Abstract
Enzymes involved in oxidation of long-chain n-alkanes are still not well known, especially those in Gram-positive bacteria. This work describes the alkane degradation system of the n-alkane degrader actinobacterium Gordonia sp. strain SoCg, which is able to grow on n-alkanes from dodecane (C12) to hexatriacontane (C36) as the sole C source. SoCg harbors in its chromosome a single alk locus carrying six open reading frames (ORFs), which shows 78 to 79% identity with the alkane hydroxylase (AH)-encoding systems of other alkane-degrading actinobacteria. Quantitative reverse transcription-PCR showed that the genes encoding AlkB (alkane 1-monooxygenase), RubA3 (rubredoxin), RubA4 (rubredoxin), and RubB (rubredoxin reductase) were induced by both n-hexadecane and n-triacontane, which were chosen as representative long-chain liquid and solid n-alkane molecules, respectively. Biotransformation of n-hexadecane into the corresponding 1-hexadecanol was detected by solid-phase microextraction coupled with gas chromatography-mass spectrometry (SPME/GC-MS) analysis. The Gordonia SoCg alkB was heterologously expressed in Escherichia coli BL21 and in Streptomyces coelicolor M145, and both hosts acquired the ability to transform n-hexadecane into 1-hexadecanol, but the corresponding long-chain alcohol was never detected on n-triacontane. However, the recombinant S. coelicolor M145-AH, expressing the Gordonia alkB gene, was able to grow on n-triacontane as the sole C source. A SoCg alkB disruption mutant that is completely unable to grow on n-triacontane was obtained, demonstrating the role of an AlkB-type AH system in degradation of solid n-alkanes.
Alkanes are saturated, linear hydrocarbons whose chain length can vary from 1 (in methane) to more than 50 carbon atoms. Alkanes constitute about 20 to 50% of crude oil, depending on the source of the oil, but living organisms, such as bacteria, plants, and some animals, also produce them as pheromones (4). As a result, alkanes are widespread in nature, and many microorganisms have evolved enzymes to use them as a carbon source. Alkanes, however, are chemically inert and must be activated before they can be metabolized. Under aerobic conditions, activation is usually achieved by oxidation of one of the terminal methyl groups to generate the corresponding primary alcohol by alkane hydroxylases (AHs) (18, 26). Although many microorganisms are capable of degrading aliphatic hydrocarbons and they are readily isolated from contaminated and noncontaminated sites, relatively little is known about the molecular characteristics of their alkane-degradative systems. Indeed, until recently, the alkane-degradative genes of only a small number of Gram-negative bacteria, namely, Pseudomonas, Acinetobacter, and Alkanivorax, had been described in detail. Among these, the alk system found in Pseudomonas putida GPo1, which degrades C5 to C12 n-alkanes, remains the most extensively characterized alkane hydroxylase system (27). The initial terminal oxidation of the alkane substrate to a 1-alkanol is catalyzed by a three-component alkane hydroxylase complex consisting of a particulate nonheme integral membrane alkane monooxygenase (AlkB) and two soluble proteins, rubredoxin (AlkG) and rubredoxin reductase (AlkT). The Pseudomonas putida alk genes are located in two different loci (alkBFGHJKL and alkST) on the OCT plasmid, separated by 10 kb of DNA (27). Five chromosomal genes (alkM, rubA, rubB, alkR, and xcpR) in at least three different loci are required for degradation of C12 to C18 alkanes in Acinetobacter sp. strain ADP1 (17). Similar to the case for P. putida GPo1, the initial terminal alkane oxidation is also catalyzed by a three-component alkane hydroxylase system, which comprises an alkane monooxygenase (AlkM), rubredoxin (RubA), and rubredoxin reductase (RubB). Acinetobacter sp. strain M-1 was shown to possess two alkane monooxygenase genes (alkMa and alkMb) as well as single copies of rubA and rubB, located in three different loci. AlkMa is involved in degradation of long-chain n-alkanes up to C16 and AlkMb in degradation of very-long-chain n-alkanes up to C30 (23). More recently, a flavin-binding monooxygenase, AlmA, was found involved in oxidation of very long-chain n-alkanes up to C32 in Acinetobacter sp. strain DSM17874 (25).
Much less is known about the alkane-degradative systems of Gram-positive bacteria. Homologs of alkB were amplified from Rhodococcus erythropolis NRRL B-16531, Amycolatopsis rugosa NRRL B-2295, and Mycobacterium tuberculosis H37Rv (22) and from Nocardiodes sp. strain CF8 (7). The M. tuberculosis alkB homologs could be functionally expressed in an alkB knockout derivative of Pseudomonas fluorescens CHA0 and in P. putida GPo1 and were shown to oxidize alkanes ranging from C10 to C16 (21). Four alkane monooxygenase homologs (two as part of alkane gene clusters and two occurring as separate genes) were identified in two closely related Rhodococcus strains and analyzed by functional heterologous expression in Escherichia coli and Pseudomonas spp. (31). Moreover, genes encoding an AH system (alkane 1-monooxygenase, rubredoxins, and rubredoxin reductase) from Gordonia sp. strain TF6 were cloned, sequenced, and expressed in E. coli, where they were found to be the minimum components required to confer alkane hydroxylase activity in this strain on n-alkanes up to C13 (6). Alkanes longer than C16 support growth of many microorganisms, but the identity of enzymes involved in their oxidation is known for only a restricted number of isolates (27, 30), among which is only one Gram-positive strain, belonging to the genus Geobacillus (LadA) (5). Rhodococcus and other closely related G+C-rich, mycolic acid-containing actinomycetes, such as Corynebacterium, Gordonia, and Nocardia, are increasingly recognized as ideal candidates for the biodegradation of hydrocarbons because of their ability to degrade a wide range of organic compounds, hydrophobic cell surfaces, production of biosurfactants, and robustness and ubiquity in the environment (12).
Recently, a Gram-positive bacterium identified as Gordonia sp. strain SoCg, which is able to grow on and to degrade long and solid n-alkanes up to hexatriacontane, was isolated (16). In this work the alkane degradation system of Gordonia SoCg was investigated by cloning and sequencing the alk locus; gene expression was analyzed in relation to the time course of n-alkane consumption, and the metabolic intermediate of n-hexadecane hydroxylation was identified. Functional expression in heterologous hosts unable to use n-alkanes and the Gordonia alkB disruption mutant confirmed the role of an actinobacterial di-iron nonheme integral membrane alkane monooxygenase in degradation of n-alkanes longer than C16.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and general methods.
Gordonia sp. strain SoCg was isolated from a hydrocarbon-contaminated Mediterranean shoreline (16). It grows on the mineral medium Bushnell-Haas (BH) medium (Difco) with a wide range of n-alkanes, from C12 up to C36, as the sole carbon source but does not grow on short-chain n-alkanes. Gordonia was routinely grown on JM medium (14) or on liquid mineral BH medium supplemented with 10 mM n-alkanes directly supplied in the liquid medium (n-hexadecane, C16) or supplied as finely ground powder (n-triacontane, C30). In biodegradation kinetic experiments, n-alkanes were added to BH medium as an n-hexane solution, once it was established that n-hexane is not toxic to or utilized by the strain. In solid cultures on BH agar, n-hexadecane was supplemented as vapor as described elsewhere (16).
