Abstract
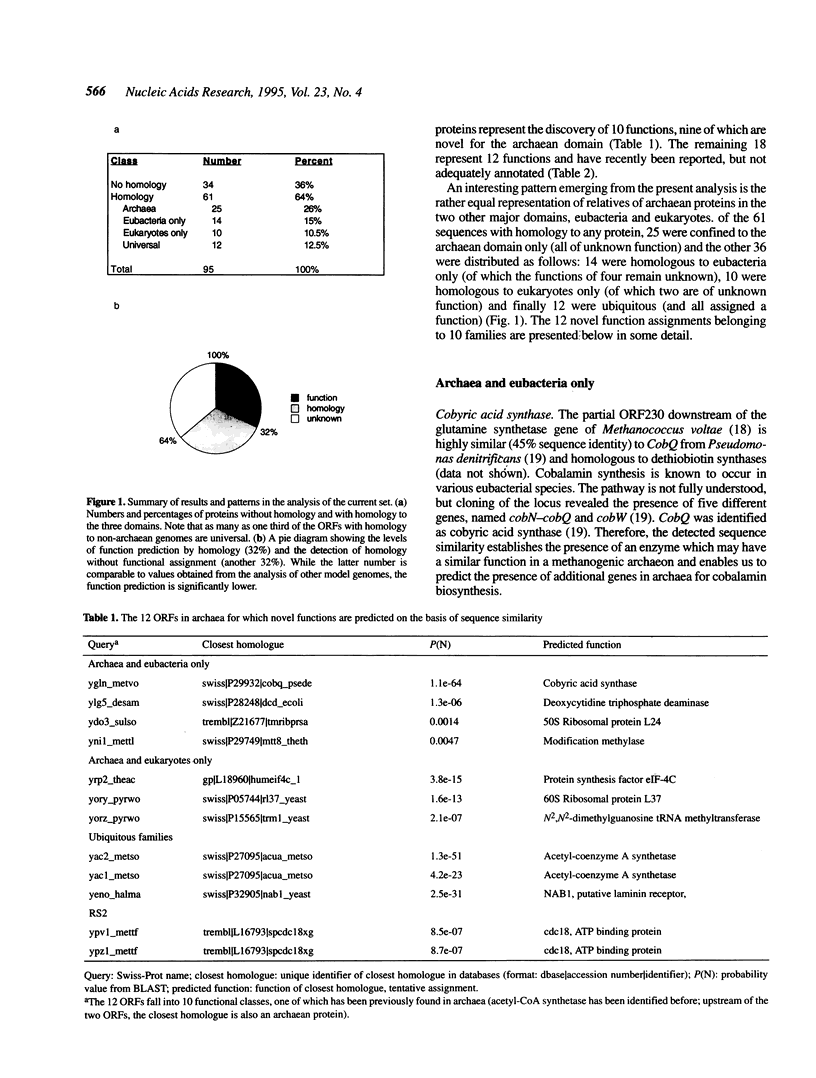
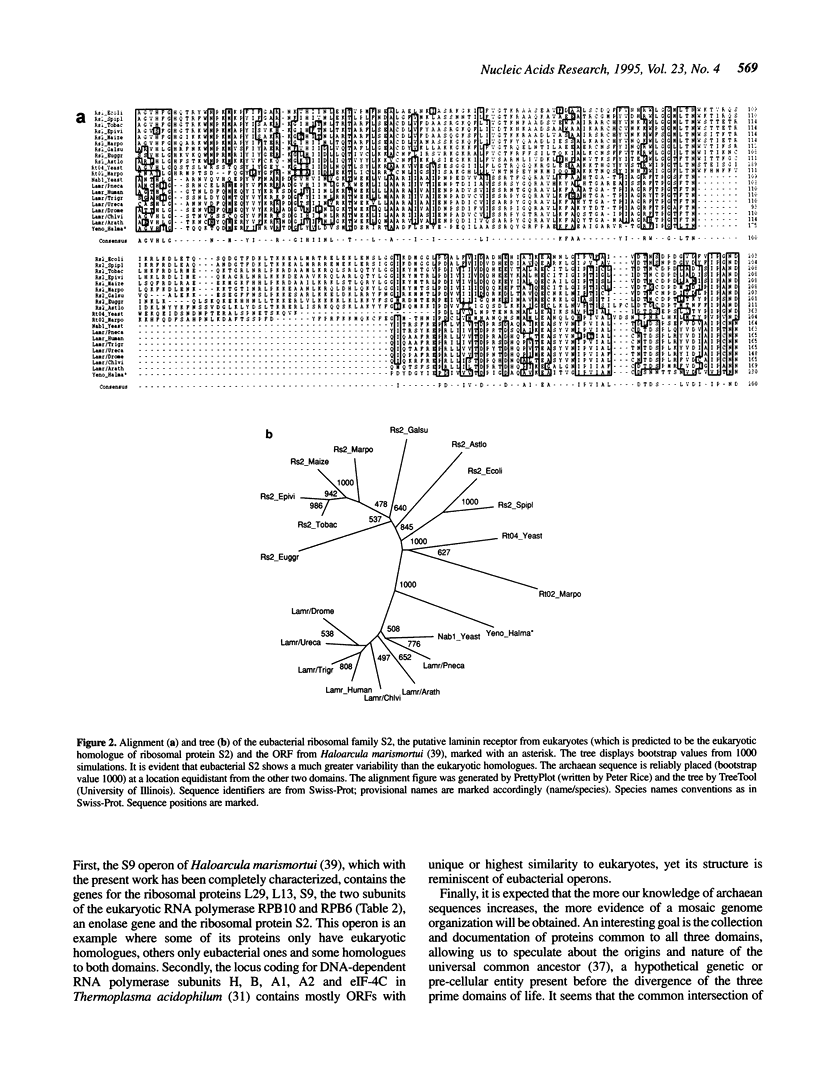
In a quest for novel functions in archaea, all archaean hypothetical open reading frames (ORFs), as annotated in the Swiss-Prot protein sequence database, were used to search the latest databases for the identification of characterized homologues. Of the 95 hypothetical archaean ORFs, 25 were found to be homologous to another hypothetical archaean ORF, while 36 were homologous to non-archaean proteins, of which as many as 30 were homologous to a characterized protein family. Thus the level of sequence similarity in this set reaches 64%, while the level of function assignment is only 32%. Of the ORFs with predicted functions, 12 homologies are reported here for the first time and represent nine new functions and one gene duplication at an acetyl-coA synthetase locus. The novel functions include components of the transcriptional and translational apparatus, such as ribosomal proteins, modification enzymes and a translation initiation factor. In addition, new enzymes are identified in archaea, such as cobyric acid synthase, dCTP deaminase and the first archaean homologues of a new subclass of ATP binding proteins found in fungi. Finally, it is shown that the putative laminin receptor family of eukaryotes and an archaean homologue belong to the previously characterized ribosomal protein family S2 from eubacteria. From the present and previous work, the major implication is that archaea seem to have a mode of expression of genetic information rather similar to eukaryotes, while eubacteria may have proceeded into unique ways of transcription and translation. In addition, with the detection of proteins in various metabolic and genetic processes in archaea, we can further predict the presence of additional proteins involved in these processes.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- An G., Bendiak D. S., Mamelak L. A., Friesen J. D. Organization and nucleotide sequence of a new ribosomal operon in Escherichia coli containing the genes for ribosomal protein S2 and elongation factor Ts. Nucleic Acids Res. 1981 Aug 25;9(16):4163–4172. doi: 10.1093/nar/9.16.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A., Boeckmann B. The SWISS-PROT protein sequence data bank. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):2247–2249. doi: 10.1093/nar/19.suppl.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A. PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res. 1992 May 11;20 (Suppl):2013–2018. doi: 10.1093/nar/20.suppl.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barany F., Danzitz M., Zebala J., Mayer A. Cloning and sequencing of genes encoding the TthHB8I restriction and modification enzymes: comparison with the isoschizomeric TaqI enzymes. Gene. 1992 Mar 1;112(1):3–12. doi: 10.1016/0378-1119(92)90296-2. [DOI] [PubMed] [Google Scholar]
