Abstract
Nitrate respiration is a common and strain-specific property in Thermus thermophilus encoded by the nitrate respiration conjugative element (NCE) that can be laterally transferred by conjugation. In contrast, nitrite respiration and further denitrification steps are restricted to a few isolates of this species. These later steps of the denitrification pathway are under the regulatory control of an NCE-encoded transcription factor, but nothing is known about their coding sequences or its putative genetic linkage to the NCE. In this study we examine the genetic linkage between nitrate and nitrite respiration through lateral gene transfer (LGT) assays and describe a cluster of genes encoding the nitrite-nitric oxide respiration in T. thermophilus PRQ25. We show that the whole denitrification pathway can be transferred from the denitrificant strain PRQ25 to an aerobic strain, HB27, and that the genes coding for nitrite and nitric oxide respiration are encoded near the NCE. Sequence data from the draft genome of PRQ25 confirmed these results and allowed us to describe the most compact nor-nir cluster known thus far and to demonstrate the expression and activities of the encoded enzymes in the HB27 denitrificant derivatives obtained by LGT. We conclude that this NCE nor-nir supercluster constitutes a whole denitrification island that can be spread by lateral transfer among Thermus thermophilus strains.
Several prokaryotes can respire nitrogen oxides by using one or more reduction steps of the denitrification pathway (NO3− > NO2− > NO > N2O > N2) (31). In the denitrificant mesophilic bacterial models studied thus far, this process starts with a heterotrimeric membrane-bound nitrate reductase (Nar). Nitrite produced in the cytoplasm is secreted and reduced to nitric oxide either by heme (NirS)- or by copper (NirK)-containing periplasmic nitrite reductases. The highly toxic product NO is immediately reduced to N2O by a membrane-bound reductase depending on either cytochrome c (cNor) or quinones (qNor) as electron donors. In many but not all of the denitrificant microorganisms, N2O is finally reduced to N2 by a periplasmic reductase (NosZ). However, N2 is also produced in organisms that encode no NosZ homologues, supporting that a different kind of N2O reductase might exist (33). The nir and nor genes are frequently clustered in the genomes of denitrificant strains and expressed in a coordinated form to avoid the separate and potentially toxic accumulation of NO, whereas nar and nos genes usually appear at separate loci in the genome (32).
Denitrification is widespread among the prokaryotic phylogeny, including the genomes of ancient bacterial and archaeal lineages such as extreme thermophiles and hyperthermophiles. This and the discrepancies existent between the phylogeny of denitrification genes and that of the 16S RNA has been used to suggest an ancient origin for this pathway and its spread through lateral gene transfer (LGT) (15, 16, 22). However, studies on the denitrification pathway and putative LGT in ancient bacterial or archaeal models are limited because of the difficulties associated with the laboratory handling of such microorganisms. The exception to this rule is Thermus thermophilus, an extreme thermophilic bacterium for which a complete genetic toolbox exists (7). The type strain and other isolates of this species are obligate aerobes, but many other strains have been isolated that can grow as facultative anaerobes through partial or, apparently, complete denitrification (9). Partial denitrificant strains such as NAR1 reduce nitrate with a final production of nitrite that accumulates in the growth medium. Nitrate reduction is carried out by a special kind of nitrate reductase (Nar) that contains a periplasmic cytochrome c in addition to the three subunits found in other nitrate reductases (30). Nar is encoded within the NCE (for nitrate conjugative element), a genetic element that can be transferred by conjugation to aerobic strains of the same species, allowing the receptor to grow anaerobically with nitrate (24). The NCE also encodes a nitrate-dependent NADH dehydrogenase, two nitrate/nitrite transporters, and genes required for their transcriptional control by oxygen and nitrate (reviewed in reference 6). In addition to the NCE, denitrificant strains such as PRQ25 are able to grow by reducing nitrite with the production of gas. However, the genetic analysis of the whole denitrification equipment of the PRQ25 strain has been hampered by the low efficiency of its natural competence system, which shows 1,000-fold-reduced plasmid transformation efficiency compared to the aerobic HB27 strain (unpublished data).
To analyze a putative genetic linkage between nitrate and nitrite respiration in T. thermophilus and concomitantly to gain genetic access to the denitrification process itself, we demonstrate the LGT of both pathways to the aerobic HB27 strain. We show that the NCE, coding for the nitrate respiration, and a compact gene cluster coding for the Nir and Nor reductases are encoded together within a variable region of a megaplasmid of T. thermophilus PRQ25 forming a denitrification supercluster. The compactness of this supercluster allows the spread by LGT of the pathway as a single unit to aerobic strains of T. thermophilus.
MATERIALS AND METHODS
Strains and growth conditions.
T. thermophilus HB27 and its derivative HB27gdh::kat grow aerobically, whereas T. thermophilus PRQ25 is a facultative strain that can grow by denitrification (9, 19). T. thermophilus strains were grown aerobically in TB liquid medium (23) at 70°C with mild shaking (150 rpm). Anaerobic growth was achieved in screw-cap tubes containing 10 ml of TB medium supplemented with potassium nitrate (40 mM) or sodium nitrite (5 to 10 mM) overlaid by mineral oil. For gas detection, the strains were grown in TB with nitrate 10 mM in Hungate tubes, leaving a headspace for the collection of gas samples with a syringe. T. thermophilus colonies were grown aerobically on TB agar (1.5% [wt/vol] agar) plates. Kanamycin (30 mg/liter) was added to liquid or solid medium for selection when required. Nitrate reduction and nitrite consumption assays were carried out in 96-multiwell plates containing 250 μl of TB with either 40 mM potassium nitrate or 5 mM sodium nitrite. The plates were covered with an aluminum adhesive plate (Greiner Bio-One, catalog no. 676090), followed by incubation at 70°C for 16 h for nitrate reduction or for 48 h for nitrite consumption.
