT CAA GG CGG TTC 3′ |
| ATG1comrev | 5′-GGGG ACA ACT TTG TAT AAT AAA GTT GCC TGG TAT CTG CAA ACC CAA TCC-3′ |
| pyrGupst | 5′-GAACGCCTCGCAGACAATGCTC 3′ |
| AMDS.rev | 5′-GCATGCCAGAAAGAGTCACC-3′ |
The underlined sequences are the attB recombination sites for the Multisite Gateway system.
Plasmid construction. (i) Construction of a P. chrysogenum atg1 deletion strain.
The atg1 deletion cassette was constructed using the Multisite Gateway cloning system. The upstream flanking region (1.7 kb) and downstream flanking region (2.1 kb) of the atg1 gene were amplified by PCR with the primers PdelATG1_Fow1 and PdelATG1_Rev1 and with TdelATG1_Fow1 and TdelATG1_Rev1, respectively, using DS54465 genomic DNA as template. The resulting PCR products were introduced into pDONRP2R-P3 (3′-flanking region of atg1) and pDONRP4-P1R (5′-flanking region of atg1) by using the Gateway BP clonase reaction, resulting in plasmids pENTR23-Tatg1 and pENTR41-Patg1, respectively. Subsequently, a Gateway LR reaction was executed with pENTR41-Patg1, pENTRY221phleo, pENTR23-Tatg1, and the destination vector pDEST R4-R3/AMDS, resulting in the formation of the atg1 deletion construct pDELatg1. This plasmid was linearized with NotI and transformed into P. chrysogenum DS54465 protoplasts. Bler transformants were selected and analyzed by colony PCR using the primers 5′ atgknout.for and Phleo.rev. Deletion of atg1 was confirmed by Southern Blotting using a 936-bp atg1 promoter-specific probe, which was generated by PCR with primers FWRprobe and REVprobe.
(ii) Construction of P. chrysogenum atg1::GFP, atg1::GFP.SKL, and DS17690 GFP strains.
The pMDB005 plasmid, which contains PgpdA-eGFP-TpenDE, was constructed as follows: plasmid pGBRH2-eGFP was digested with BamHI and SmaI, and the enhanced green fluorescent protein (eGFP)-containing fragment was cloned between the BamHI and SmaI sites of plasmid pBBK007, thereby replacing DsRed.SKL. Subsequently, pMDB005 was used as template to generate an attB1-PgpdA-eGFP-TpenDE-attB2 fragment by PCR using the primers MDB008 and LMOp012. The resulting PCR product was introduced into pDONR221 using Gateway technology, generating plasmid pMDB006. Next, the Gateway LR reaction was performed with pENTR41-5′niaD, pENTR23-3′niaD, pMDB006, and pDESTR4-R3 to obtain plasmid pMDB007. This plasmid was linearized with KpnI and transformed into P. chrysogenum atg1 protoplasts. NiaD− transformants were selected based on their ability to grow on medium supplemented with chlorate. Production of GFP was confirmed by fluorescence microscopy.
In order to generate the P. chrysogenum atg1::GFP.SKL strain, the pEXP-5′-niaD-PgpdA-GFP.SKL-TpenDE-3′niaD plasmid was linearized with KpnI and transformed into P. chrysogenum atg1 protoplasts. NiaD− transformants were selected based on their ability to grow on medium supplemented with chlorate. Production of GFP.SKL protein was confirmed by fluorescence microscopy.
In order to generate the P. chrysogenum DS17690 GFP strain, a 2.2-kb NotI fragment from pGBRH2-eGFP containing the PpcbC-eGFP-TpenDE cassette was cotransformed into P. chrysogenum DS17690 protoplasts together with a 6.2-kb NotI-SmaI DNA fragment containing the amdS selection marker from pNiGaNi. AmdS+ cotransformants that showed GFP fluorescence were selected.
Complementation of the atg1 deletion strain.
