Abstract
The presence of Cryptosporidium and Giardia in 221 fecal samples from different species of Antarctic pinnipeds was investigated by immunofluorescence microscopy and PCR. Cryptosporidium, a skunk-like genotype, was detected only in a southern elephant seal. Giardia was not detected. This is the first report of a Cryptosporidium sp. in Antarctic marine mammals.
Cryptosporidium and Giardia are ubiquitous protozoan parasites which infect a wide variety of hosts, including humans and domesticated and wild animals (27). In recent years, increasing research has been carried out in marine mammals since they may act as indicator species for environmental contamination with these waterborne parasites (1). Cryptosporidium oocysts and/or Giardia cysts have been identified in feces or intestinal contents of various animal species, including an Australian dugong (Dugong dugon), California sea lions (Zalophus californianus), ringed seals (Phoca hispida), harp seals (Phoca groenlandica), gray seals (Halichoerus grypus), hooded seals (Cyptophora cristatai), bearded seals (Erignathus barbatus), and harbor seals (Phoca vitulina), as well as right whales (Eubalaena glacialis) and bowhead whales (Balaena mysticetus) from different locations worldwide (reviewed in references 1, 5, and 13). However, no studies have been conducted on Antarctic marine mammals. Regarding the species or genotypes involved, the presence of zoonotic assemblages A and B of Giardia duodenalis has been commonly reported (1, 2, 5, 16), as have assemblages F (2) and D and novel genotypes related to the canine assemblages C and D (10). Cryptosporidium hominis, a species thought to be infective exclusively for humans, nonhuman primates, and gnotobiotic pigs (19), has been identified only in a dugong (12). Other species reported include Cryptosporidium muris and two novel genotypes, designated Cryptosporidium sp. seal 1 and 2 (2, 5, 25). These studies indicate that marine mammals could represent potential zoonotic reservoirs for Cryptosporidium and Giardia, but they also reflect that human activities may have an impact on the health of marine mammals and the environment. It is therefore important to monitor the health status of wildlife in general and identify potential sources of infection and routes of transmission or dissemination, particularly in unspoiled areas.
In the present study, we investigated the presence of the zoonotic parasites Cryptosporidium and Giardia in Antarctic pinnipeds in order to determine the occurrence of these parasites, to identify the species or genotypes involved in infection, and to evaluate whether they might be linked to anthropogenic activities.
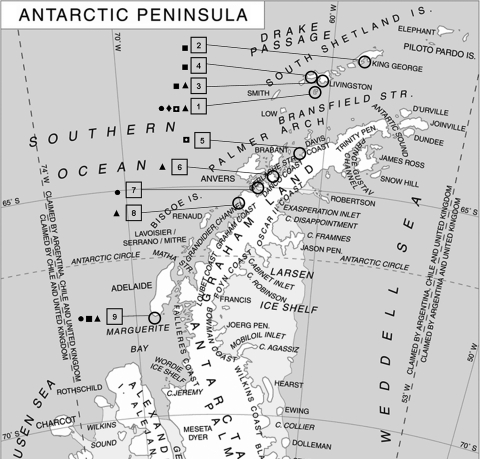
A total of 221 fresh fecal samples from different pinniped populations from different locations along the west coast of the Antarctic Peninsula (ranging from 62°15′S to 58°37′W-67°46′S and 68°43′W) (Fig. 1) were collected from the ground during the month of February in 2006 and 2007. These included samples from 31 Weddell seals (Leptonychotes weddelli), 2 crabeater seals (Lobodon carcinophagus), 4 leopard seals (Hydrurga leptonyx), 53 southern elephant seals (Mirounga leonina), and 131 Antarctic fur seals (Arctocephalus gazella).
FIG. 1.
Locations of sampling areas and animal distribution (adapted from Wikimedia Commons [Giovanni Fattori]). 1, Deception Island, South Shetland Islands; 2, King George Island, South Shetland Islands; 3, Hannah Point, Livingston Island, South Shetland Islands; 4, Byers Peninsula, Livingston Island, South Shetland Islands; 5, Cape Primavera, Antarctic Peninsula; 6, Rongé Island, Errera Channel; 7, Paradise Bay, Antarctic Peninsula; 8, Galindez Island, Argentine Islands; 9, Avian Island, Marguerite Bay, Antarctic Peninsula. •, Weddell seal (Leptonychotes weddelli); ◘, crabeater seal (Lobodon carcinophagus); ⧫, leopard seal (Hydrurga leptonyx); ▪, southern elephant seal (Mirounga leonina); ▴, Antarctic fur seal (Arctocephalus gazella).
Fecal slides were prepared on the same day of sample collection by spreading in triplicate approximately 40 μl of homogenized sample onto a microscope glass slide and fixing in methanol and were stored at −20°C. Fecal samples were kept at +4°C without preservatives for periods of up to 2 months until they were analyzed.