Escherichia coli DH5α (supE44 ΔlacU169 [φ80lacZΔM15] hsdR17 recA endA1 gyrA96 thi-1 relA1] was used for the construction of plasmids. E. coli BL21(DE3) [hsdS gal(λcIts857 ind-1 Sam7 nin-5 lacUV5-T7 gene 1)] and Rosetta-gami 2 [BL21(DE3) (lonA ompT trxB/gor pRARE); Novagen] were used for expression from T7-dependent promoters. E. coli strains were grown at 37°C on liquid or solid LB medium. Kanamycin (30 mg/liter), ampicillin (100 mg/liter), and chloramphenicol (30 mg/liter) were used when needed.
Transformation and conjugation experiments.
Total DNA from T. thermophilus PRQ25 was isolated as described previously (20). For transformation experiments, total DNA (200 ng) from the donor strain was added to 0.8-ml exponential cultures (optical density at 550 nm = 0.2) of HB27 or HB27gdh::kat aerobic strains, allowing incubation at 70°C to continue for 4 h with standard shaking. The cells were then diluted to 10 ml with preheated TB and incubated under anaerobic conditions with nitrate or nitrite for the selection of facultative anaerobes. For conjugation, cultures of the acceptor (HB27gdh::kat) and donor (PRQ25) strains grown aerobically for 16 h in TB at 70°C to stationary phase were centrifuged (5,000 × g, 5 min, room temperature), washed with 1 volume of magnesium sulfate 10 mM at room temperature, and resuspended in the same volume of this buffer solution. Then, 10 μl of the each cell suspension containing ∼107 cells was added together on top of sterile nitrocellulose filters (Whatman Protran BA85) placed on TB plates. After 16 h at 70°C, the cells on the filters were resuspended in 10 ml of preheated TB medium containing kanamycin and nitrate or nitrite. After overnight incubation under anaerobic conditions, individual colonies were isolated on kanamycin-containing TB plates and subjected to nitrate reduction/nitrite consumption assays (25). The nature of the colonies derived from HB27 was confirmed by analysis of their membrane protein profiles.
Detection of denitrification genes and proteins.
A draft sequence from T. thermophilus PRQ25 was obtained through pyrosequencing in a Roche-454 system (Lifesequencing, Valencia, Spain). The genes encoding enzymes implicated in the denitrification process were identified by BLAST sequence comparisons (1). The sequence of the nor-nir cluster was deposited in the gene bank with the accession number FN666415. The presence of denitrification genes in the HB27 derivatives was assayed by PCR with the primers indicated in Table 1.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′-3′) | Amplified gene |
|---|---|---|
| narC BcuI dir | ACTAGTGGAGGTGAGGATGGCGAGAAGGCTCCTACCC | narC |
| narC ClaI rev | ATCGATCCTCCTAAGCTGGGCGCGGATCC | narC |
| nirSΔ24NdeIdir | AAAACATATGACTCCGGAGGAGCGGG | nirS |
| nirS+stopEcoRIrev | AAAAGAATTCTCAGTAGATGTCGTGGGCGG | nirS |
| norCΔ58NdeIdir | AAAACATATGAACCAGATCCGCGAGGGCT | norC |
| norC+stopHindIIIrev | AAAAAAGCTTCTACTCCGCCGCCGCAAA | norC |
| Tth_GroES_Nco_fw3 | AAAAAACCATGGATGGCCGCGGAGGTGAAGAC | groE |
| groES_Cla_rv | AAAAAATCGATCTGCAGGACCGCAAGCAGGTC | groE |
For the detection of the NirS and NorC proteins in vivo, N-terminal deletion derivatives of these proteins were overexpressed in E. coli and purified to further raise specific rabbit antisera as described previously (23). The NirSΔ24N protein was overproduced in E. coli Rosetta-gami 2/DE3 from a pET28b (Novagen) derivative carrying between its single NdeI and EcoRI sites a nirS fragment obtained by PCR with the primers nirSΔ24NdeIdir and nirS+stopEcoRIrev (Table 1). The NorCΔ58N protein was expressed in E. coli BL21 from a pET22b (Novagen) derivative in which the PCR-amplified sequence (primers norCΔ58NdeIdir and norC+stopHindIIIrev in Table 1) was cloned between the NdeI and the HindIII sites. Expression was carried out at 37°C in 4-h induction experiments after the addition to exponential cultures of 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The proteins from the soluble (NorCΔ58N) or insoluble (NirSΔ24N) fractions were separated by SDS-PAGE, excised from the gels, and used to immunize New Zealand rabbits in four bursts, separated by 15 days each. The antisera obtained after 2 months were used for immunodetection of NirS and NorC by Western blotting. For this, proteins analyzed by SDS-PAGE were electrotransferred to polyvinylidene difluoride membranes, followed by incubation for 2 h at room temperature with specific rabbit antiserum diluted (1:5,000) in PBST buffer (137 mM NaCl, 2.7 mM KCl, 10 mM sodium phosphate dibasic, 2 mM potassium phosphate monobasic [pH 7.4], 0.1% Tween 20). After being washed with the same buffer, the antibodies bound to the membrane were identified after incubation in similar conditions with secondary mouse monoclonal anti-rabbit antibodies (1:5,000) labeled with horseradish peroxidase. Detection was carried out by using bioluminescence. The antisera used to detect NarG and DnrT were previously described (8).