The bacterial strains, commercial cloning vectors, and plasmids constructed in this study are described in Table 1. E. coli was routinely grown in Luria medium (19) and S. coelicolor in JM medium (14).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| Gordonia sp. | ||
| SoCg | Long-chain n-alkane degrader, alkB+ | 16 |
| SoCg ΩalkB | SoCg disruption mutant, alkB− Aprar | This study |
| Escherichia coli | ||
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74recA1 endA1 araD139 Δ(ara leu)7697 galU galK λ−rpsL nupG | Invitrogen |
| ET12567 | F−dam-13::Tn9 dcm-6hsdM hsdRzjj-202::Tn10recF143galK2galT22ara14 lacY1 xyl-5 leuB6 Cmlr Tetr Kanr | 9 |
| BL21(DE3)/pLysS | F−ompT hsdSB (rB− mB−) dcmgal λ(DE3) pLysS Camr | Invitrogen |
| BL21-AH | E. coli BL21 containing the recombinant expression vector pRalkB; alkB Ampr | This study |
| Streptomyces coelicolor | ||
| M145 | Wild type; SCP1− SCP2− | 9 |
| M145-AH | S. coelicolor M145 containing the recombinant integrative pIalkB; alkB Thior Aprar | This study |
| Cloning and expression vectors | ||
| pUC18 | E. coli cloning vector, Ampr | Invitrogen |
| pGEM-T Easy vector | E. coli cloning vector, Ampr | Promega |
| TOPO-TA | E. coli cloning vector, Ampr | Invitrogen |
| pRSET-B | E. coli expression vector, Ampr | Invitrogen |
| pIJ8600 | Streptomycete expression vector, Aprar, promoter induction by thiostrepton | 9 |
| pIJ773 | Streptomycete cloning vector, Aprar, used to extract apramycin resistance cassette aac(3)IV with its oriT | 9 |
| Plasmids containing DNA from Gordonia SoCg | ||
| palkCg23 | pGEM-T Easy vector derivative containing a 570-bp alkB fragment (GenBank accession no. EF437969) | 16 |
| palk68 | pUC18 derivative containing an 8-kb fragment of Gordonia SoCg including the alk cluster | This study |
| pGalkB1 | pGEM-T Easy vector derivative containing alkB (1.2 kb), amplified by PCR with primers alkNIFor and alkBHIRev | This study |
| pRalkB | pRSET-B derivative containing alkB (1.2 kb) | This study |
| pIalkB | pIJ8600 derivative containing alkB (1.2 kb) | This study |
| palkapra | palk68 derivative containing the apramycin resistance cassette cloned into a unique AleI restriction site of alkB | This study |
Plasmid and chromosomal DNA purification, enzymatic digests, ligations, and bacterial transformations were performed using standard molecular techniques (9, 19) or according to the manufacturer's instructions. All primers used for PCR amplification were synthesized by Invitrogen and are listed in Table 2. The 16S rRNA gene was amplified using primers rD1 and fD1 (29) in a 20-μl reaction mixture containing 1 μl of chromosomal DNA, 0.2 μM each primer, 0.2 mM deoxynucleoside triphosphates (dNTPs), and 1.5 U of recombinant Taq DNA polymerase (Invitrogen, Life Technologies). PCR was carried out in a Biometra thermocycler using the following program: 94°C for 5 min; 30 cycles of 1 min at 94°C, 1 min at 55°C, and 2 min at 72°C; and a final extension at 72°C for 7 min. Where not otherwise specified, the PCR was carried out under the same conditions but with an annealing temperature of 62°C.
TABLE 2.
Primers used in this study
| Primer | Sequencea | Reference or source |
|---|---|---|
| AHqRTFor | 5′-GGACCGATGCTGGTCTATGT-3′ | This study |
| AHqRTRev | 5′-CAGATAACAGGCCATGACGA-3′ | This study |
| rubA3qRTFor | 5′-CTACCGTGTCCGGTCTGTG-3′ | This study |
| rubA3qRTRev | 5′-CCAGTCGTCGGGAATGTC-3′ | This study |
| rubA4qRTFor | 5′-CTGCGAGGTCTGCGGATT-3′ | This study |
| rubA4qRTRev | 5′-GGCCACCTCGACCATCTC-3′ | This study |
| rubBqRTFor | 5′-GGGTGTTGATCCAGTTCAGG-3′ | This study |
| rubBqRTRev | 5′-TATCTGGCACATCACCAACG-3′ | This study |
| alkUqRTFor | 5′-GCGTTCACCGAGTACTTCAC-3′ | This study |
| alkUqRTRev | 5′-ATCGACAACCACGTCGACTC-3′ | This study |
| alkNIFor | 5′-AACATATGCTCGTGAGAGGAGCGTGC-3′ | This study |
| alkBHIRev | 5′-AAGGATCCCCGGACAACGGTAGGCGC-3′ | This study |
| CF | 5′-ATGTTYATHGCNATGGAYCCNC-′ | 11 |
| CR | 5′-NARNCKRTTNCCCATRCANCKRTG-′ | 11 |
| apra750FR | 5′-ATTCCGGGGATCCGTCGACC-′ | This study |
| apra750RV | 5′-TGTAGGCTGGAGCTGCTTC-′ | This study |
| ladAFR | 5′-GGCGTSTACGMCRWCTACGGYRGG-′ | This study |
| ladARV | 5′-GAYCTACCAGGYCGGGTCGTCG-′ | This study |
| alkCG341FR | 5′-CCGAGGACCCGGCGAGCTC-′ | This study |
| alkCG341RV | 5′-CTCCGGGGTGCACCGCTC-′ | This study |
Underlining indicates NdeI (for alkNIFor) and BamHI (for alkBHIRev) restriction sites.
For pulsed-field gel electrophoresis (PFGE) analysis, undigested DNA plugs of Gordonia SoCg were prepared as described by Kieser et al. (9). PFGE was performed with 0.5× Tris-borate-EDTA (TBE) as the running buffer at 14°C by using a CHEF DRII PFGE system (Bio-Rad) at 160 V, 400 mA, and a pulse time of 20 s for 18 h (2). For hybridization experiments, undigested PFGE-separated DNA was transferred to Hybond N nylon membranes (Amersham International plc, Buckinghamshire, United Kingdom), according to the protocol for large DNA fragment transfer (2).
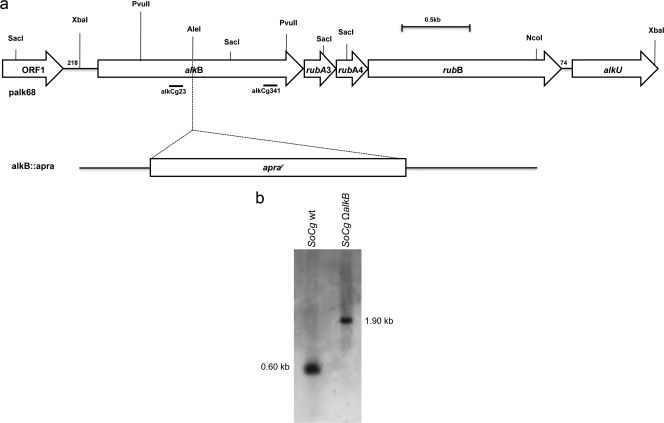
All Southern hybridizations were carried out using digoxigenin (DIG)-dUTP-labeled probes (Fig. 1) obtained by PCR and labeled using the digoxigenin system by Boehringer Manheim Biochemicals (Indianapolis, IN).
FIG. 1.