- Bazan J. F., Weaver L. H., Roderick S. L., Huber R., Matthews B. W. Sequence and structure comparison suggest that methionine aminopeptidase, prolidase, aminopeptidase P, and creatinase share a common fold. Proc Natl Acad Sci U S A. 1994 Mar 29;91(7):2473–2477. doi: 10.1073/pnas.91.7.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P., Ouzounis C., Sander C., Scharf M., Schneider R., Sonnhammer E. Comprehensive sequence analysis of the 182 predicted open reading frames of yeast chromosome III. Protein Sci. 1992 Dec;1(12):1677–1690. doi: 10.1002/pro.5560011216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno A., Russell P. Dual functions of CDC6: a yeast protein required for DNA replication also inhibits nuclear division. EMBO J. 1992 Jun;11(6):2167–2176. doi: 10.1002/j.1460-2075.1992.tb05276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouzet J., Levy-Schil S., Cameron B., Cauchois L., Rigault S., Rouyez M. C., Blanche F., Debussche L., Thibaut D. Nucleotide sequence and genetic analysis of a 13.1-kilobase-pair Pseudomonas denitrificans DNA fragment containing five cob genes and identification of structural genes encoding Cob(I)alamin adenosyltransferase, cobyric acid synthase, and bifunctional cobinamide kinase-cobinamide phosphate guanylyltransferase. J Bacteriol. 1991 Oct;173(19):6074–6087. doi: 10.1128/jb.173.19.6074-6087.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S. C., Tzagoloff A., Ellis S. R. Characterization of a yeast mitochondrial ribosomal protein structurally related to the mammalian 68-kDa high affinity laminin receptor. J Biol Chem. 1992 Mar 15;267(8):5508–5514. [PubMed] [Google Scholar]
- Dever T. E., Wei C. L., Benkowski L. A., Browning K., Merrick W. C., Hershey J. W. Determination of the amino acid sequence of rabbit, human, and wheat germ protein synthesis factor eIF-4C by cloning and chemical sequencing. J Biol Chem. 1994 Feb 4;269(5):3212–3218. [PubMed] [Google Scholar]
- Eggen R. I., Geerling A. C., Boshoven A. B., de Vos W. M. Cloning, sequence analysis, and functional expression of the acetyl coenzyme A synthetase gene from Methanothrix soehngenii in Escherichia coli. J Bacteriol. 1991 Oct;173(20):6383–6389. doi: 10.1128/jb.173.20.6383-6389.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis S. R., Hopper A. K., Martin N. C. Amino-terminal extension generated from an upstream AUG codon is not required for mitochondrial import of yeast N2,N2-dimethylguanosine-specific tRNA methyltransferase. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5172–5176. doi: 10.1073/pnas.84.15.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzold T., Argos P. SRS--an indexing and retrieval tool for flat file data libraries. Comput Appl Biosci. 1993 Feb;9(1):49–57. doi: 10.1093/bioinformatics/9.1.49. [DOI] [PubMed] [Google Scholar]
- Gribskov M., Lüthy R., Eisenberg D. Profile analysis. Methods Enzymol. 1990;183:146–159. doi: 10.1016/0076-6879(90)83011-w. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Bleasby A. J., Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992 Apr;8(2):189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- Hodgman T. C. The elucidation of protein function by sequence motif analysis. Comput Appl Biosci. 1989 Feb;5(1):1–13. doi: 10.1093/bioinformatics/5.1.1. [DOI] [PubMed] [Google Scholar]