The atg1 complementation cassette was constructed using the Multisite Gateway cloning system. The atg1 gene, containing 1,500 bp upstream of the predicted translation start site and 500 bp downstream of the stop codon, was amplified by PCR with the primers ATG1comfor and ATG1comrev, with DS54465 genomic DNA as the template. The resulting PCR product was introduced into pDONR P2R-P3 by using the Gateway BP clonase reaction, resulting in plasmid pENTR23-ATG1. Subsequently, a Gateway LR reaction was performed with pENTR23-ATG1, pENTR221/AMDS, LMOPE41pyrG, and the destination vector pDEST R4-R3, resulting in the formation of the atg1 complementation construct pCOMatg1. This plasmid was transformed in circular form into atg1::GFP P. chrysogenum protoplasts, thereby promoting single crossover at the pyrG locus. AmdS+ transformants were selected. Correct integration was confirmed by colony PCR, using the primers pyrGupst and AMDS.rev.
Bioassays.
PEN bioassays using M. luteus ATCC 9341 as an indicator organism were carried out as described previously (6) using the spent medium of cultures grown on PEN induction medium supplemented with phenoxyacetic acid. Dry weights of the cultures were determined and used as a reference.
Biochemical analysis.
Crude extracts of P. chrysogenum cells were prepared as described previously (13). Protein concentrations were measured using the RC DC assay system (Bio-Rad) with bovine serum albumin as a standard. SDS-PAGE and Western blotting were carried out in accordance with established procedures. Blots were decorated with polyclonal antibodies raised against P. chrysogenum IPNS, IAT, and transcription elongation factor EF1-α (7, 12, 42).
Microscopy.
Wide-field fluorescence microscopy was carried out using an Axio Observer Z1 fluorescence microscope (Zeiss). Images were taken using an EC-Plan Neofluar 100×/1.3 numerical aperture objective and a Coolsnap HQ2 camera (Roper Scientific Inc.). GFP fluorescence was visualized with a 470/40-nm band-pass excitation filter, a 495-nm dichromatic mirror, and a 525/50-nm band-pass emission filter. FM4-64 (Invitrogen) fluorescence was analyzed with a 545/25-nm band-pass excitation filter, a 570-nm dichromatic mirror, and a 605/70-nm band-pass emission filter. Z-stack images were made with an interval of 1 μm. High-resolution overall images were generated from individual tiles by using the MosaiX module. P. chrysogenum mycelia were stained with fluorescent probes prior to microscopy investigation, in accordance with manuals provided by the suppliers (Molecular Probes and Invitrogen).
For electron microscopy cells were fixed and prepared as described previously (44).
NMR analysis of β-lactams.
The quantitative 1H nuclear magnetic resonance (NMR) analysis of P. chrysogenum fermentation products was performed using a Bruker Avance 600 spectrometer at 600 MHz. The filtrate was mixed with a known amount of maleic acid as the internal standard and lyophilized. The lyophilizate was dissolved in D2O and measured at 300 K. The 30-s delay between scans was more than five times the T1 of all compounds, and so the exact amounts of analyzed compounds were measured by comparing the ratios between the integrals of the analyzed chemicals and the integral of the internal standard.
RESULTS
Deletion of the atg1 gene results in inhibition of autophagy in P. chrysogenum.
The Saccharomyces cerevisiae atg1 gene encodes a serine-threonine kinase, which is involved in the early steps of autophagy induction (5). Inactivation of the Atg1 protein in yeast or its ortholog in filamentous fungi results in impairment of autophagy (11, 20, 31).
To determine the significance of the P. chrysogenum atg1 gene in PEN production, multiple independent atg1 deletion strains were constructed and analyzed. Correct integration of the deletion cassette was confirmed by Southern blotting (Fig. 1 A). The atg1 null mutants did not display any growth defect but showed a strong reduction in conidiospore formation (Fig. 1B).
FIG. 1.
Disruption of the atg1 gene in P. chrysogenum. (A) Strategy for the integration of the phleomycin cassette in the atg1 locus (left). Southern blot analysis of HindIII-digested genomic DNA by using the indicated 936-bp probe identified the expected 1.23-kb band in wild-type DNA and the 6.6-kb band in DNA from the atg1 mutants (right). (B) Morphology of colonies of the WT and the atg1 mutant strain after 7 days of growth on R-agar plates at 25°C. The atg1 null mutant generates white colonies because of its reduced sporulation ability. The colony of the WT control normally forms green spores.