Schematic representation of the 4,472-bp region of Gordonia sp. strain SoCg carried by palk68. (a) Genetic organization of the gene cluster and restriction map. The orientations of identified genes are indicated by arrows. Probes alkCg23 and alkCg341 were used in Southern hybridization experiments for alkB localization and disruption mutant analysis, respectively. The 3.0-kb alkB::apra fragment was used to disrupt the alkB gene in Gordonia SoCg by double-crossover homologous recombination. The apramycin resistance cassette (1,342 bp) was inserted into the unique AleI site of the alkB gene sequence. (b) Southern analysis using DIG-labeled alkCg341 as a probe of the PvuII-digested genomic DNA extracted from the disruption mutant Gordonia SoCg ΩalkB, and wild-type (wt) Gordonia SoCg.
Growth curves on n-alkanes.
Gram-positive strains were grown in 30 ml JM medium (14) in 250-ml baffled flasks for 48 h at 30°C. Cells were washed three times with BH medium and suspended in the same medium to give an optical density at 600 nm (OD600) of 1.0. Afterwards, 1 ml (about 1 mg [dry weight]) of bacterial suspension was inoculated in 1-liter baffled flasks containing 120 ml BH supplemented with 10 mM n-alkane. The flasks were incubated at 30°C on a rotary shaker (200 rpm), and at each time point 1-ml aliquots were sampled and centrifuged at 4,000 × g and the pellet dried at 65°C to a constant weight.
Time course of n-alkane consumption.
To determine Gordonia SoCg n-alkane utilization, 300 μl of washed cells suspended in BH medium to an OD600 of 1.0 as described above was inoculated into 100-ml glass tubes containing 10 ml BH medium supplemented with 10 mM n-hexadecane or n-triacontane in an n-hexane solution. A total of 48 tubes were incubated at 30°C under shaking conditions; 12 tubes for each n-alkane were inoculated with Gordonia SoCg, and 12 were left uninoculated to be used as abiotic controls. Residual long-chain n-alkanes were n-hexane extracted as described elsewhere (16) from the whole tube content after 22, 46, 62, and 96 h of incubation, in triplicate, and analyzed by the gas chromatography-mass spectrometry (GC-MS) analytical technique with a Hewlett-Packard 5890 GC system interfaced with an HP 5973 quadrupole mass spectrometer detector. As the stationary phase, an HP5-MS capillary column (5% diphenyl-95% dimethylpolysiloxane; 30 m by 0.2 mm; 0.25-μm film thickness [J&W Scientific]) was used. The GC oven temperature program was as follows: 40°C for 5.00 min, increase of 10°C min−1 to 280°C, and holding for 20 min. Helium was used as the carrier gas with a constant flow rate of 1 ml min−1. Electron impact ionization spectra were obtained at 70 eV, with recording of mass spectra from 42 to 550 amu, which allows 3.5 scans s−1. The time course of consumption was expressed as n-alkane residue with respect to abiotic controls.
Analysis of the metabolic intermediates from the n-alkane oxidation pathways.
The metabolic intermediates resulting from incubation of SoCg, M145-AH, and BL21-AH (expressing the Gordonia alkB gene) on C16 and C30 were analyzed by solid-phase microextraction (SPME) coupled with GC-MS. Gordonia SoCg, Gordonia SoCg ΩalkB, S. coelicolor M145-AH, and S. coelicolor M145 carrying the empty pIJ8600 were grown in JM medium, washed, and resuspended in BH medium to a final OD600 of 1.0 as described above. E. coli BL21-AH and E. coli BL21 carrying the empty pRSET-B were grown overnight in LB medium, washed three times with phosphate buffer (pH 7.2), and suspended in the same volume. One milliliter of cells was inoculated in 100-ml glass tubes with 10 mM each n-alkane in the presence of inducers (isopropyl-β-d-thiogalactopyranoside [IPTG] in E. coli tubes according to the instructions for the Ni-nitrilotriacetic acid [NTA] purification system [Invitrogen] and thiostrepton [10 ng ml−1] in Streptomyces tubes) and incubated at 37°C for 6 h with shaking. Abiotic controls were incubated under the same conditions and analyzed in parallel.
The entire suspensions were analyzed by immersing the SPME fiber, which was coated with 85-μm polyacrylate (PA) and equipped with a holder for manual injection. The time needed to reach equilibrium between the amount of analyte adsorbed by the polymeric film and the initial concentration of the analyte in the sample matrix during the SPME sampling is dependent on the properties of both the analyte and the matrix (4) and in our study was 20 min at 45°C. Prior to use, the fiber was conditioned at 300°C for 2 h in the GC injector port. An HP-5MS 5% phenyl methyl siloxane capillary column was used to perform the gas chromatographic separations. The initial oven temperature was 80°C with a constant helium flow, corresponding to the nominal head pressure of 9.37 lb/in2. The temperature increase was 5°C min−1 to 280°C, and then the temperature was held for 20 min. The ionization spectra were obtained as described above. Analytical identification and quantifications were carried out using standard-grade compounds purchased from Sigma-Aldrich and the commercial NIST 2005 mass spectrum library search database.
Cloning and sequence analysis of Gordonia SoCg alkane hydroxylase genes.
The probe alkCg23 was obtained by PCR from palkCg23 (Table 1) using the pair of primers AH+for and AH+rev (16) and was used in a Southern analysis to identify suitable restriction fragments in BamHI/BglII-digested SoCg chromosomal DNA (Fig. 1). Fragments in the range of 8 to 10 kb were cut out from a preparative agarose gel, purified, and ligated into BamHI-digested and dephosphorylated pUC18. The ligation mixture was used to transform E. coli DH10B (Invitrogen) by electroporation. E. coli transformants were selected on Luria agar supplemented with IPTG, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), and ampicillin (19). The transformants were screened by colony hybridization with the same probe. The recombinant plasmid designated palk68 was isolated from an alk+ clone using the Miniprep column kit (Qiagen) and analyzed by restriction fragment length polymorphism (RFLP) to estimate the insert size. A 4,472-bp region of the insert was commercially sequenced on both strands by primer walking. Nucleotide and deduced amino acid sequences were compared with EMBL/SwissProt/GenBank databases using BLASTN and BLASTX at NCBI, and open reading frames (ORFs) were identified using Chromas Pro 2.33.
A LadA-related monooxygenase gene in the SoCg genome was searched for by PCR using a degenerate primer pair (ladAFR and ladARV [Table 2]) designed from the consensus sequences of the ladA genes from Geobacillus thermodenitrificans NG80-2 (NCBI accession no. YP_001127577.1) (5), Mycobacterium smegmatis MC2 155 (NCBI accession no. YP_886406.1), Mycobacterium avium 104 (NCBI accession no. 883212.1), and Nocardia farcinica IFM 10152 (NCBI accession no. YP_117411.1). A touch-down PCR protocol was carried out with the following program: an initial denaturation step at 94°C for 2 min; 16 cycles of 45 s at 94°C, 1 min at annealing temperatures decreasing from 72 to 58°C (with a 2°C decremental step from cycle 2 to 8), and 1 min at 72°C; 26 cycles of 1 min at 94°C, 1 min at 56°C, and 1 min at 72°C; and a final 7-min extension at 72°C.
Genes encoding cytochrome P450 alkane hydroxylases were searched for using primers CF and CR (Table 2) designed from the conserved domains in the N-terminal and C-terminal regions of CYP153A family enzymes.
Cloning of alkB and construction of expression plasmids.
