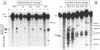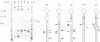Abstract
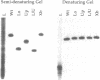
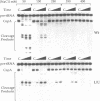
Bulged-out nucleotides or internal loops are present in the stem-loop structures of several antisense RNAs. We have used the antisense/target RNA system (CopA/CopT) that controls the copy number of plasmid R1 to examine the possible biological function of bulged-out nucleotides. Two regions within the major stem-loop of the antisense RNA, CopA, carry bulged-out nucleotides. Base pairing in either one or both of these regions of the stem was restored by site-specific mutagenesis and in one case a new internal loop was introduced. The set of mutant and wild-type CopA variants was characterized structurally in vitro. The results reported here indicate a possible function of the bulges: their presence protects CopA RNA from being a substrate for the double-strand-specific enzyme RNase III. In vitro cleavage rates were drastically increased when either the lower or both bulges were absent. This is paralleled by a similar, but not identical, effect of the bulges on metabolic stability of the CopA RNAs in vivo. The degradation pathways of wild-type and mutant CopA in various strain backgrounds are discussed. In the accompanying paper, we address the significance of bulges in CopA for binding to the target RNA in vitro and for its inhibitory efficiency in vivo.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BERTANI G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951 Sep;62(3):293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomberg P., Engdahl H. M., Malmgren C., Romby P., Wagner E. G. Replication control of plasmid R1: disruption of an inhibitory RNA structure that sequesters the repA ribosome-binding site permits tap-independent RepA synthesis. Mol Microbiol. 1994 Apr;12(1):49–60. doi: 10.1111/j.1365-2958.1994.tb00994.x. [DOI] [PubMed] [Google Scholar]
- Blomberg P., Nordström K., Wagner E. G. Replication control of plasmid R1: RepA synthesis is regulated by CopA RNA through inhibition of leader peptide translation. EMBO J. 1992 Jul;11(7):2675–2683. doi: 10.1002/j.1460-2075.1992.tb05333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomberg P., Wagner E. G., Nordström K. Control of replication of plasmid R1: the duplex between the antisense RNA, CopA, and its target, CopT, is processed specifically in vivo and in vitro by RNase III. EMBO J. 1990 Jul;9(7):2331–2340. doi: 10.1002/j.1460-2075.1990.tb07405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Chelladurai B. S., Li H., Nicholson A. W. A conserved sequence element in ribonuclease III processing signals is not required for accurate in vitro enzymatic cleavage. Nucleic Acids Res. 1991 Apr 25;19(8):1759–1766. doi: 10.1093/nar/19.8.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelladurai B., Li H., Zhang K., Nicholson A. W. Mutational analysis of a ribonuclease III processing signal. Biochemistry. 1993 Jul 27;32(29):7549–7558. doi: 10.1021/bi00080a029. [DOI] [PubMed] [Google Scholar]
- Ehresmann C., Baudin F., Mougel M., Romby P., Ebel J. P., Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987 Nov 25;15(22):9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart E., Wagner H., Nordström K. Structural analysis of an RNA molecule involved in replication control of plasmid R1. Nucleic Acids Res. 1986 Mar 25;14(6):2523–2538. doi: 10.1093/nar/14.6.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gornicki P., Baudin F., Romby P., Wiewiorowski M., Kryzosiak W., Ebel J. P., Ehresmann C., Ehresmann B. Use of lead(II) to probe the structure of large RNA's. Conformation of the 3' terminal domain of E. coli 16S rRNA and its involvement in building the tRNA binding sites. J Biomol Struct Dyn. 1989 Apr;6(5):971–984. doi: 10.1080/07391102.1989.10506525. [DOI] [PubMed] [Google Scholar]
- Gough J. A., Murray N. E. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J Mol Biol. 1983 May 5;166(1):1–19. doi: 10.1016/s0022-2836(83)80047-3. [DOI] [PubMed] [Google Scholar]
- Hardt W. D., Schlegl J., Erdmann V. A., Hartmann R. K. Role of the D arm and the anticodon arm in tRNA recognition by eubacterial and eukaryotic RNase P enzymes. Biochemistry. 1993 Dec 7;32(48):13046–13053. doi: 10.1021/bi00211a014. [DOI] [PubMed] [Google Scholar]
- He L., Söderbom F., Wagner E. G., Binnie U., Binns N., Masters M. PcnB is required for the rapid degradation of RNAI, the antisense RNA that controls the copy number of ColE1-related plasmids. Mol Microbiol. 1993 Sep;9(6):1131–1142. doi: 10.1111/j.1365-2958.1993.tb01243.x. [DOI] [PubMed] [Google Scholar]