Nitrate reductase activity and N2O detection.
Nitrate reductase activity was measured by the amount of nitrite produced (25) with reduced methyl viologen as electron donor. Nitrous oxide was measured from gas samples taken from the headspace of cultures grown in Hungate tubes at 70°C after 5 days of incubation. Gas samples were injected into a Perkin-Elmer Clarus 500 gas chromatograph with an Elite-Plot Q 30-m, 0.53-mm (inner diameter) column installed. The injector temperature was maintained at 115°C, the column was maintained at 90°C, and the electron capture detector was maintained at 350°C. The N2O peak has a retention time of 5.4 min (± 3%), and its concentration was calculated by using a standard curve with 0.4-, 100-, and 1,000-ppm standards (Stg gas mixtures).
The putative production of N2 by the strains was studied by using 30% of 15N-labeled sodium nitrate 5 mM in the growth medium. Gas samples from the headspace of cultures grown in Hungate tubes for 5 days at 70°C were analyzed by using an isotope ratio mass spectrometer (SerCon, Ltd.) at the University of Aberdeen, Aberdeen, Scotland, and the amounts of N2 and N2O accumulated were measured as described previously (3).
RESULTS
LGT of the denitrification pathway through natural competence.
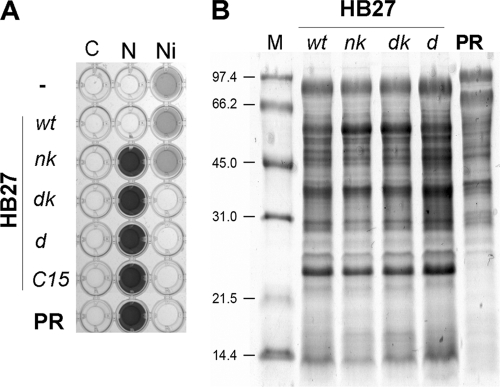
The aerobic strain T. thermophilus HB27 expresses constitutively an extremely efficient natural competence system (2, 17). In order to check whether the genes encoding the denitrification pathway in T. thermophilus PRQ25 could be transferred by this system, we added genomic DNA (200 ng/ml) from this denitrificant strain to exponential cultures of a kanamycin-resistant derivative of HB27 (HB27gdh::kat) (8). After overnight selection for anaerobic growth at 70°C in the presence of nitrate (40 mM) (Materials and Methods), the colonies that grew on kanamycin plates were subjected to nitrate reductase presence assays. From 25 colonies assayed, 22 were able to reduce nitrate to nitrite (derivative nk, Fig. 1A), showing that the NCE can be the subject of LGT by natural competence. However, none of these nitrate-reducing colonies was able to consume (reduce) nitrite, despite several repetitions of the experiment were performed.
FIG. 1.
LGT of the denitrification pathway. (A) Nitrite detection after 48 h of anaerobic incubation in the presence of nitrate 40 mM (column N), nitrite 5 mM (column Ni), or without electron acceptor (column C). Note that nitrate reduction results in the production of high nitrite concentrations (dark color), whereas nitrite consumption is detected as the lack of color. (B) SDS-PAGE of membrane proteins from the indicated strains stained with Coomassie blue. Strains: wild-type HB27 (lane wt); PRQ25 (lane PR); HB27 derivatives HB27nkat (lane nk), HB27dkat (lane dk), HB27d (lane d), and HB27C15 (lane C15).
When overnight selection was carried out with nitrite as electron acceptor, none of the colonies analyzed were able to reduce nitrite in nitrite consumption assays. Thus, to check whether our inability to isolate nitrite-respiring strains was related to the absence of the NCE in the receptor strain, we transformed the nitrate-respiring (NCE-containing) nk derivative with DNA from PRQ25. In this case, the overnight enrichment by anaerobic growth with nitrite led to the selection of a high number (25 of 48) of nitrite-reducing clones (dk in Fig. 1). Since the nk derivatives are kanamycin resistant, as is their parental HB27gdh::kat strain, we repeated the isolation process with the HB27 strain as acceptor to obtain a kanamycin-sensitive denitrificant derivative for future genetic manipulations (strain d in Fig. 1). To verify that all of the nitrate and nitrite respiring strains isolated were derived from HB27, comparison of the respective membrane protein profiles to that of the parental HB27 (wt) and the PRQ25 strains was performed. As shown in Fig. 1B, the protein profiles of the isolates are indistinguishable from that of HB27 and are quite different from the protein profile of PRQ25.
LGT of the denitrification pathway by conjugation.
The transformation experiments described above demonstrated that nitrate and nitrite respiration cannot be cotransferred within a single 20- to 25-kbp DNA fragment of the genome but did not indicate how distant the genes encoding these properties were located in the genome. Since conjugation between strains of T. thermophilus has been addressed (24), we checked whether the PRQ25 strain could transfer the denitrification pathway to the HB27 strain by this method. For this, we incubated overnight the PRQ25 and HB27gdh::kat strains together on top of nitrocellulose filters placed on TB plates and then selected overnight by anaerobic growth on nitrate in kanamycin-containing medium (see Materials and Methods). After plating, we noted a high number of nitrate-reducing colonies, supporting that PRQ25 was also able to transfer DNA to the HB27 strain by conjugation. Interestingly, many of these nitrate-reducing colonies (20 of 50 examined) were also able to reduce nitrite (derivative C15 in Fig. 1A). This genetic linkage between nitrate and nitrite reduction indicates that both properties are clustered within the genome of the PRQ25 strain, thus supporting the existence of a supercluster encoding the denitrification pathway. As expected, all of the nitrite-respiring derivatives isolated after selection for anaerobic growth with nitrite were also able to use nitrate, supporting again the NCE dependence of the nitrite respiration. The membrane protein profiles of some of these colonies were analyzed and found indistinguishable from that of the HB27 strain (not shown).