In order to analyze the significance of the Atg1 protein in turnover of cytoplasmic components, we constructed atg1 strains that either produced GFP.SKL to mark peroxisomes (atg1::GFP.SKL) or GFP without any sorting sequence to visualize the cytosol (atg1::GFP). Wild-type (WT) and atg1 strains producing GFP.SKL or GFP were grown for 24 h on PEN induction medium and were subsequently subjected to nitrogen starvation conditions to induce autophagy. Under these conditions, in WT cells a strong accumulation of GFP (Fig. 2 B) and GFP.SKL (Fig. 2D) fluorescence was observed in the vacuolar lumen, indicative of uptake and degradation of these components. This vacuolar fluorescence was below the limit of detection in cells prior to the shift (Fig. 2A). In contrast, in the atg1::GFP and atg1::GFP.SKL strains, vacuolar GFP fluorescence was never observed under nitrogen limitation conditions (Fig. 2C and E). As a control, an atg1 strain was used that was complemented with the atg1 gene under the control of its own promoter. In this strain the N-limited-induced autophagy was fully restored (see Fig. S1A in the supplemental material). Also, this strain regained the ability to produce green spores (see Fig. S1B).
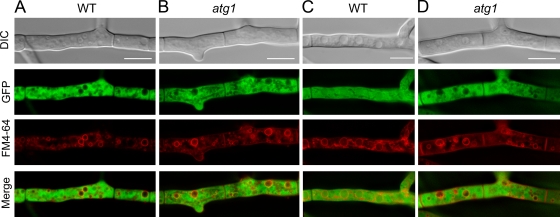
FIG. 2.
Autophagy is impaired in P. chrysogenum atg1 null mutants. Mycelia of P. chrysogenum strains expressing GFP or GFP.SKL were cultivated under PEN-inducing conditions for 24 h, harvested by centrifugation, and shifted to PEN-inducing medium in the absence of a nitrogen source to induce autophagy. Vacuoles are marked red by FM4-64. (A) GFP fluorescence inside the vacuolar lumen of WT cells was below the limit of detection at the onset of the experiment. (B and C) Degradation of cytosolic components was evident in WT cells after 24 h of growth under nitrogen starvation conditions (B) but not in atg1 cells (C). (D and E) In WT cells, also GFP.SKL was observed in the vacuoles due to pexophagy (D), but not in atg1 cells (E). Bars, 5 μm.
These data suggest that autophagy is impaired in the P. chrysogenum atg1 null mutant.
Autophagic turnover of cytosolic components and peroxisomes occurs in subapical regions.
The kinetics of constitutive autophagy in P. chrysogenum strains cultivated under PEN-inducing conditions was analyzed using fluorescence microscopy. P. chrysogenum strains producing GFP were used to monitor autophagy-related degradation of cytosolic components. In WT cells, accumulation of GFP inside the vacuole was not observed at early time points of cultivation (0 to 24 h after inoculation of spores), suggesting that recycling of cytosolic components via autophagy was below the limit of detection in young hyphal cells (Fig. 3 A). However, after 48 h of growth, GFP accumulation in vacuoles of subapical cells was evident (Fig. 3C). This phenomenon was not observed in atg1 deletion cells, indicating that accumulation of GFP inside the vacuole in P. chrysogenum WT cells represents an autophagic process (Fig. 3B and D). Comparable results were observed for peroxisomes, marked with GFP.SKL. However, accumulation of GFP.SKL in the vacuole was retarded relative to uptake of its cytosolic counterpart, but it was massively detected predominantly in late subapical cells at approximately 70 h of cultivation (Fig. 4 A). During prolonged cultivation, these phenomena occurred consecutively in time, in successive cells of the hypha (Fig. 4B and 5 A). In contrast, vacuolar GFP.SKL fluorescence was not observed in atg1 cells (Fig. 5B). Morphometric analysis revealed that atg1 mutant cells contained significantly enhanced numbers of peroxisomes in late subapical cells relative to WT (Fig. 5C). These data imply that continuous turnover of cytosol and peroxisomes occurs via autophagy-related processes in subapical compartments of hyphae during cultivation of P. chrysogenum under PEN-inducing conditions.
FIG. 3.