- Hjalt T. A., Wagner E. G. Bulged-out nucleotides in an antisense RNA are required for rapid target RNA binding in vitro and inhibition in vivo. Nucleic Acids Res. 1995 Feb 25;23(4):580–587. doi: 10.1093/nar/23.4.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjalt T., Wagner E. G. The effect of loop size in antisense and target RNAs on the efficiency of antisense RNA control. Nucleic Acids Res. 1992 Dec 25;20(24):6723–6732. doi: 10.1093/nar/20.24.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittle J. D., Simons R. W., Lee J., Kleckner N. Insertion sequence IS10 anti-sense pairing initiates by an interaction between the 5' end of the target RNA and a loop in the anti-sense RNA. J Mol Biol. 1989 Dec 5;210(3):561–572. doi: 10.1016/0022-2836(89)90132-0. [DOI] [PubMed] [Google Scholar]
- Krinke L., Wulff D. L. The cleavage specificity of RNase III. Nucleic Acids Res. 1990 Aug 25;18(16):4809–4815. doi: 10.1093/nar/18.16.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacatena R. M., Cesareni G. Interaction between RNA1 and the primer precursor in the regulation of Co1E1 replication. J Mol Biol. 1983 Nov 5;170(3):635–650. doi: 10.1016/s0022-2836(83)80125-9. [DOI] [PubMed] [Google Scholar]
- Li H. L., Chelladurai B. S., Zhang K., Nicholson A. W. Ribonuclease III cleavage of a bacteriophage T7 processing signal. Divalent cation specificity, and specific anion effects. Nucleic Acids Res. 1993 Apr 25;21(8):1919–1925. doi: 10.1093/nar/21.8.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters M., Colloms M. D., Oliver I. R., He L., Macnaughton E. J., Charters Y. The pcnB gene of Escherichia coli, which is required for ColE1 copy number maintenance, is dispensable. J Bacteriol. 1993 Jul;175(14):4405–4413. doi: 10.1128/jb.175.14.4405-4413.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström K., Molin S., Light J. Control of replication of bacterial plasmids: genetics, molecular biology, and physiology of the plasmid R1 system. Plasmid. 1984 Sep;12(2):71–90. doi: 10.1016/0147-619x(84)90054-4. [DOI] [PubMed] [Google Scholar]
- Nordström K., Wagner E. G. Kinetic aspects of control of plasmid replication by antisense RNA. Trends Biochem Sci. 1994 Jul;19(7):294–300. doi: 10.1016/0968-0004(94)90008-6. [DOI] [PubMed] [Google Scholar]
- Peattie D. A., Douthwaite S., Garrett R. A., Noller H. F. A "bulged" double helix in a RNA-protein contact site. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7331–7335. doi: 10.1073/pnas.78.12.7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepe C. M., Maslesa-Galić S., Simons R. W. Decay of the IS10 antisense RNA by 3' exoribonucleases: evidence that RNase II stabilizes RNA-OUT against PNPase attack. Mol Microbiol. 1994 Sep;13(6):1133–1142. doi: 10.1111/j.1365-2958.1994.tb00504.x. [DOI] [PubMed] [Google Scholar]
- Persson C., Wagner E. G., Nordström K. Control of replication of plasmid R1: kinetics of in vitro interaction between the antisense RNA, CopA, and its target, CopT. EMBO J. 1988 Oct;7(10):3279–3288. doi: 10.1002/j.1460-2075.1988.tb03195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson C., Wagner E. G., Nordström K. Control of replication of plasmid R1: structures and sequences of the antisense RNA, CopA, required for its binding to the target RNA, CopT. EMBO J. 1990 Nov;9(11):3767–3775. doi: 10.1002/j.1460-2075.1990.tb07590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumbridge J. A., Dondon J., Nakamura Y., Grunberg-Manago M. Effect of NusA protein on expression of the nusA,infB operon in E. coli. Nucleic Acids Res. 1985 May 10;13(9):3371–3388. doi: 10.1093/nar/13.9.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portier C., Dondon L., Grunberg-Manago M., Régnier P. The first step in the functional inactivation of the Escherichia coli polynucleotide phosphorylase messenger is a ribonuclease III processing at the 5' end. EMBO J. 1987 Jul;6(7):2165–2170. doi: 10.1002/j.1460-2075.1987.tb02484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder T. B., Davidson D. B., Rosen J. I., Ohtsubo E., Ohtsubo H. Analysis of plasmid genome evolution based on nucleotide-sequence comparison of two related plasmids of Escherichia coli. Gene. 1982 Mar;17(3):299–310. doi: 10.1016/0378-1119(82)90146-9. [DOI] [PubMed] [Google Scholar]
- Taylor J. W., Ott J., Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. W., Schmidt W., Cosstick R., Okruszek A., Eckstein F. The use of phosphorothioate-modified DNA in restriction enzyme reactions to prepare nicked DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8749–8764. doi: 10.1093/nar/13.24.8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J. Control of ColE1 plasmid replication: the process of binding of RNA I to the primer transcript. Cell. 1984 Oct;38(3):861–870. doi: 10.1016/0092-8674(84)90281-2. [DOI] [PubMed] [Google Scholar]
- Wagner E. G., Simons R. W. Antisense RNA control in bacteria, phages, and plasmids. Annu Rev Microbiol. 1994;48:713–742. doi: 10.1146/annurev.mi.48.100194.003433. [DOI] [PubMed] [Google Scholar]
- Xu F., Lin-Chao S., Cohen S. N. The Escherichia coli pcnB gene promotes adenylylation of antisense RNAI of ColE1-type plasmids in vivo and degradation of RNAI decay intermediates. Proc Natl Acad Sci U S A. 1993 Jul 15;90(14):6756–6760. doi: 10.1073/pnas.90.14.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]