- Huet J., Schnabel R., Sentenac A., Zillig W. Archaebacteria and eukaryotes possess DNA-dependent RNA polymerases of a common type. EMBO J. 1983;2(8):1291–1294. doi: 10.1002/j.1460-2075.1983.tb01583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaine B. P., Mehr I. J., Woese C. R. The sequence, and its evolutionary implications, of a Thermococcus celer protein associated with transcription. Proc Natl Acad Sci U S A. 1994 Apr 26;91(9):3854–3856. doi: 10.1073/pnas.91.9.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T. J., Martin G. S., Forsburg S. L., Stephen R. J., Russo A., Nurse P. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell. 1993 Jul 30;74(2):371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- Klenk H. P., Doolittle W. F. Evolution. Archaea and eukaryotes versus bacteria? Curr Biol. 1994 Oct 1;4(10):920–922. doi: 10.1016/s0960-9822(00)00206-2. [DOI] [PubMed] [Google Scholar]
- Klenk H. P., Renner O., Schwass V., Zillig W. Nucleotide sequence of the genes encoding the subunits H, B, A' and A'' of the DNA-dependent RNA polymerase and the initiator tRNA from Thermoplasma acidophilum. Nucleic Acids Res. 1992 Oct 11;20(19):5226–5226. doi: 10.1093/nar/20.19.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kletzin A. Molecular characterisation of a DNA ligase gene of the extremely thermophilic archaeon Desulfurolobus ambivalens shows close phylogenetic relationship to eukaryotic ligases. Nucleic Acids Res. 1992 Oct 25;20(20):5389–5396. doi: 10.1093/nar/20.20.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong H., Morgan R. D., Maunus R. E., Schildkraut I. A unique restriction endonuclease, BcgI, from Bacillus coagulans. Nucleic Acids Res. 1993 Feb 25;21(4):987–991. doi: 10.1093/nar/21.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krömer W. J., Arndt E. Halobacterial S9 operon. Three ribosomal protein genes are cotranscribed with genes encoding a tRNA(Leu), the enolase, and a putative membrane protein in the archaebacterium Haloarcula (Halobacterium) marismortui. J Biol Chem. 1991 Dec 25;266(36):24573–24579. [PubMed] [Google Scholar]
- Langer D., Zillig W. Putative tfIIs gene of Sulfolobus acidocaldarius encoding an archaeal transcription elongation factor is situated directly downstream of the gene for a small subunit of DNA-dependent RNA polymerase. Nucleic Acids Res. 1993 May 11;21(9):2251–2251. doi: 10.1093/nar/21.9.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh T. L., Reich C. I., Whitelock R. B., Olsen G. J. Transcription factor IID in the Archaea: sequences in the Thermococcus celer genome would encode a product closely related to the TATA-binding protein of eukaryotes. Proc Natl Acad Sci U S A. 1994 May 10;91(10):4180–4184. doi: 10.1073/pnas.91.10.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nölling J., van Eeden F. J., Eggen R. I., de Vos W. M. Modular organization of related Archaeal plasmids encoding different restriction-modification systems in Methanobacterium thermoformicicum. Nucleic Acids Res. 1992 Dec 25;20(24):6501–6507. doi: 10.1093/nar/20.24.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohama T., Muto A., Osawa S. Spectinomycin operon of Micrococcus luteus: evolutionary implications of organization and novel codon usage. J Mol Evol. 1989 Nov;29(5):381–395. doi: 10.1007/BF02602908. [DOI] [PubMed] [Google Scholar]