Constitutive turnover of cytosolic components under PEN-inducing conditions. Fluorescence microscopy images show time-dependent degradation of cytosolic components via autophagy in cells of the WT strain grown under PEN-inducing conditions. This process was impaired in the atg1 strain. Vacuoles were stained by FM4-64. Degradation of cytosol was below the limit of detection after 24 h in both WT (A) and atg1 cells (B) but prominent after 48 h of cultivation in WT cells (C), but not in atg1 cells (D). Bars, 5 μm.
FIG. 4.
Turnover of peroxisomes under PEN-inducing conditions. Fluorescence microscopy images show degradation of peroxisomes over time in the WT strain grown under penicillin-inducing conditions. (A) Accumulation of GFP.SKL fluorescence inside the vacuolar lumen (arrows), indicating turnover of peroxisomes, was observed after approximately 70 h of cultivation, predominantly in late subapical cells. (B) During prolonged growth (96 h), degradation of peroxisomes inside the vacuole was observed in successively formed cells of the hyphae. The asterisk indicates an apical compartment. Bars, 20 μm.
FIG. 5.
atg1 cells contain enhanced numbers of peroxisomes. Mycelia were cultivated under PEN-inducing conditions for 130 h. Peroxisomes were labeled with GFP.SKL. (A) Significant turnover of peroxisomes was present in almost all cells along the hyphae of WT P. chrysogenum. (B) An accumulation of peroxisomes and absence of GFP.SKL fluorescence inside vacuoles were observed in the atg1 null mutant. Bars, 20 μm. (C) Peroxisome numbers in late subapical cells. Fluorescent spots were counted from grouped Z-stacks. The numbers of peroxisomes per μm of hyphae are presented. The atg1 null mutant strain is characterized by a significantly increased number of peroxisomes in late subapical compartments (Student's t test, P = 0.0004). At least 65 subapical hyphal cells were randomly counted in three independent experiments. Scale bars represent the standard errors of means.
Autophagy contributes to cell deterioration in old subapical compartments.
The substantial degradation of cytosolic components and peroxisomes in subapical cells during cultivation of WT P. chrysogenum was particularly evident at later stages of growth. In order to determine the effect of extensive autophagy on the morphology of stationary-phase cells (after 130 h of cultivation), the autophagy-deficient atg1 and WT P. chrysogenum strains were analyzed by electron microscopy. These analyses revealed that old hyphal compartments of the WT strain were characterized by extensive autophagy and cell deterioration, in contrast to the corresponding compartments in atg1 cells (Fig. 6). These data suggest that inhibition of autophagy in P. chrysogenum may be associated with a delay in progressive deterioration of cells at late stages of growth.
FIG. 6.
Morphological analysis of P. chrysogenum WT and atg1 cells. Electron micrographs of KMnO4-fixed P. chrysogenum hyphae are shown in WT (A) and atg1 (B) cells. Subapical compartments of WT but not atg1 hyphae showed extensive autophagy characterized by large vacuoles containing autophagic bodies and reduced numbers of organelles relative to atg1 cells. M, mitochondrion; P, peroxisome; V, vacuole; AV, autophagosome. Bars, 2 μm.
Inhibition of autophagy-related processes in P. chrysogenum results in enhanced levels of PEN biosynthesis enzymes and enhanced PEN production.
To determine the effect of atg1 deletion on the levels of the enzymes involved in β-lactam production, we cultured WT and atg1 mutant cells under PEN-inducing conditions and analyzed protein abundance levels of the cytosolic enzyme IPNS and the peroxisomal enzyme IAT over time via Western blotting. This analysis revealed that both IAT and IPNS protein levels were enhanced in crude extracts of atg1 cells relative to those in WT extracts at time points at which WT cells showed significant induction of autophagy in subapical compartments of the hyphae (Fig. 7 A). In order to analyze whether the enhanced amounts of PEN biosynthesis enzymes are accompanied by elevated PEN production levels, bioassays using Microccocus luteus as the indicator strain were performed. The data showed a significantly larger clearing surface when culture supernatants of atg1 cells were used, relative to WT (Fig. 7B), implying that deletion of atg1 leads to enhanced production of antibacterial agents by P. chrysogenum. Both increased antibacterial activity and enhanced levels of PEN biosynthesis enzymes were observed for multiple independent atg1 mutants (see Fig. S2 in the supplemental material).
FIG. 7.