The 1.2-kb alkB gene was amplified from Gordonia SoCg genomic DNA using primers alkNIfor and alkBHIrev (Table 2) (containing an NdeI restriction site immediately upstream of the putative ATG start codon of alkB and a BamHI restriction site downstream of the putative TGA stop codon, respectively) using a touch-down PCR protocol as described previously. The amplicon was purified using NucleoSpin extract II (Macherey-Nagel GmbH & Co. KG) and ligated with pGEM- T-Easy vector (Promega). The resulting plasmid, pGalkB1 (Table 1), was checked by sequencing and the alkB insert was ligated as a PstI-NcoI fragment into the pRSET-B expression vector (Invitrogen) in frame with the T7 RNA polymerase promoter. The ligation mixture was used to transform E. coli BL21(DE3)/pLysS (Invitrogen), and the recombinant clones were selected on LB supplemented with ampicillin (200 mg ml−1). Clone E. coli BL21-AH containing the recombinant plasmid pRalkB (Table 1) was selected and used for alkB expression analysis.
To clone alkB in S. coelicolor M145, the entire gene from pGalkB1 was cloned as an NdeI-BamHI fragment into pIJ8600 (Table 1) in frame with the tipA promoter. The ligation mixture was used to transform E. coli DH10B; the derived pIalkB (Table 1) was isolated and then transformed into E. coli ET12567 by electroporation. From the recombinant clone of E. coli ET12567, pIalkB was transferred to S. coelicolor M145 by conjugation (9). The resulting exconjugants were selected on apramycin (50 mg ml−1) and thiostrepton (200 mg ml−1), and the correct integration was verified by Southern hybridization. BamHI-digested genomic DNA was probed using the 1.2-kb alkB fragment (PCR amplified from pGalkBI with primers alkNIfor and alkBHIrev) and the 750-bp apramycin resistance cassette fragment (PCR amplified from pIJ773 using primers apra750FR and apra750RV).
Heterologous expression analysis.
Crude extracts of E. coli BL21-AH and E. coli BL21 transformed with the empty vector pRSET-B were collected after incubation in LB medium supplemented with ampicillin to an OD600 of 0.6 in the presence or absence of IPTG as an inducer, according to the Ni-NTA purification system instructions (Invitrogen). About 0.5 μg of each soluble and insoluble protein fraction from each extract was loaded for SDS-PAGE and run in a Mini-Protean Tetra cell (Bio-Rad) at 20 mA and 150 V for 50 min using the SeeBluePlus2 prestained standard (Invitrogen) as a molecular size marker. After electrophoresis, proteins were electrotransferred from the gel to the Hybond-C Extra membrane (Amersham) using a Hoefer mini-VE semidry blotting apparatus (Amersham Pharmacia Biotech) at 150 V and 20 mA for 1 h. Immunostaining was carried out using alkaline phosphatase-conjugated anti-His tag monoclonal antibodies (Invitrogen) followed by detection with nitroblue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolylphosphate (BCIP) (Invitrogen).
To evaluate alkB gene expression in S. coelicolor M145-AH, total RNA was extracted and used in a reverse transcription-PCR (RT-PCR) assay as described below.
RNA isolation, RT-PCR analysis, and absolute quantitative RT-PCR.
For RNA isolation, SoCg was grown for 22 h in BH medium supplemented with 10 mM hexadecane, triacontane, and fructose as described above. M145-AH was grown in JM medium supplemented with thiostrepton as an inducer. After incubation for 22 h at 30°C with shaking, cells were suspended in P buffer (9) and lysed using lysozyme (6 mg ml−1). Total RNA was extracted using the RNeasy midikit (Qiagen). DNase I (Roche) treatment was performed at 37°C for 1 h, and after ethanol precipitation and a washing step with 70% ethanol, the air-dried RNA pellet was resuspended in 50 μl of sterile distilled water.
RT-PCR was performed by using the Superscript one-step RT-PCR kit (Invitrogen) with about 0.1 μg of total RNA as a template, the primer pair AHqRTfor and AHqRTRev designed from the internal region of alkB (Table 1), and the conditions indicated by the supplier, routinely using 35 PCR cycles. For each reaction, a negative control with Taq polymerase and without reverse transcriptase was included in order to exclude DNA contamination.
The expression of the Gordonia SoCg alk cluster genes was analyzed by quantitative reverse transcription-PCR using the Applied Biosystems 7300 real-time PCR system (Applied Biosystems). A high-capacity cDNA archive kit (Applied Biosystems) was used, according to the manufacturer's instructions, to reverse transcribe 5 μg of total DNA-free RNA. Then, 3 μl of the cDNA was mixed with 10 μl of SYBR green PCR master mix (Applied Biosystems) and 5 pmol of each primer in a final volume of 20 μl. In addition to AHqRTfor and AHqRTRev, the primer pairs rubA3qRTFor/rubA3qRTRev, rubA4qRTFor/rubA4qRTRev, rubBqRTFor/rubBqRTRev, and alkUqRTFor/alkUqRTRev were specifically designed from the genes rubA3, rubA4, rubB, and alkU, respectively. PCR was performed, in triplicate for each gene, under the following conditions: 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 62°C. Eventually, a dissociation reaction was performed under the following conditions: a 1-min step with a temperature gradient increase of 1°C per step from 55 to 99°C. A negative control (distilled water) was included in all real-time PCR assays. Standards for the alk genes were constructed from purified palk68 that was quantified using a Qbit fluorometer (Invitrogen) and diluted in a 10-fold series to create a six-point standard curve (0.5 × 102 to 0.5 × 106 molecules) that was run in duplicate with each set of samples. The number of copies per microliter was calculated as follows: molecular mass of palk68 (standard template) = 11,433 bp × 660 Da = 7.54 × 106 g mol−1; 1 molecule or 1 copy of fragment = 7.54 × 106/6.02 × 1023 = 1.25 × 10−17 g; therefore, 10 ng of template contains 10 × 10−9/1.25 × 10−23 copies = 8 ×1014 molecules.
Construction of Gordonia SoCg alkB disruption mutant.
In order to obtain Gordonia SoCg electrocompetent cells 2 mg (wet weight) of cells was inoculated and left for 3 days in 25 ml YEME (9) with 2 g liter−1 of glycine at 30°C. The rich biomass was collected at the bottom of the 50-ml tube and pretreated for 15 min in an ultrasonic bath. The pellet obtained after centrifugation at 5,500 × g for 30 min was immediately incubated with 10 g liter−1 chilled glycerol on ice for 1 h. The cells were then washed three times with 10 g liter−1 chilled glycerol and finally resuspended in 3 ml of 10 g liter−1 glycerol, aliquoted into 200-μl samples, and stored at −80°C.
The apramycin resistance cassette, including its own promoter and oriT, was extracted from plasmid pIJ773 by digestion with EcoRI and HindIII and cloned into pUC18. The correct recombinant plasmid was checked by sequencing, and the cassette was excised using EcoRI and filled in, using the Klenow fragment enzyme (Roche), to obtain blunt ends. The apramycin resistance cassette was inserted into the unique AleI site within the alkB gene of palk68. The resulting plasmid (palkapra [Table 1]) was XbaI-NcoI digested to obtain an alkB::apra linear fragment (Fig. 1), which was introduced by electroporation into Gordonia SoCg electrocompetent cells. Apramycin-resistant transformants were selected on apramycin, and gene disruption by double-crossover homologous recombination was confirmed by Southern analysis using the DIG-labeled alkCg341 as a probe.
Nucleotide sequence accession numbers.
The Gordonia SoCg 16S rRNA gene sequence and the 4,472-bp palk68 insert have been submitted to GenBank under accession no. AY496285.2 and HQ026811, respectively.
RESULTS
Identification and properties of strain SoCg.