Anaerobic growth of HB27 derivatives.
Figure 2A shows that all of the HB27 derivatives that were able to reduce nitrate in Fig. 1A were also able to grow anaerobically with this electron acceptor, whereas the parental HB27 strain showed only residual growth likely due to the oxygen remaining in the tubes from the start of the experiment. As expected, the denitrificant derivatives displayed a better cell yield than the nitrate-respiring strains, likely due to an increase in energy yield per mol of nitrate expected from the subsequent denitrification steps. Actually, the nitrite-reducing strains grew on nitrite only to slightly lesser cell yields than the PRQ25 strain (Fig. 2B). As previously stated, only residual growth due to residual oxygen was detected for the aerobic HB27 and its nitrate-respiring derivative nk.
FIG. 2.
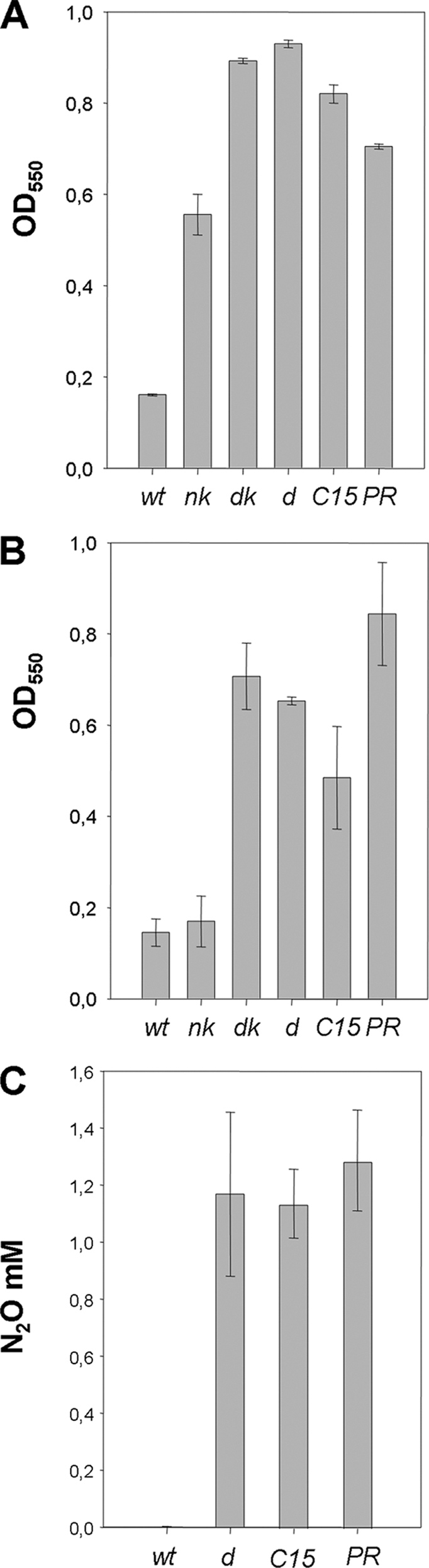
Denitrification in HB27 derivatives. (A) Anaerobic growth of the indicated strains on nitrate (40 mM) after 48 h at 70°C. (B) Anaerobic growth with nitrite (10 mM) after 48 h at the same temperature. (C) N2O production by the indicated strains after 5 days of anaerobic growth with nitrate (10 mM). Strains are named as in Fig. 1. The standard deviations are indicated.
In order to know whether nitrite respiration was also followed by NO reduction, we analyzed the production of N2O on anaerobic cultures with nitrate. As shown in Fig. 2C, the transformant HB27d, the exconjugant HB27C15, and the PRQ25 strain produced similar amounts of N2O. In contrast, no gas was detected in the parental HB27 strain incubated under the same conditions. Interestingly, the amount of N2O detected (1.2 to 1.4 mM) was less than the maximum expected if all of the nitrate used was reduced to N2O (5 mM), suggesting that most N2O was reduced to N2. To address this point, we carried out anaerobic growth assays in the presence of 15N-labeled nitrate and analyzed the amounts of 15N-labeled N2 and N2O formed (see Materials and Methods). Our data showed that no N2 was formed from nitrate, despite the total amount of N2O detected (ca. 0.35 and 0.2 mM for the PRQ25 and the HB27d strains, respectively) was less than expected if all nitrate added to the culture was used for denitrification (2.5 mM). We concluded that NO reduction was the last step of the denitrification pathway in these strains.
nor-nir cluster of T. thermophilus PRQ25.
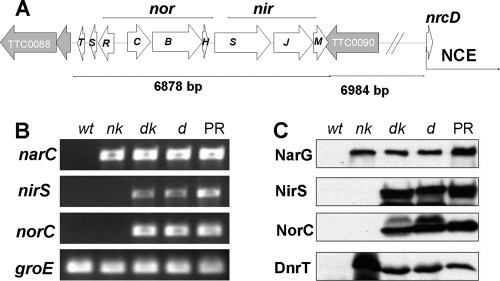
The data from Fig. 2C showing the production of N2O in HB27 derivatives selected for anaerobic growth on nitrate (C15) or nitrite (dk or d) suggest that the genes encoding the NO reductase are clustered close to those encoding for nitrate and nitrite respiration in the PRQ25 genome. To demonstrate this, we obtained a draft sequence of the PRQ25 strain through pyrosequencing. As schematized in Fig. 3A, we identified a 6.9-kbp gene cluster encoding homologues to nitrite and nitric oxide reductases from different organisms (nor-nir cluster). This cluster was surrounded by genes encoding homologues to proteins TTP0088 and TTP0090 from the megaplasmid of the aerobic strain HB27. No homologues to putative multi-copper oxidases were coded around the nor-nir cluster that could play a role in nitrous oxide reduction. However, the nor-nir cluster was located 7 kbp upstream from the first gene (nrcD) of the 23-kbp NCE. Therefore, we confirmed by sequencing the presence in the PRQ25 strain of a 37-kbp denitrification cluster encoding the nitrate, nitrite, and nitric oxide reductases.