Detection of Cryptosporidium and Giardia.
Immunofluorescence staining was performed using the Crypto/Giardia Cel IF test (Cellabs Pty. Ltd., Brookvale, Australia) on fecal slides. The numbers of oocysts/cysts on slides were determined at magnification ×400, and the means for 20 fields were calculated. If no oocysts/cysts were seen in 20 fields, the entire slide was examined. To approximately calculate the number of oocysts, the following categories were established: no oocysts; <1 oocyst per field; 1 to 10 oocysts per field; 11 to 100 oocysts per field; and >100 oocysts per field, which corresponded to approximately 0, <103, 103 to 104, 104 to 105, and >105 oocysts per g (or per ml) of feces, respectively, performing spiking trials with control C. parvum oocysts in negative seal fecal samples. Fecal slides were prepared as described above.
DNA purification was performed using 200 to 300 μl of homogenized feces and comprised oocyst/cyst disruption with zirconia beads in the presence of guanidinium thiocyanate, followed by purification with activated silica as previously described (17). Positive (both positive fecal samples, bovine and canine, and control oocysts/cysts of C. parvum and G. duodenalis assemblage D) and negative controls were included in each batch.
For Cryptosporidium, a nested PCR procedure was performed for amplification of an 827- to 840-bp polymorphic fragment of the 18S ribosomal DNA (rDNA) (28). In addition, a 446-bp fragment of the HSP70 gene was amplified using the primers HSPF4 and HSPR4 (20). For Giardia, a nested procedure was performed to amplify a 511-bp fragment of the beta-giardin gene (15). Positive and negative controls were included for all PCRs.
The presence of Cryptosporidium oocysts was detected by immunofluorescence and PCR only in one sample (0.45%) from a Southern elephant seal collected in the southernmost sampling area, Avian Island, in 2006. The presence of Giardia was not detected by either method in any of the samples analyzed. These results suggest that the presence of these parasites in these regions is rare. The detection methods used in this study are widely applied and have proven very sensitive. However, we did not perform concentration of the fecal material or purification of oocysts/cysts, and therefore samples with very low numbers of oocysts/cysts might not have been detected. Nevertheless, we consider the application of both immunofluorescence microscopy and PCR to enhance the detection power. To our knowledge, our study constitutes the first report of the presence of Cryptosporidium in Antarctic marine mammals. Few studies have been conducted in this respect; Fayer (6) has indicated that Antarctica was the only continent in which the presence of Cryptosporidium had not been reported. However, recently the presence of Cryptosporidium oocysts in Antarctic adelie (Pygoscelis adeliae) and gentoo penguins (Pygoscelis papua) from Ardley Island, South Shetlands (62°13′S, 58°54′W) has been described (7, 9), although other studies in different locations have reported the absence of Cryptosporidium and/or Giardia in gentoo and adelie penguins and in chinstrap penguins (Pygoscelis antarctica) (8, 22). In contrast to the results presented here, prevalence rates of Cryptosporidium in pinnipeds from other less-preserved areas range from 16 to 24% (2, 4, 12, 13, 25), whereas for Giardia, they range from 12 to 64.5% (2, 13, 18, 21). This indicates that the Antarctic fauna has suffered from a lower level of exposure to these agents, which is in agreement with the relative geographical and biological isolation of the Antarctic continent. However, further studies are needed to investigate their potential sources of infection and to monitor their possible introduction and dissemination in this singular environment.
The number of oocysts observed per field was 5, which approximately corresponded to 103 to 104 oocysts per g of feces, suggesting infection in this animal rather than passive transfer. In contrast to other animal species analyzed in this study, whose migratory and foraging ranges seem to be confined to the Antarctic region, the southern elephant seal is widely distributed in the Southern hemisphere. Therefore, infection in this animal might have been acquired outside Antarctica and introduced into the area. Nevertheless, this might have important implications for the Antarctic fauna, since these animals can act as reservoirs of the disease to those in close vicinity and also disseminate these pathogens to different geographic locations in the marine and terrestrial environments.
Molecular characterization of the Cryptosporidium isolate.