The n-alkane degrader Gordonia sp. strain SoCg was isolated from a hydrocarbon-contaminated Mediterranean shoreline. This strain is able to grow on n-alkanes of different lengths, from dodecane (C12) to hexatriacontane (C36), as the sole C source (16); it is unable to grow on n-octane or shorter n-alkanes, but it is not inhibited by short-chain n-alkanes, e.g., n-hexane. The analysis of the complete 16S rRNA gene sequence of SoCg showed the highest similarity (98% identity, 1,442/1,470 nucleotides) to the DNA sequence of Gordonia amicalis strain T3 (GenBank accession no. EU427321.1), which is a tert-amyl methyl ether degrader recently isolated from a hydrocarbon-contaminated soil (15). The presence of an alkB homolog gene in the SoCg genome has been previously demonstrated by PCR using degenerate primers (16). Pulsed-field gel electrophoresis (PFGE) of undigested DNA extracted from strain SoCg revealed the presence of a large cryptic plasmid; the PFGE-separated DNA was probed with DIG-labeled probe alkCg23 (Fig. 1) in a Southern hybridization, and the alkB gene was localized on the chromosome of Gordonia SoCg (data not shown). The same probe hybridized to only one band in the genomic DNA digested with various restriction enzymes (data not shown). Southern hybridization analysis confirmed that strain SoCg harbors in its chromosome a single copy of the alkB gene, as previously suggested by sequencing of cloned PCR fragments (16). As SoCg degrades a large range of long-chain n-alkanes, we also tried to find in its genome other, alkB-unrelated putative genes involved in long-chain alkane degradation, using PCR. The primer pair designed from the consensus sequence of ladA from Geobacillus thermodenitrificans NG80-2 and other Gram-positive strains gave two different amplification products whose sequences were unrelated to known alkane hydroxylase genes. A second pair of primers, CF and CR (11), used to amplify the conserved region of the p450-CYP153 family genes gave no amplification product.
SoCg grows on long-chain n-alkanes and rapidly degrades them.
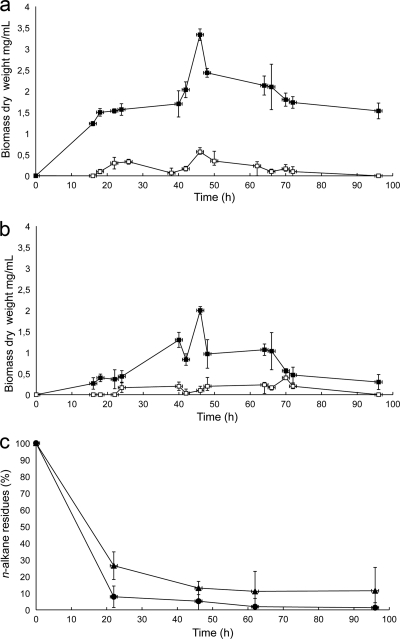
Growth of Gordonia sp. strain SoCg in mineral broth supplemented with n-hexadecane or n-triacontane as the sole carbon source was followed for 96 h. An increasing biomass accumulation was observed on both n-alkanes up to 46 h, followed by a growth decline (Fig. 2 a and b). Comparison of the two curves reveals that n-hexadecane supports higher biomass accumulation (3.4 mg ml−1) than n-triacontane (2 mg ml−1), as measured after 46 h of growth. GC-MS analysis of n-alkane residues showed that substrate consumption is followed by biomass increase. Both n-alkanes, in fact, almost completely disappeared within 62 h (with 11.1% n-hexadecane residue and 1.9% n-triacontane residue), with a rapid decline in the first 22 h (Fig. 2c). Consumption of n-triacontane is more rapid than that of n-hexadecane, in contrast with lower biomass accumulation on the longest alkane. It appears that the efficiency of the AH system is reduced with increasing chain length, as previously reported for other strains (5, 20, 21, 24, 28).
FIG. 2.
Growth on long-chain n-alkanes and degradation kinetics of Gordonia sp. strain SoCg. (a and b) Time courses of growth of wild-type Gordonia sp. strain SoCg (▪) and Gordonia sp. strain SoCg ΩalkB (□) on mineral BH medium supplemented with 10 mM n-hexadecane (a) and n-triacontane (b) as the sole carbon source; growth was measured as an increase in dry biomass in cultures over time. (c) Time courses of consumption of hexadecane (▴) and n-triacontane (•, determined by GC-MS and expressed as n-alkane residues with respect to abiotic controls. Standard errors were calculated from three independent determinations.
SoCg encodes an AlkB-type alkane hydroxylase system.
In order to isolate the alkane hydroxylase gene and its flanking region, a BamHI-BglII fragment was cloned into pUC18, giving the plasmid palk68 (Table 1). An internal fragment of palk68 was completely sequenced, and a 4,472-bp nucleotide sequence that exhibited overall identities of 79% with the AH-encoding system of Gordonia sp. strain TF6 (6) and 78% with those of Rhodococcus sp. strains Q15 and NRRL B-16531 (31) was obtained. Sequence analysis revealed six consecutive open reading frames (ORFs) (Table 3) which were designated as encoding Orf1 (a conserved hypothetical protein), AlkB (alkane 1-monooxygenase), RubA3 (rubredoxin), RubA4 (rubredoxin), RubB (rubredoxin reductase), and AlkU (putative TetR-like regulator) according to the sequence homology with other known genes (6, 31).
TABLE 3.
Genes identified and sequence similarities in the Gordonia sp. strain SoCg alk locus
| Gene | Product length (amino acids) | Best BLASTP alignment (accession no.) | Overlap (% identity) |
|---|---|---|---|
| orf1 | 124 | Conserved hypothetical protein, Rhodococcusequi ATCC 33707 (06829834.1) | 48/124 (38) |
| alkB | 411 | Alkane 1-monooxygenase, Gordonia sp. strain TF6 (BAD67020.1) | 287/323 (88) |
| rubA3 | 55 | Rubredoxin 3, Gordonia sp. strain TF6 (BAD67021.1) | 45/54 (83) |
| rubA4 | 61 | Rubredoxin 4, Gordonia sp. strain TF6 (BAD67022.1) | 34/59 (57) |
| rubB | 400 | Rubredoxin reductase, Gordonia sp. strain TF6 (BAD67023.1) | 249/349 (71) |
| alkU | 160 | Putative transcriptional regulator, TetR family, Mycobacteriumabscessus (YP 001704325.1) | 87/168 (51) |
Expression of SoCg alk genes is induced by long-chain n-alkanes.
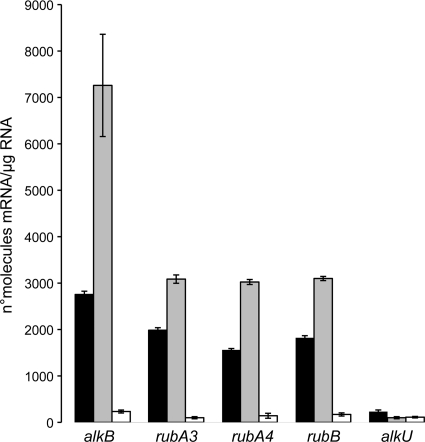
The expression of alk genes in the presence of long-chain and solid n-alkanes was analyzed by quantitative reverse transcription-PCR. Total RNA was extracted from Gordonia SoCg cultures after 22 h of growth at 30°C in mineral broth supplemented with n-hexadecane or n-triacontane as the sole carbon source. Total mRNA from fructose-grown cells was extracted and analyzed as a control. cDNA molecules were reverse transcribed from the total DNA-free RNA and used as templates to quantify alkB, rubA3, rubA4, rubB, and alkU transcripts. The amounts of alkB, rubA3, rubA4, and rubB transcripts were larger on n-alkanes than on fructose (Fig. 3) and larger on n-triacontane than on n-hexadecane. The results show that both long-chain n-alkanes induce the expression of all the alk genes except alkU. alkU has been found downstream of the alkB-rubA-rubB cluster in Gram-positive n-alkane degraders (7, 21, 31) and also in the genome of Nocardia farcinica (8); it possesses helix-turn-helix DNA-binding motifs and shows deduced amino acid similarity to putative regulatory proteins of the TetR family; however, its expression is not influenced by n-alkanes, and further investigations are needed to assess its involvement in n-alkane degradation.