Effects of autophagy impairment on antibiotic production. (A) Western blot analysis of crude extracts of P. chrysogenum WT and atg1 strains. Mycelia were cultivated under PEN-inducing conditions. Equal amounts of protein were loaded per lane. Polyclonal antibodies against IPNS, IAT proteins, and transcription elongation factor EF1-α (loading control) were used to decorate the blots. Enhanced levels of both IPNS and IAT were detected in atg1 extracts relative to those in WT cells at all time points studied. (B) Spent media of atg1 and WT strains cultivated under PEN-inducing conditions were diluted and loaded into wells of a bioassay plate that had been overlaid with the PEN-sensitive indicator strain M. luteus. Cell densities (in terms of the dry weight per liter) of both WT and atg1 cultures were comparable at all time points studied. After 24 h of incubation at 30°C, the sizes of the growth inhibition zones (halos) were determined. The data indicate that the atg1 strain produced increased amounts of antibacterial agents relative to the WT strain at all time points studied. (C) NMR quantitative determination of secreted PenV levels. Spores of atg1 and WT strains were inoculated in PEN induction medium supplemented with phenoxyacetic acid. Samples were taken after 7 days of cultivation. At this time point the sugars were completely consumed and cell densities (in terms of dry weight per liter) of both cultures were comparable (data not shown). The amount of detected extracellular β-lactam antibiotic, PenV, was significantly increased in the atg1 null mutant relative to the parental strain (Student's t test, P = 0.033). Scale bars represent the standard errors of means of three independent experiments.
In order to analyze whether the increased antibacterial activity of atg1 culture supernatants was caused by enhanced levels of PEN produced, quantitative NMR determinations of secreted penicillin V (PenV) were performed. The parental high-PEN-producing strain (DS54465) and the isogenic atg1 mutant strain were cultivated on PEN induction medium for 7 days. Under these conditions the atg1 mutant produced 37% more penicillin V (average, 1.3 g/liter) than the corresponding parental strain (average, 0.95 g/liter; P = 0.033 [Student's t test]). At this time point the sugars were completely consumed, and cell densities (in terms of dry weight per liter) of both cultures were comparable (Fig. 7C). Thus, impairment of autophagy-related processes in a high-PEN-producing strain results in significantly increased production of PenV.
DISCUSSION
We have studied the role of autophagy in P. chrysogenum and have provided evidence that blocking autophagy promotes PEN production by this fungus. Autophagy has been implicated in many cellular processes, including survival under nutrient starvation conditions, cell remodeling (25), removal of damaged organelles and proteins, and protection against oxidative stress (45). The core machinery executing autophagy is conserved among eukaryotes (25), including filamentous fungi (21, 30). Inhibition of autophagy in filamentous fungi by disruption of specific ATG genes has provided evidence that this process is involved in starvation-induced differentiation, including the formation of aerial hyphae (31), conidiophores (15), and sexual reproductive structures (19, 29).
In our studies the role of autophagy in the β-lactam antibiotic-producing fungus P. chrysogenum was analyzed. We showed that disruption of the atg1 gene in P. chrysogenum impairs autophagy and causes sporulation defects, as in other fungi (20, 22, 28, 31). Interestingly, we observed that in P. chrysogenum the degradation of cytosolic components preceded the turnover of peroxisomes under nitrogen starvation conditions. These data, together with previous observations (18), suggest that bulk autophagy, although considered a nonselective process, may degrade cellular components in a specific order. A more nonspecific autophagic response under nitrogen starvation conditions could be disadvantageous for the cell, as it may degrade portions of essential subcellular components required for basic cell function at the initiation of the recycling process.
The mycelium of ascomycetous fungi is composed of interconnected hyphae that are divided by perforated septa into multinucleated compartments, which allows bulk flow of cytosol and organelles throughout the hyphae. Recycling of cellular components of mature hyphal compartments via autophagy and autolysis has been suggested to support apical cell growth under nutrient limitation conditions (35, 36). It is probable that at a late stage of industrial production of PEN in batch-fed cultures the organism experiences nutrient limitations that result in induction of autophagy in mature hyphal compartments.