FIG. 3.
Denitrification genes and proteins in HB27 derivatives. (A) Gene map of the nor-nir cluster and the surrounding genetic environment. (B) PCR amplification of narC, nirS, norC, and groE (housekeeping gene) using similar amounts of total DNA from the indicated strains as templates and the primer pair indicated in Table 1. (C) Western blots to detect NarG (nitrate reductase alpha subunit), NirS (putative cytochrome cd1 nitrite reductase), NorC (putative nitric oxide reductase cytochrome c subunit), and DnrT (NCE transcription factor) on the indicated strains grown under anaerobic conditions with nitrate for 16 h. The strains are labeled as in Fig. 1.
In the nor-nir cluster, NorC (221 amino acids) and NorB (476 amino acids) are homologues to the subunits of cytochrome c-dependent nitric oxide reductases (cNor). Both coding genes are separated by 10 bases and preceded by a ribosome-binding site (RBS). As expected from their similarities to the subunits of the cNor, the topology predictions indicated that NorC is a periplasmic mono-heme c cytochrome anchored to the membrane through two transmembrane helices located near its N-terminal region. On the other hand, NorB is a polytopic (12 transmembrane helices) integral membrane protein. Overlapping norB and preceded by its RBS, we identified a putative gene (norH) that encodes a 98-amino-acid membrane protein, for which three transmembrane helices are predicted. A putative Rho-independent transcription terminator is found downstream of norH, suggesting that norCBH constitutes an operon. Interestingly, from a phylogenetic point of view, this nor operon and its encoded proteins are highly conserved in Hydrogenobacter thermophilus TK-6 and other members of the ancient Aquificae phylum. The corresponding proteins from this organism are 61, 67, and 54% identical to T. thermophilus PRQ25 NorC, NorB, and NorH, respectively.
At a position 120 bp downstream from norH, the nor-nir cluster contains a gene, nirS, which encodes a 536-amino-acid homologue to the cytochrome cd1 nitrite reductases found in many denitrifiers. Like its homologues, the NirS precursor includes a sec-dependent signal peptide with a cutting site (LA-Q) common to several periplasmic proteins from T. thermophilus, including the cytochrome c552 precursor. Sequence alignments and structure models based on the NirS protein from Paracoccus pantotrophus (PDB entries 1e2r, 1gq1, and 1hj5) confirm that mature NirS protein from T. thermophilus contains the amino acids implicated in the binding of heme c and heme d1 groups (12). A putative rho-independent terminator follows nirS, supporting that this gene is transcribed as a monocistronic unit. Downstream and separated by 81 bp from the stop codon of nirS, a gene was found (nirJ) that encodes a 375-amino-acid membrane protein belonging to the radical SAM protein family. This protein has homologues of unknown function in most nir clusters from other origins. Downstream of nirJ and separated by 5-bp starts nirM, a gene that encodes a 144-amino-acid protein. The putative NirM precursor includes a sec-dependent signal peptide that ends with the LA-Q cutting site, and a heme c binding motif (CSSCH) near the N terminus of the mature form of the protein. Sequence comparison and topology predictions support that NirM is a soluble periplasmic cytochrome c homologue to cytochrome c552 from Thermus sp. and to other periplasmic cytochromes c of unknown function clustered with nirS on different denitrificant bacteria. As above, each of the nir genes start codons is preceded by an RBS, supporting their actual translation in T. thermophilus.
A putative operon located upstream from norC and transcribed in opposite direction contains three small open reading frames (ORFs). The first ORF of this putative operon (norR) encodes a 141-amino-acid protein that shows similarities to transcription factors from the MarR family. The protein contains an N-terminal HTH DNA-binding motif and three cysteines at its C-terminal domain. A homologue of this putative protein—NsrR from Streptomyces coelicolor—has been implicated in NO sensing through a [2Fe-2S] cluster (29). Downstream of norR, two small proteins are encoded (NorS [78 amino acids] and NorT [98 amino acids]) that are homologues to uncharacterized proteins conserved in several bacteria. On the other hand, a search for homologues of NosZ in the PRQ25 genome draft yielded no significant hits.
Expression of the denitrification genes in HB27 derivatives.
Figure 3B confirms through PCR that selected genes implicated in nitrate, nitrite, and nitric oxide respiration were transferred to the HB27 derivatives. The narC gene from the NCE was positively amplified in all of the facultative strains (PRQ25 and HB27 derivatives), whereas nirS and norC were found only in the nitrite-respiring strains and not in the nitrate-respiring ones. In the parental HB27 strain, only the housekeeping groE gene, used as a PCR control, was detected. To confirm the actual expression of the main genes of the nor-nir cluster in the receptor strain, we expressed NirS and NorC fragments in E. coli and prepared antisera against them (see Materials and Methods). As seen in Fig. 3C, the NirS and NorC proteins were detected only in the denitrificant strains upon anaerobic incubation with nitrate, whereas the major subunit of the nitrate reductase (NarG) and the DnrT transcription factor were expressed in all of the facultative strains. None of these proteins were detected in the aerobic HB27 parental strain. It is important to note that the NirS and NorC proteins sizes corresponded to that expected for their respective amino acid sequences.