FIG. 3.
Absolute real-time RT-PCR analysis of Gordonia SoCg alk genes. mRNA levels after 22 h of incubation in the presence of n-hexadecane (black bars), n-triacontane (gray bars), or fructose (white bars) are expressed as number of molecules μg−1 total RNA. Standard errors were calculated from three independent determinations of mRNA abundance in each sample.
The SoCg AH system is functional on liquid long-chain n-alkanes.
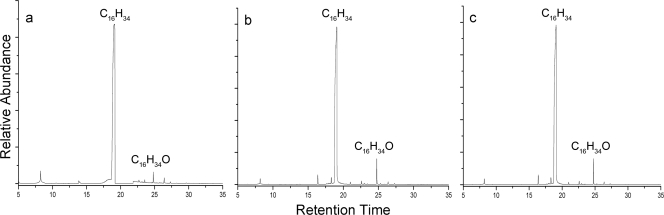
Metabolic intermediates were extracted by SPME from SoCg cells incubated with n-alkanes and identified by GC-MS. After 6 h of incubation in the presence of n-hexadecane as the sole C source, 1-hexadecanol was identified by comparing its Kovats index and the electron impact mass spectra with those obtained by the injection of the authentic standards (Fig. 4a). When SoCg was grown on n-triacontane, in contrast, we were unable to determine the corresponding primary long-chain alcohol. A longer incubation time or other extraction methods (i.e., hexane extraction) did not lead to triacontanol detection (data not shown). Almost all alkane hydroxylase activities described so far have been analyzed indirectly by mineralization of 14C labeled n-alkanes (31) and growth assays (21, 25). Only a few authors have reported the detection of metabolic intermediates of long-chain n-alkanes metabolism in a naturally occurring n-alkane degrader (32) or in a heterologous host (6) or the activity of the purified protein (5). Here, biotransformation activity on hexadecane to the corresponding alcohol by a strain of Gordonia is reported. These results clearly indicate that n-hexadecane is metabolized via the terminal oxidation pathway, like in other n-alkane degrading bacteria (6). As no long-chain alcohol could be detected on triacontane, it can be hypothesized that it is immediately used in the following reactions (10) or that it is undetectable because of its insolubility (23).
FIG. 4.
Hydroxylation of n-hexadecane by Gordonia SoCg (a), E. coli AH (b), and S. coelicolor M145 AH (c). GC chromatographs show conversion of hexadecane to 1-hexadecanol.
The Gordonia SoCg alkB is heterologously expressed in E. coli and S. coelicolor.
The unique alkB gene was heterologously expressed in E. coli BL21 using the expression vector pRSET-B. The expression of His-tagged AlkB in the resulting recombinant strain E. coli BL21-AH (Table 1) was confirmed by Western blotting using alkaline phosphatase-conjugated anti-His tag monoclonal antibodies. The protein was detected mainly in the insoluble fraction of a crude extract of E. coli BL21-AH after 4 h of induction with IPTG (data not shown). SPME/GC-MS analysis of bacterial cultures that were IPTG induced for 4 h revealed that E. coli BL21-AH was able to transform n-hexadecane into 1-hexadecanol (Fig. 4b). No hexadecanol or other products were detected using E. coli carrying pRSET-B. Although alkB-related alkane hydroxylase activity is known to be rubredoxin and NAD(P)H dependent (26), hexadecane hydroxylation was obtained in E. coli expressing only alkB. Fujii and colleagues defined alkB, rubA3, rubA4, and rubB as the minimum component genes of the alkane hydroxylase systems (6). In fact, those authors obtained biotransformation of n-octane to 1-octanol in E. coli TOP10 carrying plasmid pAL526, which contained the Gordonia TF6 alk cluster composed of the four genes. However, the relative AlkB activity was not completely eliminated in the absence of the other alkane hydroxylase system components. Similarly, two E. coli recombinants which expressed the Rhodococcus opacus B-4 alkB1 and alkB2 genes were able to convert n-alkanes (C5 to C16) to their corresponding alcohols in anhydrous organic solvents (20).
When n-triacontane was used as a substrate for E. coli BL21-AH, the corresponding primary long-chain alcohol could not be revealed using SPME/GC-MS analysis. As E. coli may not be the appropriate host, S. coelicolor M145 was used to express the Gordonia alkB gene. This strain does not grow on n-alkanes, but n-hexadecane-degrading Streptomyces species have recently been isolated (1). Using the integrative plasmid pIJ8600, the recombinant strain S. coelicolor M145-AH was obtained, in which the expression of alkB confers the ability to grow on n-hexadecane (data not shown). SPME/GC-MS analysis showed the presence of 1-hexadecanol in S. coelicolor M145-AH cultures on n-hexadecane (Fig. 4c). This is the first study to achieve biotransformation of n-hexadecane to 1-hexadecanol using S. coelicolor expressing an alkane hydroxylase gene. Wild-type S. coelicolor M145 and S. coelicolor carrying pIJ8600 were unable to biotransform n-hexadecane (data not shown). However, using S. coelicolor M145-AH, the corresponding primary long-chain alcohol n-triacontanol could not be detected.
S. coelicolor M145-AH expressing SoCg alkB grows on n-triacontane.
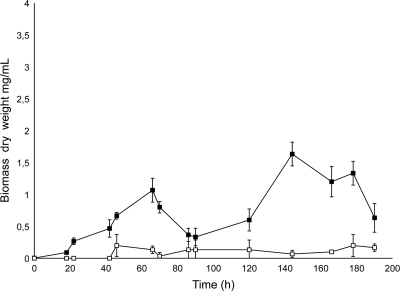
In order to analyze the activity of Gordonia AlkB on solid n-alkanes, a growth assay was set using S. coelicolor M145-AH in the presence of n-triacontane. E. coli was not used for growth assays because it lacks the metabolic pathway for alcohol metabolization. The growth curve of M145-AH on triacontane (Fig. 5) shows that Gordonia alkB confers the ability to grow on solid n-alkanes and also that this strain possesses in its own genome the genes involved in utilization of fatty alcohols. S. coelicolor M145 was thus revealed to be a good system to express the alkane hydroxylase genes from long-chain alkane degraders.
FIG. 5.
Growth curves of the recombinant strain Streptomyces coelicolor M145-AH (▪) and wild-type S. coelicolor M145 (□) in BH mineral medium supplemented with 10 mM n-triacontane as the sole C source. The solid n-alkane was added as finely ground powder, and growth was measured as increase in dry biomass in cultures over time. Standard errors were calculated from three independent determinations.
The SoCg alkB disruption mutant is unable to grow on n-triacontane.