In P. chrysogenum, the biosynthetic pathway of β-lactam antibiotics is compartmentalized and is suggested to take place in mature, vacuolated subapical hyphal compartments (10, 27). Extensive autophagy-related degradation of cytosolic components and peroxisomes in vacuolated, late hyphal elements was observed in these studies, which implies that autophagy could have an impact on the efficiency of PEN production. Indeed, our studies demonstrated that impairment of autophagy in P. chrysogenum led to significantly increased PenV production, possibly related to the increased amounts of PEN biosynthesis enzymes in the cells.
The elevated amounts of β-lactam antibiotic biosynthesis enzymes might positively affect PEN production (2, 38, 39, 43), but these enzyme levels are not the only factor that may impact productivity of this secondary metabolite. The number of peroxisomes may also play an important role in efficient PEN production (14, 24, 41). Indeed, the disruption of atg1 results in an increased number of peroxisomes, since the autophagy-dependent removal of these organelles is impaired (1). Hence, it is likely that the presence of elevated numbers of peroxisomes in subapical hyphal cells is one of the reasons for increased PEN production in cells of the atg1 strain.
The high-PEN-producing strains used now in industrial penicillin fermentations are the result of time-consuming, extensive strain improvement programs. Application of genetic engineering has opened the possibility for targeted, comprehensive design of strains. We show here that a promising way of improving PEN productivity in P. chrysogenum may be modulation of autophagy-related processes.
Supplementary Material
Acknowledgments
This project was carried out within the research program of the Kluyver Centre for Genomics of Industrial Fermentation, which is part of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research. J.A.K.W.K. was supported by a grant from DSM, Delft, Netherlands.
We thank Rinse de Boer, Susan Fekken, and Wiebe H. Meijer for excellent technical assistance.
Footnotes
Published ahead of print on 17 December 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aksam, E. B., et al. 2007. A peroxisomal lon protease and peroxisome degradation by autophagy play key roles in vitality of Hansenula polymorpha cells. Autophagy 3:96-105. [DOI] [PubMed] [Google Scholar]
- 2.Brakhage, A. A. 1998. Molecular regulation of beta-lactam biosynthesis in filamentous fungi. Microbiol. Mol. Biol. Rev. 62:547-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brakhage, A. A., et al. 2004. Regulation of penicillin biosynthesis in filamentous fungi. Adv. Biochem. Eng. Biotechnol. 88:45-90. [DOI] [PubMed] [Google Scholar]
- 4.Cantoral, J. M., B. D. Díez, J. L. Barredo, E. Alvarez, and J. F. Martín. 1987. High-frequency transformation of Penicillium chrysogenum. Biotechnology (NY) 5:494-497. [Google Scholar]
- 5.Cheong, H., and D. J. Klionsky. 2008. Dual role of Atg1 in regulation of autophagy-specific PAS assembly in Saccharomyces cerevisiae. Autophagy 4:724-726. [DOI] [PubMed] [Google Scholar]
- 6.Gidijala, L., et al. 2008. Production of functionally active Penicillium chrysogenum isopenicillin N synthase in the yeast Hansenula polymorpha. BMC Biotechnol. 8:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gidijala, L., I. J. van der Klei, M. Veenhuis, and J. A. Kiel. 2007. Reprogramming Hansenula polymorpha for penicillin production: expression of the Penicillium chrysogenum pcl gene. FEMS Yeast Res. 7:1160-1167. [DOI] [PubMed] [Google Scholar]
- 8.Harris, D. M., et al. 2006. Enzymic analysis of NADPH metabolism in beta-lactam-producing Penicillium chrysogenum: presence of a mitochondrial NADPH dehydrogenase. Metab. Eng. 8:91-101. [DOI] [PubMed] [Google Scholar]
- 9.Hillenga, D. J., H. J. Versantvoort, A. J. Driessen, and W. N. Konings. 1994. Structural and functional properties of plasma membranes from the filamentous fungus Penicillium chrysogenum. Eur. J. Biochem. 224:581-587. [DOI] [PubMed] [Google Scholar]