The denitrification supercluster is surrounded by genes from a megaplasmid.
Downstream (267 bp) of norT, we identified a DNA sequence almost identical to that encoding the C terminus of the TTC0089 ORF from the pTT27 megaplasmid of the HB27 strain. At the 3′ extreme of the nor-nir cluster, the last 26 bases of NirM are complementary to a sequence encoding the C terminus of TTC0090, also from pTT27. Therefore, it appears as though in PRQ25 the nor-nir cluster was inserted within a group of preexisting genes from its megaplasmid. It is interesting that synteny comparisons between the pTT27 megaplasmids from the aerobic strains T. thermophilus HB27 and HB8 reveal that this region is highly variable (5). On the other hand, the first gene of the NCE (nrcD), located 6,984 bp downstream from nirM, is also preceded by genes homologous to those found in the pTT27 megaplasmid, including a putative transposase found downstream the NCE (data not shown). In conclusion, the NCE and the nor-nir cluster are inserted within homologues to genes belonging to a highly variable region of the pTT27 megaplasmid. Actually, our draft sequence shows that most of the core genes of the HB27 and HB8 pTT27 megaplasmid are conserved in PRQ25, strongly supporting the existence of an equivalent megaplasmid in PRQ25.
DISCUSSION
The availability of hundreds of genome sequences has revealed that the denitrification pathway is not associated with specific taxa but heterogeneously distributed among members of several phylogenetic phyla. Such analysis has led to the suggestion that this metabolic ability has been spread through several LGT events (15, 16, 22). Our data demonstrate that LGT of the denitrification pathway occurs between T. thermophilus strains.
The aerobic HB27 strain used as a receptor in our LGT experiments is well known for the high efficiency of its natural competence system, allowing individual cells to incorporate linear DNA at rates of ∼40 kbp/s, irrespective of its origin (2). However, the processivity of the system, i.e., the size of a DNA fragment that can be incorporated into the cell in a continuous form by each competence apparatus, is not known. In our transformation experiments, we observed the efficient transfer of the whole NCE (23 kbp) when selection was carried out on nitrate, thus confirming that the competence system can handle DNA fragments of at least this size. Having in mind that the size of the nor-nir cluster was smaller than the NCE, the inability to obtain nitrite-respiring strains through a single transformation step confirms the dependence of this process from transcription factors encoded by the NCE (9). On the other hand, these data support that the transfer of a single DNA fragment encoding both properties—at least 37 kbp, as shown by sequencing—is not frequently achieved under the conditions used in our assays. In any case, it is likely that the natural competence system could incorporate the whole denitrification cluster if challenged with larger PRQ25 DNA fragments than those used in our assays.
In contrast to transformation, conjugation can handle even a whole chromosome (13, 18). Actually, the NAR1 strain can transfer chromosomal markers in a mating time-dependent way (24), supporting the existence of an integrated F-like conjugative plasmid wearing the NCE that leads the DNA transfer as in Hfr (high frequency of recombination) strains of E. coli (18). In the conjugation assays with PRQ25 as a donor, we detected a highly efficient transfer of the denitrification equipment, indicating that in this strain, too, the denitrification cluster is part of a conjugative element. Because the sequence data support that in NAR1 (not shown) and in PRQ25 the denitrification enzymes are integrated in the same region of the pTT27 megaplasmid, it is tempting to speculate that the conjugative element itself is the megaplasmid and that the transfer of chromosome-associated markers observed in the NAR1 strain is the consequence of its integration into the chromosome through any of the several insertion sequences common to both replicons (14) as happens with the F or R plasmids of E. coli (18). However, we did not find around the denitrification cluster any homologues to proteins implicated in conjugation in other bacteria, suggesting that the conjugative capability is not part of the denitrification cluster but likely is associated with separate genes from the megaplasmid. In this sense, our sequence shows the presence of genes encoding homologues to components of type IV pili but not to any other conserved protein implicated in conjugation in other organisms.
Though it was demonstrated that the whole denitrification capability could be transferred by natural competence and by conjugation, the question regarding the phylogenetic origin of the supercluster remains. The calculation of anomalous GC content and the codon adaptation index (11, 21) supports the idea that neither the nor-nir cluster nor the NCE had been acquired recently by T. thermophilus because these values are in good agreement with those for the housekeeping genes of the genus. However, protein sequence comparisons revealed high similarities between the sequences of the Nor and Nir proteins from T. thermophilus and those of Hydrogenobacter thermophilus (27, 28) and other members of the Aquificae phylum (e.g., Persephonella maritima and Hydrogenovirga sp.), which share with T. thermophilus a thermophilic lifestyle. In addition, the use of bipartition dissimilarity analysis (4) suggests an old LGT event from the Aquificae to Thermus as the origin of the nor-nir cluster. In this sense, it is important to note that nor-nir and the NCE clusters are likely inserted within a highly variable region from a pTT27-like megaplasmid of PRQ25. Therefore, the most likely phylogenetic analysis indicates that the NCE and the nor-nir clusters have been inserted recently in the genome of PRQ25, perhaps from another Thermus sp. or from a closely related species, where it has been adapted to fit the appropriate codon usage and the regulatory components that make it able to be expressed in response to the appropriate external signals.