To investigate the relevance of AlkB for degradation of long-chain n-alkanes by Gordonia SoCg, alkB was inactivated by introducing an apramycin resistance cassette in a unique AleI restriction site that is present at position 576 of its nucleotide sequence (Fig. 1a). The recombinant strains were selected on apramycin, and site-specific apramycin cassette insertion was first analyzed by PCR amplification of an internal fragment with primers apra750FR and apra750VR (Table 2) (data not shown). Finally, the PvuII-digested genomic DNAs of four positive clones were analyzed in a Southern hybridization experiment using the DIG-labeled alkCg341 probe (Fig. 1b). One strain showing the correct insertion of the cassette was named Gordonia SoCg ΩalkB and used for further experiments. Strain SoCg ΩalkB exhibited poor growth on n-hexadecane (7-fold lower than that of the wild-type strain) and, interestingly, no growth on triacontane, making it evident that alkB disruption had a negative effect on solid n-alkanes metabolic pathway. When the disruption mutant was incubated in the presence of 1-triacontanol as the sole C source it was able to grow even better than the wild type on triacontane (data not shown). On the other hand, Gordonia SoCg ΩalkB was still able to transform n-hexadecane into the corresponding alcohol (data not shown), suggesting that oxidation of n-hexadecane in the absence of alkB must be carried out by an unknown oxidation system that in any case does not allow efficient growth of the strain.
DISCUSSION
Many bacteria capable of degrading long-chain alkanes have been isolated, and the enzyme systems that oxidize long-chain n-alkanes up to C16 have been characterized (see references 18, 26, and 30 for reviews). Although long-chain alkanes are more persistent in the environment than shorter-chain alkanes, genes involved in degradation of n-alkanes longer than C16 had not been reported prior to the work of Throne-Holst et al. (25) and Feng et al. (5). A flavin-binding monooxygenase involved in oxidation of very-long-chain n-alkanes up to C32 has been characterized in Acinetobacter sp. strain DSM17874 (25), and LadA from Geobacillus thermodenitrificans NG80-2 is the first long-chain n-alkane monooxygenase functional on alkanes in the range from C15 to C36 to be cloned and structurally characterized from a Gram-positive strain (5). Both enzymes show little or no homology with the widespread and well-characterized AlkB-type alkane hydroxylases usually reported as being functional on long-chain n-alkanes up to C16 in Gram-positive and Gram-negative strains (21, 30, 31).
Here the unique functional AlkB-type alkane hydroxylase system that allows growth on long-chain liquid and solid n-alkanes in the Gram-positive Gordonia strain SoCg is described. To date the only long-chain alkane hydroxylase system of this genus that has been characterized is that of Gordonia TF6, which was found to be functional on n-alkanes from C5 to C13 (6).
The ability of Gordonia SoCg AlkB to biotransform n-hexadecane into the corresponding primary alcohol was assessed by SPME/GC-MS analysis in SoCg and in two heterologous hosts expressing the SoCg alkB gene. S. coelicolor M145 was successfully used as a heterologous host for an alkane hydroxylase gene. Although the n-triacontane biotransformation product, triacontanol, could not be detected in any of the heterologous systems, the role of SoCg AlkB in triacontane metabolization was demonstrated by growth assays. S. coelicolor M145-AH expressing the Gordonia alkB gene acquired the ability to grow on triacontane, while the disruption mutant SoCg ΩalkB lost this ability. Moreover, the SoCg alkane hydroxylase-encoding genes are induced by both liquid and solid n-alkanes, which is in accordance with the ability of this strain to grow on and rapidly metabolize n-alkanes up to C36 (16).
Taken together, these results suggest that the identified alkane oxidation system plays a central role in the degradation of long-chain and solid n-alkanes by Gordonia SoCg. Moreover, at least one other, less efficient enzyme that is responsible for oxidation of n-hexadecane exists. This second AH system seems to be unrelated to other known alkane hydroxylase systems characterized so far.
Many bacterial strains contain multiple, and quite divergent, integral membrane AlkBs (31) that have different substrate ranges (23, 24) or are activated during different growth phases (13). The strategy of Gordonia SoCg seems to be based on a single alkB gene, which is induced by a wide range of long and solid n-alkanes throughout the time course of growth (L. Lo Piccolo, unpublished results), encoding an enzyme with highest activity on hexadecane and reduced activity on triacontane. Growth of SoCg on triacontane would be poorer that that on hexadecane for this reason and also because a second, unknown system, that is functional on C16 but not on C30, would contribute to overcome the limiting step of n-alkane degradation on C16. The alkane hydroxylase, in fact, catalyzes the initial attack and determines the size range of n-alkanes to be degraded; its specific activity is generally reduced with increasing chain length (5, 20, 21, 24, 28).
The relationship between the AlkB protein structure and its function has been investigated; it has been proposed that AlkB is made of six transmembrane helices that are assembled in a hexagonal structure forming a deep hydrophobic pocket where four conserved histidine residues that chelate the iron atoms necessary for its activity are located on the cytoplasm surface (28). The alkane molecule should slide into the pocket until the terminal methyl group is correctly positioned relative to the His residues. Amino acids with bulky side chains protruding into the pocket would limit the size of the n-alkane to be hydroxylated, while less bulky side chain amino acids allow longer alkanes to deeper enter into the hydrophobic pocket (28). Pseudomonas putida GPo1 and Alkanivorax borkumensis AP1 AlkB mutant derivatives oxidize alkanes longer than C12 when tryptophan is replaced by serine, cysteine, or other small amino acids at position 55 or 58 of the two proteins (28). Amino acid sequence alignment of AlkB proteins showed a valine residue in the corresponding amino acid position of Gordonia SoCg AlkB, confirming the possibility of accepting long-chain alkanes in the active site, although other residues/mechanisms could be involved in n-alkane recognition.
Bacteria appear to degrade chemicals only when they are dissolved in water, and dissolution of solid substrates is generally considered a prerequisite for their biodegradation (33). Long-chain and solid n-alkanes are insoluble in water and, although we know how n-alkanes are oxidized, we still poorly know how they are recognized and how they enter the cells, especially when they are in the solid state. Two mechanisms for accessing medium and long-chain liquid alkanes have been recognized in bacteria: (i) biosurfactant-mediated accession by cell contact with emulsified hydrocarbons and (ii) interfacial accession by direct contact of the cell surface with the hydrocarbon (3). Gordonia belongs to the Corynebacterium/Mycobacterium/Nocardia (CMN) complex, which is characterized by mycolic acid-containing cell walls that confer hydrophobicity to these bacteria and allow cell adherence to the n-alkanes by direct contact of cells with hydrocarbons, generally with no or low biosurfactant production. Our observations confirm that the strategy of SoCg for accessing liquid hydrocarbons is by direct contact and that this strategy is also used for solid alkanes. In fact, a massive adhesion of SoCg cells to triacontane (supplemented as finely ground powder) was observed, while the culture liquid phase was almost clear for a long period of growth. Direct contact with the solid substrate might favor growth of Gordonia, as it can have direct access to the substrate without its previous solubilization in the aqueous environment.
The recent first report of expression of Rhodococcus alkB genes in anhydrous organic solvents corroborates these observations (20) and suggests new biotechnological applications in water-free environments.
The alkane hydroxylase from Gordonia SoCg is active on a wide range of long-chain liquid and solid n-alkanes and is able to use other electron transfer systems in the absence of its two specific components, rubredoxin and rubredoxin reductase. Gordonia sp. strain SoCg is the first actinobacterial strain that is able to grow on solid n-alkanes to be characterized.
Acknowledgments
We thank Stefano Colazza for scientific collaboration, Valentina Catania and Valentina Imparato for cloning experiments, and Sandra Marineo for valuable technical suggestions.
This study was partly supported by the Italian Ministry of Education, University and Research (fondi MIUR ex 60%, 2007).
Footnotes
Published ahead of print on 23 December 2010.