- 10.Justen, P., G. C. Paul, A. W. Nienow, and C. R. Thomas. 1998. Dependence of penicillium chrysogenum growth, morphology, vacuolation, and productivity in fed-batch fermentations on impeller type and agitation intensity. Biotechnol. Bioeng. 59:762-775. [DOI] [PubMed] [Google Scholar]
- 11.Kamada, Y., T. Sekito, and Y. Ohsumi. 2004. Autophagy in yeast: a TOR-mediated response to nutrient starvation. Curr. Top. Microbiol. Immunol. 279:73-84. [DOI] [PubMed] [Google Scholar]
- 12.Kiel, J. A., V. I. Titorenko, I. J. van der Klei, and M. Veenhuis. 2007. Overproduction of translation elongation factor 1-alpha (eEF1A) suppresses the peroxisome biogenesis defect in a Hansenula polymorpha pex3 mutant via translational read-through. FEMS Yeast Res. 7:1114-1125. [DOI] [PubMed] [Google Scholar]
- 13.Kiel, J. A., et al. 2009. Matching the proteome to the genome: the microbody of penicillin-producing Penicillium chrysogenum cells. Funct. Integr. Genomics 9:167-184. [DOI] [PubMed] [Google Scholar]
- 14.Kiel, J. A., I. J. van der Klei, M. A. van den Berg, R. A. Bovenberg, and M. Veenhuis. 2005. Overproduction of a single protein, Pc-Pex11p, results in 2-fold enhanced penicillin production by Penicillium chrysogenum. Fungal Genet. Biol. 42:154-164. [DOI] [PubMed] [Google Scholar]
- 15.Kikuma, T., M. Ohneda, M. Arioka, and K. Kitamoto. 2006. Functional analysis of the ATG8 homologue Aoatg8 and role of autophagy in differentiation and germination in Aspergillus oryzae. Eukaryot. Cell 5:1328-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koetsier, M. J., et al. 2010. The Penicillium chrysogenum aclA gene encodes a broad-substrate-specificity acyl-coenzyme A ligase involved in activation of adipic acid, a side-chain precursor for cephem antibiotics. Fungal Genet. Biol. 47:33-42. [DOI] [PubMed] [Google Scholar]
- 17.Kolar, M., P. J. Punt, C. A. van den Hondel, and H. Schwab. 1988. Transformation of Penicillium chrysogenum using dominant selection markers and expression of an Escherichia coli lacZ fusion gene. Gene 62:127-134. [DOI] [PubMed] [Google Scholar]
- 18.Kristensen, A. R., et al. 2008. Ordered organelle degradation during starvation-induced autophagy. Mol. Cell Proteomics 7:2419-2428. [DOI] [PubMed] [Google Scholar]
- 19.Liu, X. H., J. P. Lu, and F. C. Lin. 2007. Autophagy during conidiation, conidial germination and turgor generation in Magnaporthe grisea. Autophagy 3:472-473. [DOI] [PubMed] [Google Scholar]
- 20.Liu, X. H., et al. 2007. Involvement of a Magnaporthe grisea serine/threonine kinase gene, MgATG1, in appressorium turgor and pathogenesis. Eukaryot. Cell 6:997-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meijer, W. H., I. J. van der Klei, M. Veenhuis, and J. A. Kiel. 2007. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy 3:106-116. [DOI] [PubMed] [Google Scholar]
- 22.Mukaiyama, H., et al. 2009. Autophagy-deficient Schizosaccharomyces pombe mutants undergo partial sporulation during nitrogen starvation. Microbiology 155:3816-3826. [DOI] [PubMed] [Google Scholar]
- 23.Muller, W. H., et al. 1992. Involvement of microbodies in penicillin biosynthesis. Biochim. Biophys. Acta 1116:210-213. [DOI] [PubMed] [Google Scholar]
- 24.Muller, W. H., et al. 1991. Localization of the pathway of the penicillin biosynthesis in Penicillium chrysogenum. EMBO J. 10:489-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakatogawa, H., K. Suzuki, Y. Kamada, and Y. Ohsumi. 2009. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10:458-467. [DOI] [PubMed] [Google Scholar]
- 26.Opalinski, L., J. A. Kiel, T. G. Homan, M. Veenhuis, and I. J. van der Klei. Penicillium chrysogenum Pex14/17p: a novel component of the peroxisomal membrane that is important for penicillin production. FEBS J. 277:3203-3218. [DOI] [PubMed]