The association between the presence of these norC and nirS genes and their expression and activity in the HB27 derivatives shows a direct relationship between the presence of these genes and anaerobic growth by denitrification. Thus, there are no doubts about the roles as reductases of the products of the norCB and nirS genes. However, there are different genes in the cluster for which we can only speculate about their role. For example, the norH gene codes for a small membrane protein conserved in nor operons from the thermophilic members of the Aquificae group (27), supporting its putative role as a third unit or as a dedicated chaperone of the corresponding enzyme. On the other hand, the nirJM putative operon constitutes the smaller cluster of nirS-associated genes thus far described. The actual roles of NirJ, a radical SAM protein, and NirM, a periplasmic cytochrome c, are not known, but both have homologues in nir clusters from several bacteria. However, what seems to be quite unusual in the nir cluster of T. thermophilus is the absence of homologues to genes putatively implicated in the synthesis of heme d1. As an example, all of the nir clusters thus far described contain a gene (nirE) coding for uroporphyrinogen III methyltransferase (EC 2.1.1.107) (26). Its absence in the cluster of T. thermophilus could be explained by the presence of a homologue in the chromosome of T. thermophilus (TTC0308 in HB27). Therefore, it is likely that most of the enzymes required for the synthesis of the heme d1 are encoded by the chromosome of the HB27 and other T. thermophilus strains, whereas only those specifically required for the maturation or the activity of NirS remain clustered with its coding gene.
In relation to the regulation of the expression of these nor-nir genes, there is a group of three proteins encoded upstream of norC that could be implicated in the process, as suggested by the similarities between NorR and the NO-dependent regulator NsrR (29). Details regarding the transcriptional control of nor and nir operons by these proteins are still to be unveiled, and only a combination of in vitro and in vivo analyses could answer this question. For this, the availability of a denitrificant derivative of HB27 would be extremely helpful for obtaining the corresponding mutants.
Despite the absence of any homologue to NosZ in the PRQ25 genome, a doubt remained about the completeness or not of the denitrification pathway in this strain because our nitrogen balance revealed that only part (1/4 approximately) of the nitrate was being reduced to N2O (Fig. 2C). The putative presence of unusual nitrous oxide-reducing activity in hyperthermophiles has been suggested (31). Actually, a laccase from Pyrobaculum aerophilum for which a homologue exists in T. thermophilus HB27 (protein TTC1370) has been shown to produce N2 actively (10). However, the experiments with [15N]nitrate demonstrated the absence of any significant N2 production in PRQ25 and HB27 derivatives, leading to the conclusion that the final product of the denitrification process in these strains was N2O. It is not known, however, if this is a general rule for other denitrificant strains of T. thermophilus or a specific trait of the PRQ25 donor strain. In any case, our data support that if this was the case, the nitrous oxide reductase will likely be encoded as a separate unit and not as part of the denitrification cluster described here. The final destination of most of the nitrogen from the nitrate used for anaerobic growth in T. thermophilus PRQ25 is also uncertain. In this sense, it is tempting to speculate about the presence of an ammonia-forming nitrite reductase by an enzyme (PRQ25_1681) 98% identical to TTC0313 from the HB27 strain. This protein has been annotated as a ferredoxin-nitrite reductase (14), despite its clustering with enzymes such as phosphoadenosine-phosphosulfate reductase (PRQ25_1678). Although this enzyme more likely represents a sulfite reductase implicated in sulfate assimilation, the possibility exists that it could reduce nitrite to ammonia, masking the nitrogen balance. Future genetic and biochemical work will allow us to confirm or to discard definitively this possibility.
Finally, an important result of the present study is the isolation of a kanamycin-sensitive denitrificant HB27 derivative (isolate d), whose ease for genetic manipulation compared to the original PRQ25 strain advances our knowledge of the denitrification pathway and its regulation at high temperatures.
Acknowledgments
This study was supported by a grant of code BIO2007-60245 from the Ministerio de Ciencia e Innovación. L.A. and C.B. are supported by fellowships from the Consejo Superior de Investigaciones Científicas and the Ministerio de Educación, respectively. An institutional grant from the Fundación Ramón Areces to CBMSO is also acknowledged.
We thank D. Richardson, A. J. Gates, and H. Felgate for their help and advice.
Footnotes
Published ahead of print on 17 December 2010.