REFERENCES
- 1.Barabás, G., et al. 2001. n-Alkane uptake and utilisation by Streptomyces strains. Antonie Van Leeuwenhoek 79:269-276. [DOI] [PubMed] [Google Scholar]
- 2.Birren, B., and E. Lai. 1993. Pulsed field gel electrophoresis. A practical guide. Academic Press, Inc., San Diego, CA.
- 3.Bouchez Naïtali, M., H. Rakatozafy, R. Marchal, J. Y. Leveau, and J. P. Vandecasteele. 1999. Diversity of bacterial strains degrading hexadecane in relation to the mode of substrate uptake. J. Appl. Microbiol. 86:421-428. [DOI] [PubMed] [Google Scholar]
- 4.De Pasquale, C., S. Guarino, E. Peri, G. Alonzo, and S. Colazza. 2007. Investigation of cuticular hydrocarbons from Bagrada hilaris genders by SPME/GC-MS. Anal. Bioanal. Chem. 389:1259-1265. [DOI] [PubMed] [Google Scholar]
- 5.Feng, L., et al. 2007. Genome and proteome of long-chain alkane degrading Geobacillus thermodenitrificans NG80-2 isolated from a deep-subsurface oil reservoir. Proc. Natl. Acad. Sci. U. S. A. 104:5602-5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujii, T., T. Narikawa, K. Takeda, and J. Kato. 2004. Biotransformation of various alkanes using the Escherichia coli expressing an alkane hydroxylase system from Gordonia sp. TF6. Biosci. Biotechnol. Biochem. 68:2171-2177. [DOI] [PubMed] [Google Scholar]
- 7.Hamamura, N., C. M. Yeager, and D. J. Arp. 2001. Two distinct monooxygenases for alkane oxidation in Nocardioides sp. strain CF8. Appl. Environ. Microbiol. 67:4992-4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishikawa, J., et al. 2004. The complete genomic sequence of Nocardia farcinica IFM 10152. Proc. Natl. Acad. Sci. U. S. A. 101:14925-14930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 10.Koma, D., et al. 2001. Biodegradation of long-chain n-paraffins from waste oil of car engine by Acinetobacter sp. J. Biosci. Bioeng. 91:94-96. [DOI] [PubMed] [Google Scholar]
- 11.Kubota, M., et al. 2005. Isolation and functional analysis of cytochrome P450 CYP153A genes from various environments. Biosci. Biotechnol. Biochem. 69:2421-2430. [DOI] [PubMed] [Google Scholar]
- 12.Larkin, M. J., L. A. Kulakov, and C. C. Allen. 2005. Biodegradation and Rhodococcus—masters of catabolic versatility. Curr. Opin. Biotechnol. 16:282-290. [DOI] [PubMed] [Google Scholar]
- 13.Marin, M., L. Yuste, and F. Rojo. 2003. Differential expression of the components of the two alkane hydroxylases from Pseudomonas aeruginosa. J. Bacteriol. 185:3232-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puglia, A. M., J. Vohradsky, and C. J. Thompson. 1995. Developmental control of the heat-shock stress regulon in Streptomyces coelicolor. Mol. Microbiol. 17:737-746. [DOI] [PubMed] [Google Scholar]
- 15.Purswani, J., C. Pozo, M. Rodríguez-Díaz, and J. González-López. 2008. Selection and identification of bacterial strains with methyl-tert-butyl ether, ethyl-tert-butyl ether, and tert-amyl methyl ether degrading capacities. Environ. Toxicol. Chem. 27:2296-2303. [DOI] [PubMed] [Google Scholar]
- 16.Quatrini, P., G. Scaglione, C. De Pasquale, S. Riela, and A. M. Puglia. 2008. Isolation of Gram-positive n-alkane degraders from a hydrocarbon-contaminated Mediterranean shoreline. J. Appl. Microbiol. 104:251-259. [DOI] [PubMed] [Google Scholar]
- 17.Ratajczak, A., W. Geissdörfer, and W. Hillen. 1998. Expression of alkane hydroxylase from Acinetobacter sp. strain ADP1 is induced by a broad range of n-alkanes and requires the transcriptional activator AlkR. J. Bacteriol. 180:5822-5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rojo, F. 2009. Degradation of alkanes by bacteria. Environ. Microbiol. 11:2477-2490. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook, J., E. F. Fritsch, and J. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 20.Sameshima, Y., K. Honda, J. Kato, T. Omasa, and H. Ohtake. 2008. Expression of Rhodococcus opacus alkB genes in anhydrous organic solvents. J. Biosci. Bioeng. 106:199-203. [DOI] [PubMed] [Google Scholar]
- 21.Smits, T. H., S. B. Balada, B. Witholt, and J. B. van Beilen. 2002. Functional analysis of alkane hydroxylases from gram-negative and gram-positive bacteria. J. Bacteriol. 184:1733-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smits, T. H., M. Röthlisberger, B. Witholt, and J. B. van Beilen. 1999. Molecular screening for alkane hydroxylase genes in Gram-negative and Gram-positive strains. Environ. Microbiol. 1:307-317. [DOI] [PubMed] [Google Scholar]
- 23.Tani, A., T. Ishige, Y. Sakai, and N. Kato. 2001. Gene structures and regulation of the alkane hydroxylase complex in Acinetobacter sp. strain M-1. J. Bacteriol. 183:1819-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Throne-Holst, M., et al. 2006. Utilization of n-alkanes by a newly isolated strain of Acinetobacter venetianus: the role of two AlkB-type alkane hydroxylases. Appl. Microbiol. Biotechnol. 72:353-360. [DOI] [PubMed] [Google Scholar]
- 25.Throne-Holst, M., A. Wentzel, T. E. Ellingsen, H. K. Kotlar, and S. B. Zotchev. 2007. Identification of novel genes involved in long-chain n-alkane degradation by Acinetobacter sp. strain DSM 17874. Appl. Environ. Microbiol. 73:3327-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Beilen, J. B., and E. G. Funhoff. 2007. Alkane hydroxylases involved in microbial alkane degradation. Appl. Microbiol. Biotechnol. 74:13-21. [DOI] [PubMed] [Google Scholar]
- 27.van Beilen, J. B., et al. 2001. Analysis of Pseudomonas putida alkane-degradation gene clusters and flanking insertion sequences: evolution and regulation of the alk genes. Microbiology. 147:1621-1630. [DOI] [PubMed] [Google Scholar]
- 28.van Beilen, J. B., et al. 2005. Identification of an amino acid position that determines the substrate range of integral membrane alkane hydroxylases. J. Bacteriol. 187:85-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weisburg, W., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wentzel, A., T. E. Ellingsen, H. K. Kotlar, S. B. Zotchev, and M. Throne-Holst. 2007. Bacterial metabolism of long-chain n-alkanes. Appl. Microbiol. Biotechnol. 76:1209-1221. [DOI] [PubMed] [Google Scholar]
- 31.Whyte, L. G., et al. 2002. Gene cloning and characterization of multiple alkane hydroxylase systems in Rhodococcus strains Q15 and NRRL B-16531. Appl. Environ. Microbiol. 68:5933-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whyte, L. G., et al. 1998. Biodegradation of variable-chain-length alkanes at low temperatures by a psychrotrophic Rhodococcus sp. Appl. Environ. Microbiol. 64:2578-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wick, L. Y., T. Colangelo, and H. Harms. 2001. Kinetics of mass transfer-limited bacterial growth on solid PAHs. Environ. Sci. Technol. 15:354-361. [DOI] [PubMed] [Google Scholar]