- Ouzounis C. A., Blencowe B. J. Bacterial DNA replication initiation factor priA is related to proteins belonging to the 'DEAD-box' family. Nucleic Acids Res. 1991 Dec 25;19(24):6953–6953. doi: 10.1093/nar/19.24.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouzounis C. A., Kyrpides N. C. On the evolution of arginases and related enzymes. J Mol Evol. 1994 Jul;39(1):101–104. doi: 10.1007/BF00178255. [DOI] [PubMed] [Google Scholar]
- Ouzounis C., Sander C. TFIIB, an evolutionary link between the transcription machineries of archaebacteria and eukaryotes. Cell. 1992 Oct 16;71(2):189–190. doi: 10.1016/0092-8674(92)90347-f. [DOI] [PubMed] [Google Scholar]
- Possot O., Sibold L., Aubert J. P. Nucleotide sequence and expression of the glutamine synthetase structural gene, glnA, of the archaebacterium Methanococcus voltae. Res Microbiol. 1989 Jul-Aug;140(6):355–371. doi: 10.1016/0923-2508(89)90012-0. [DOI] [PubMed] [Google Scholar]
- Ramírez C., Matheson A. T. A gene in the archaebacterium Sulfolobus solfataricus that codes for a protein equivalent to the alpha subunits of the signal recognition particle receptor in eukaryotes. Mol Microbiol. 1991 Jul;5(7):1687–1693. doi: 10.1111/j.1365-2958.1991.tb01916.x. [DOI] [PubMed] [Google Scholar]
- Rowlands T., Baumann P., Jackson S. P. The TATA-binding protein: a general transcription factor in eukaryotes and archaebacteria. Science. 1994 May 27;264(5163):1326–1329. doi: 10.1126/science.8191287. [DOI] [PubMed] [Google Scholar]
- Sander C., Schneider R. Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins. 1991;9(1):56–68. doi: 10.1002/prot.340090107. [DOI] [PubMed] [Google Scholar]
- Santangelo G. M., Tornow J., McLaughlin C. S., Moldave K. Screening a yeast promoter library leads to the isolation of the RP29/L32 and SNR17B/RPL37A divergent promoters and the discovery of a gene encoding ribosomal protein L37. Gene. 1991 Aug 30;105(1):137–138. doi: 10.1016/0378-1119(91)90526-h. [DOI] [PubMed] [Google Scholar]
- Souillard N., Magot M., Possot O., Sibold L. Nucleotide sequence of regions homologous to nifH (nitrogenase Fe protein) from the nitrogen-fixing archaebacteria Methanococcus thermolithotrophicus and Methanobacterium ivanovii: evolutionary implications. J Mol Evol. 1988;27(1):65–76. doi: 10.1007/BF02099731. [DOI] [PubMed] [Google Scholar]
- Tohgo A., Takasawa S., Munakata H., Yonekura H., Hayashi N., Okamoto H. Structural determination and characterization of a 40 kDa protein isolated from rat 40 S ribosomal subunit. FEBS Lett. 1994 Feb 28;340(1-2):133–138. doi: 10.1016/0014-5793(94)80188-6. [DOI] [PubMed] [Google Scholar]
- Wang L., Weiss B. dcd (dCTP deaminase) gene of Escherichia coli: mapping, cloning, sequencing, and identification as a locus of suppressors of lethal dut (dUTPase) mutations. J Bacteriol. 1992 Sep;174(17):5647–5653. doi: 10.1128/jb.174.17.5647-5653.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Kandler O., Wheelis M. L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl P., Fabry S., Bogedain C., Haas A., Hensel R. Glyceraldehyde-3-phosphate dehydrogenase from the hyperthermophilic archaebacterium Pyrococcus woesei: characterization of the enzyme, cloning and sequencing of the gene, and expression in Escherichia coli. J Bacteriol. 1990 Aug;172(8):4329–4338. doi: 10.1128/jb.172.8.4329-4338.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]