REFERENCES
- 1.Altschul, S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Averhoff, B. 2009. Shuffling genes around in hot environments: the unique DNA transporter of Thermus thermophilus. FEMS Microbiol. Rev. 33:611-626. [DOI] [PubMed] [Google Scholar]
- 3.Baggs, E. M., C. L. Smales, and E. J. Bateman. 2010. Changing pH shifts the microbial source as well as the magnitude of N2O emission from soil. Biol. Fertil. Soils 46:793-805. [Google Scholar]
- 4.Boc, A., H. A. Philippe, and V. Makarenkov. 2010. Inferring and validating horizontal gene transfer events using bipartition dissimilarity. Syst. Biol. 59:195-211. [DOI] [PubMed] [Google Scholar]
- 5.Bruggemann, H., and C. Chen. 2006. Comparative genomics of Thermus thermophilus: plasticity of the megaplasmid and its contribution to a thermophilic lifestyle. J. Biotechnol. 124:654-661. [DOI] [PubMed] [Google Scholar]
- 6.Cava, F., and J. Berenguer. 2006. Biochemical and regulatory properties of a respiratory island encoded by a conjugative plasmid in the extreme thermophile Thermus thermophilus. Biochem. Soc. Trans. 34:97-100. [DOI] [PubMed] [Google Scholar]
- 7.Cava, F., A. Hidalgo, and J. Berenguer. 2009. Thermus thermophilus as biological model. Extremophiles 13:213-231. [DOI] [PubMed] [Google Scholar]
- 8.Cava, F., et al. 2007. Control of the respiratory metabolism of Thermus thermophilus by the nitrate respiration conjugative element NCE. Mol. Microbiol. 64:630-646. [DOI] [PubMed] [Google Scholar]
- 9.Cava, F., O. Zafra, M. S. da Costa, and J. Berenguer. 2008. The role of the nitrate respiration element of Thermus thermophilus in the control and activity of the denitrification apparatus. Environ. Microbiol. 10:522-533. [DOI] [PubMed] [Google Scholar]
- 10.Fernandes, A. T., et al. The multicopper oxidase from the archaeon Pyrobaculum aerophilum shows nitrous oxide reductase activity. FEBS J. 277:3176-3189. [DOI] [PubMed]
- 11.Garcia-Vallve, S., A. Romeu, and J. Palau. 2000. Horizontal gene transfer in bacterial and archaeal complete genomes. Genome Res. 10:1719-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordon, E. H., et al. 2003. Structure and kinetic properties of Paracoccus pantotrophus cytochrome cd1 nitrite reductase with the d1 heme active site ligand tyrosine 25 replaced by serine. J. Biol. Chem. 278:11773-11781. [DOI] [PubMed] [Google Scholar]
- 13.Hayes, W. 1953. The mechanism of genetic recombination in Escherichia coli. Cold Spring Harbor Symp. Quant. Biol. 18:75-93. [DOI] [PubMed] [Google Scholar]
- 14.Henne, A., et al. 2004. The genome sequence of the extreme thermophile Thermus thermophilus. Nat. Biotechnol. 22:547-553. [DOI] [PubMed] [Google Scholar]
- 15.Heylen, K., et al. 2007. Nitric oxide reductase (norB) gene sequence analysis reveals discrepancies with nitrite reductase (nir) gene phylogeny in cultivated denitrifiers. Environ. Microbiol. 9:1072-1077. [DOI] [PubMed] [Google Scholar]
- 16.Jones, C. M., B. Stres, M. Rosenquist, and S. Hallin. 2008. Phylogenetic analysis of nitrite, nitric oxide, and nitrous oxide respiratory enzymes reveal a complex evolutionary history for denitrification. Mol. Biol. Evol. 25:1955-1966. [DOI] [PubMed] [Google Scholar]
- 17.Koyama, Y., T. Hoshino, N. Tomizuka, and K. Furukawa. 1986. Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J. Bacteriol. 166:338-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Low, K. 1996. Hfr strains of Escherichia coli K-12, p. 2402-2405. In F. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 2. ASM Press, Washington, DC. [Google Scholar]
- 19.Manaia, C. M., et al. 1994. Halotolerant Thermus strains from marine and terrestrial hot springs belong to Thermus thermophilus (ex Oshima and Imahori, 1974) nom. rev. emend. Syst. Appl. Microbiol. 17:526-532. [Google Scholar]
- 20.Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 21.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 22.Philippot, L. 2002. Denitrifying genes in bacterial and archaeal genomes. Biochim. Biophys. Acta 1577:355-376. [DOI] [PubMed] [Google Scholar]
- 23.Ramirez-Arcos, S., L. A. Fernandez-Herrero, and J. Berenguer. 1998. A thermophilic nitrate reductase is responsible for the strain specific anaerobic growth of Thermus thermophilus HB8. Biochim. Biophys. Acta 1396:215-227. [DOI] [PubMed] [Google Scholar]
- 24.Ramirez-Arcos, S., L. A. Fernandez-Herrero, I. Marin, and J. Berenguer. 1998. Anaerobic growth, a property horizontally transferred by an Hfr-like mechanism among extreme thermophiles. J. Bacteriol. 180:3137-3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snell, F. D., and C. T. Snell. 1949. Colorimetric methods of analysis. Van Nostrand, New York, NY.
- 26.Storbeck, S., et al. 2009. The Pseudomonas aeruginosa nirE gene encodes the S-adenosyl-l-methionine-dependent uroporphyrinogen III methyltransferase required for heme d(1) biosynthesis. FEBS J. 276:5973-5982. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki, M., H. Arai, M. Ishii, and Y. Igarashi. 2006. Gene structure and expression profile of cytochrome bc nitric oxide reductase from Hydrogenobacter thermophilus TK-6. Biosci. Biotechnol. Biochem. 70:1666-1671. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki, M., T. Hirai, H. Arai, M. Ishii, and Y. Igarashi. 2006. Purification, characterization, and gene cloning of thermophilic cytochrome cd1 nitrite reductase from Hydrogenobacter thermophilus TK-6. J. Biosci. Bioeng. 101:391-397. [DOI] [PubMed] [Google Scholar]
- 29.Tucker, N. P., et al. 2008. The transcriptional repressor protein NsrR senses nitric oxide directly via a [2Fe-2S] cluster. PLoS One 3:e3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zafra, O., et al. 2002. A cytochrome c encoded by the nar operon is required for the synthesis of active respiratory nitrate reductase in Thermus thermophilus. FEBS Lett. 523:99-102. [DOI] [PubMed] [Google Scholar]
- 31.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zumft, W. G., and H. Korner. 1997. Enzyme diversity and mosaic gene organization in denitrification. Antonie Van Leeuwenhoek 71:43-58. [DOI] [PubMed] [Google Scholar]
- 33.Zumft, W. G., and P. M. Kroneck. 2007. Respiratory transformation of nitrous oxide (N2O) to dinitrogen by Bacteria and Archaea. Adv. Microb. Physiol. 52:107-227. [DOI] [PubMed] [Google Scholar]