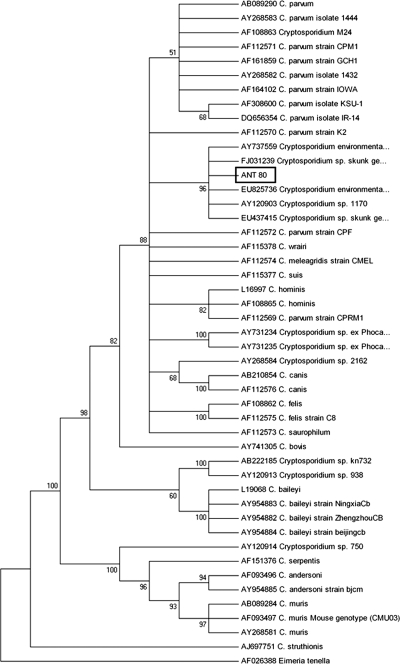
18S rDNA and HSP70-positive amplicons were directly sequenced in both directions at the Unidad Genómica del Parque Científico de Madrid. Sequences were analyzed using the BioEdit Sequence Alignment Editor software program, v.7.0.1 (7, 11). Multiple alignments were performed using the ClustalW software program, and neighbor-joining trees were constructed from the aligned sequences using the MEGA software program, version 4 (26). Analysis of the 828-bp 18S rDNA fragment revealed a 99.5% to 99.6% similarity to the sequences of the Cryptosporidium skunk genotype published in GenBank, isolated from a skunk (accession no. AY120903), from environmental samples (AY737559 and EU825736), and from a human patient (EU437415). The sequence obtained for this isolate showed the deletion of a T base at position 285 with respect to the sequence under accession no. AY120903 and the insertion of a T base at positions 456, 457, and 508 with respect to all four sequences. The neighbor-joining analysis of the multiple alignment performed with Cryptosporidium sequences retrieved from the GenBank database (Fig. 2) showed that this genotype clusters closely with other intestinal Cryptosporidium species, such as C. parvum, C. hominis, C. wrairi, C. meleagridis, and C. suis, but constitutes a separate, distinct group.
FIG. 2.
Phylogenetic relationships between the southern elephant seal isolate ANT 80 (in box) and published Cryptosporidium species or genotypes, inferred by neighbor-joining analysis of the 18S rDNA fragment. Evolutionary distances were calculated by the Kimura-2 parameter model using Eimeria tenella as an outgroup.
Sequence and phylogenetic analysis of the HSP70 gene confirmed these results. The highest similarities, 99.8%, were observed with the Cryptosporidium skunk genotype isolated from a skunk (accession no. AY120917) and from a human patient (EU437414). The sequence obtained in this study varied by a T/C substitution at position 75 and an A/G substitution at position 240 with respect to the sequence under accession no. AY120917 and EU437414, respectively. Previously, the Cryptosporidium skunk genotype had been isolated from skunk, raccoon, eastern squirrel, opossum, river otter (27), environmental samples (14, 23), and, also recently, from humans (3, 24). It was initially suggested that this genotype might be a fur-bearing wild mammal host-adapted type with no significance for public health (27). However, the identification of this genotype in a human patient who had suffered from diarrhea (24) demonstrates that it is capable of causing infection in other hosts and could disseminate through different routes of transmission. More molecular data identifying the species and genotypes present in marine mammals are needed to compare with new and existing data from humans and other terrestrial animals in order to evaluate the potential impact of human activities on these populations.
Nucleotide sequence accession numbers.
The nucleotide sequences generated in this study have been deposited in the GenBank database under accession numbers GQ421425 and GQ421426.
Acknowledgments
This work was funded by the Spanish Ministry of Education and Science (CGL-2004-22025-E/ANT, CGL-2005-25073-E/ANT, and CTM2008-00570).
We thank the military personnel at the Spanish Antarctic base Gabriel de Castilla for their help and assistance and the Marine Technology Unit (CSIC) and Spanish Navy's Oceanographic Research Ship Las Palmas for logistics and transport.
Footnotes
Published ahead of print on 17 December 2010.
REFERENCES
- 1.Appelbee, A. J., R. C. Thompson, and M. E. Olson. 2005. Giardia and Cryptosporidium in mammalian wildlife—current status and future needs. Trends Parasitol. 21:370-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogomolni, A. L., et al. 2008. Victims or vectors: a survey of marine vertebrate zoonoses from coastal waters of the Northwest Atlantic. Dis. Aquat. Organ. 81:13-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies, A. P., et al. 2009. Asymptomatic carriage of protozoan parasites in children in day care centers in the United Kingdom. Pediatr. Infect. Dis. J. 28:838-840. [DOI] [PubMed] [Google Scholar]
- 4.Deng, M. Q., R. P. Peterson, and D. O. Cliver. 2000. First findings of Cryptosporidium and Giardia in California sea lions (Zalophus californianus). J. Parasitol. 86:490-494. [DOI] [PubMed] [Google Scholar]
- 5.Dixon, B. R., et al. 2008. Giardia duodenalis and Cryptosporidium spp. in the intestinal contents of ringed seals (Phoca hispida) and bearded seals (Erignathus barbatus) in Nunavik, Quebec, Canada. J. Parasitol. 94:1161-1163. [DOI] [PubMed] [Google Scholar]
- 6.Fayer, R. 2008. General biology, p. 1-42. In R. Fayer and L. Xiao (ed.), Cryptosporidium and Cryptosporidiosis, 2nd ed. CRC Press, Boca Raton, FL.