- 27.Paul, G. C., and C. R. Thomas. 1996. A structured model for hyphal differentiation and penicillin production using Penicillium chrysogenum. Biotechnol. Bioeng. 51:558-572. [DOI] [PubMed] [Google Scholar]
- 28.Pinan-Lucarre, B., A. Balguerie, and C. Clave. 2005. Accelerated cell death in Podospora autophagy mutants. Eukaryot. Cell 4:1765-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinan-Lucarre, B., M. Paoletti, K. Dementhon, B. Coulary-Salin, and C. Clave. 2003. Autophagy is induced during cell death by incompatibility and is essential for differentiation in the filamentous fungus Podospora anserina. Mol. Microbiol. 47:321-333. [DOI] [PubMed] [Google Scholar]
- 30.Pollack, J. K., S. D. Harris, and M. R. Marten. 2009. Autophagy in filamentous fungi. Fungal Genet. Biol. 46:1-8. [DOI] [PubMed] [Google Scholar]
- 31.Richie, D. L., et al. 2007. Unexpected link between metal ion deficiency and autophagy in Aspergillus fumigatus. Eukaryot. Cell 6:2437-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samara, C., P. Syntichaki, and N. Tavernarakis. 2008. Autophagy is required for necrotic cell death in Caenorhabditis elegans. Cell Death Differ. 15:105-112. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J. F., and T. Maniatis. 1989. Molecular cloning, a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 34.Scott, R. C., G. Juhasz, and T. P. Neufeld. 2007. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr. Biol. 17:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shoji, J. Y., M. Arioka, and K. Kitamoto. 2006. Possible involvement of pleiomorphic vacuolar networks in nutrient recycling in filamentous fungi. Autophagy 2:226-227. [DOI] [PubMed] [Google Scholar]
- 36.Shoji, J. Y., M. Arioka, and K. Kitamoto. 2006. Vacuolar membrane dynamics in the filamentous fungus Aspergillus oryzae. Eukaryot. Cell 5:411-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Snoek, I. S., et al. 2009. Construction of an hdfA Penicillium chrysogenum strain impaired in non-homologous end-joining and analysis of its potential for functional analysis studies. Fungal Genet. Biol. 46:418-426. [DOI] [PubMed] [Google Scholar]
- 38.Theilgaard, H., M. van Den Berg, C. Mulder, R. Bovenberg, and J. Nielsen. 2001. Quantitative analysis of Penicillium chrysogenum Wis54-1255 transformants overexpressing the penicillin biosynthetic genes. Biotechnol. Bioeng. 72:379-388. [DOI] [PubMed] [Google Scholar]
- 39.Thykaer, J., and J. Nielsen. 2003. Metabolic engineering of beta-lactam production. Metab. Eng. 5:56-69. [DOI] [PubMed] [Google Scholar]
- 40.Todde, V., M. Veenhuis, and I. J. van der Klei. 2009. Autophagy: principles and significance in health and disease. Biochim. Biophys. Acta 1792:3-13. [DOI] [PubMed] [Google Scholar]
- 41.van den Berg, M. A., et al. 2008. Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nat. Biotechnol. 26:1161-1168. [DOI] [PubMed] [Google Scholar]
- 42.van der Lende, T. R., et al. 2002. δ-(l-α-Aminoadipyl)-l-cysteinyl-d-valine synthetase, that mediates the first committed step in penicillin biosynthesis, is a cytosolic enzyme. Fungal Genet. Biol. 37:49-55. [DOI] [PubMed] [Google Scholar]
- 43.Veenstra, A. E., P. van Solingen, R. A. Bovenberg, and L. H. van der Voort. 1991. Strain improvement of Penicillium chrysogenum by recombinant DNA techniques. J. Biotechnol. 17:81-90. [DOI] [PubMed] [Google Scholar]
- 44.Waterham, H. R., et al. 1994. The Hansenula polymorpha PER1 gene is essential for peroxisome biogenesis and encodes a peroxisomal matrix protein with both carboxy- and amino-terminal targeting signals. J. Cell Biol. 127:737-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yorimitsu, T., and D. J. Klionsky. 2005. Autophagy: molecular machinery for self-eating. Cell Death Differ. 12(Suppl. 2):1542-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu, L., et al. 2006. Autophagic programmed cell death by selective catalase degradation. Proc. Natl. Acad. Sci. U. S. A. 103:4952-4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.