- 7.Fredes, F., A. Diaz, E. Raffo, and P. Muñoz. 2008. Cryptosporidium spp. oocysts detected using acid-fast stain in faeces of gentoo penguins (Pygoscelis papua) in Antarctica. Antarct. Sci. 20:495-496. [Google Scholar]
- 8.Fredes, F., et al. 2007. Gastrointestinal parasite fauna of gentoo penguins (Pygoscelis papua) from the Peninsula Munita, Bahia Paraiso, Antarctica. Antarct. Sci. 19:93-94. [Google Scholar]
- 9.Fredes, F., E. Raffo, and P. Muñoz. 2007. First report of Cryptosporidium spp. oocysts in stool of Adelie penguin from the Antarctic using acid-fast stain. Antarct. Sci. 19:437-438. [Google Scholar]
- 10.Gaydos, J. K., et al. 2008. Novel and canine genotypes of Giardia duodenalis in harbor seals (Phoca vitulina richardsi). J. Parasitol. 94:1264-1268. [DOI] [PubMed] [Google Scholar]
- 11.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 12.Hill, B. D., I. R. Fraser, and H. C. Prior. 1997. Cryptosporidium infection in a dugong (Dugong dugon). Aust. Vet. J. 75:670-671. [DOI] [PubMed] [Google Scholar]
- 13.Hughes-Hanks, J. M., et al. 2005. Prevalence of Cryptosporidium spp. and Giardia spp. in five marine mammal species. J. Parasitol. 91:1225-1228. [DOI] [PubMed] [Google Scholar]
- 14.Jellison, K. L., A. E. Lynch, and J. M. Ziemann. 2009. Source tracking identifies deer and geese as vectors of human-infectious Cryptosporidium genotypes in an urban/suburban watershed. Environ. Sci. Technol. 43:4267-4272. [DOI] [PubMed] [Google Scholar]
- 15.Lalle, M., et al. 2005. Genetic heterogeneity at the beta-giardin locus among human and animal isolates of Giardia duodenalis and identification of potentially zoonotic subgenotypes. Int. J. Parasitol. 35:207-213. [DOI] [PubMed] [Google Scholar]
- 16.Lasek-Nesselquist, E., et al. 2008. Molecular characterization of Giardia intestinalis haplotypes in marine animals: variation and zoonotic potential. Dis. Aquat. Organ. 81:39-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLauchlin, J., S. Pedraza-Díaz, C. Amar-Hoetzeneder, and G. L. Nichols. 1999. Genetic characterization of Cryptosporidium strains from 218 patients with diarrhea diagnosed as having sporadic cryptosporidiosis. J. Clin. Microbiol. 37:3153-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Measures, L. N., and M. E. Olson. 1999. Giardiasis in pinnipeds from eastern Canada. J. Wildl. Dis. 35:779-782. [DOI] [PubMed] [Google Scholar]
- 19.Morgan, U. M., et al. 2000. Detection of the Cryptosporidium parvum “human” genotype in a dugong (Dugong dugon). J. Parasitol. 86:1352-1354. [DOI] [PubMed] [Google Scholar]
- 20.Morgan, U. M., et al. 2001. Molecular and phylogenetic characterisation of Cryptosporidium from birds. Int. J. Parasitol. 31:289-296. [DOI] [PubMed] [Google Scholar]
- 21.Olson, M. E., P. D. Roach, M. Stabler, and W. Chan. 1997. Giardiasis in ringed seals from the western Arctic. J. Wildl. Dis. 33:646-648. [DOI] [PubMed] [Google Scholar]
- 22.Palacios, M. J., et al. 2010. Absence of Cryptosporidium, Giardia and Toxoplasma gondii in three species of penguins along Antarctic Peninsula. Antarct. Sci. 22:265-270. [Google Scholar]
- 23.Perz, J. F., and S. M. Le Blancq. 2001. Cryptosporidium parvum infection involving novel genotypes in wildlife from lower New York State. Appl. Environ. Microbiol. 67:1154-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson, G., K. Elwin, and R. M. Chalmers. 2008. Unusual Cryptosporidium genotypes in human cases of diarrhea. Emerg. Infect. Dis. 14:1800-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santín, M., B. R. Dixon, and R. Fayer. 2005. Genetic characterization of Cryptosporidium isolates from ringed seals (Phoca hispida) in Northern Quebec, Canada. J. Parasitol. 91:712-716. [DOI] [PubMed] [Google Scholar]
- 26.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 27.Xiao, L., and R. Fayer. 2008. Molecular characterisation of species and genotypes of Cryptosporidium and Giardia and assessment of zoonotic transmission. Int. J. Parasitol. 38:1239-1255. [DOI] [PubMed] [Google Scholar]
- 28.Xiao, L., et al. 1999. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 65:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]