Abstract
Internalin A (InlA; encoded by inlA) facilitates the crossing of the intestinal barrier by Listeria monocytogenes. Mutations leading to a premature stop codon (PMSC) in inlA and thus attenuated mammalian virulence have been reported. We recently characterized 502 L. monocytogenes food isolates from a retail survey and 507 human clinical isolates from multiple U.S. states with respect to the presence/absence of inlA mutations. The objective of this study was to investigate the hypothesis that dose responses for human listeriosis vary between L. monocytogenes strains with and those without a PMSC in inlA. Subtype-specific prevalence and concentration distributions in food, along with epidemiologic and consumption data, were input into established dose-response models to generate an r value (probability of a cell causing illness). Under the conservative assumption that L. monocytogenes levels at retail represent levels consumed, mean log10 r values were −8.1 and −10.7 for L. monocytogenes subtypes with genes encoding a full-length and a truncated InlA, respectively. L. monocytogenes carrying a 5′ frameshift mutation in a homopolymeric tract showed a mean log10 r value of −12.1. Confidence intervals for the r values and their differences varied depending on subtypes. When the increase in concentration of L. monocytogenes subtypes between retail and consumption was considered, mean log10 r values were reduced to −10.4, −13.8, and −12.8 for the subtypes with genes encoding a full-length InlA, for the subtypes carrying a PMSC in inlA, and for all L. monocytogenes isolates regardless of subtype, respectively. Our study provides further quantitative evidence that L. monocytogenes subtypes vary in abilities and relative likelihoods of causing human disease, which were mechanistically related to defined genetic markers.
Listeria monocytogenes continues to represent a major challenge for the ready-to-eat (RTE) food industry due to the ubiquitous presence of this pathogen along the food supply continuum, its food-borne route of transmission, and the exceptionally high hospitalization (90%) and mortality (20 to 30%) rates associated with invasive listeriosis (17, 22, 45). L. monocytogenes isolates can be classified into four genetic lineages, including two common ones termed lineages I and II (43); lineage I isolates have been reported to be overrepresented among isolates from human clinical cases in the United States, while lineage II isolates appear overrepresented among isolates from contaminated foods (13). Studies combining molecular subtyping and in vitro virulence phenotype assays provided initial evidence for heterogeneity in virulence among L. monocytogenes molecular subtypes (e.g., ribotypes) beyond the genetic lineage level (13, 25, 46). Specifically, a subpopulation of highly clonal strains (termed epidemic clones) has been linked to most listeriosis outbreaks worldwide and epidemic clone strains appear to be overrepresented among sporadic listeriosis cases in the United States (13, 17, 40, 42). On the other hand, recent studies support the idea that a significant proportion (approximately 45%) of L. monocytogenes isolates from RTE foods carry unique virulence-attenuating mutations leading to a premature stop codon (PMSC) in inlA (25, 40, 42). The virulence factor internalin A (InlA; encoded by inlA) is sufficient for L. monocytogenes to invade intestinal epithelial cells expressing certain isoforms of the cellular receptor E-cadherin and thus facilitates the crossing of the intestinal barrier during the establishment of a systemic infection (19, 20). Consequently, PMSCs in inlA, which typically lead to production of a truncated and secreted InlA but can also completely abolish InlA production (if mutations occur early in the open reading frame), significantly reduce the ability of L. monocytogenes to invade human intestinal epithelial cells or to cause a systemic infection in a guinea pig model (24, 25, 31).
Substantial progress has been made in the last decade in developing dose-response models for L. monocytogenes. Buchanan et al. (2) used epidemiologic data on the incidence of human listeriosis in Germany and food survey data (from RTE smoked fish) to develop an intentionally conservative exponential dose-response relationship for L. monocytogenes. This approach was also applied by Lindqvist and Westöö (21) to develop an exponential dose-response model using disease incidence and food survey data from Sweden (two RTE food categories) and by Chen et al. (7) using data from the United States (eight RTE food categories). This approach assumes that each L. monocytogenes cell is able to cause disease and generates an r value (the exponential model parameter), which is essentially the probability of illness from a single L. monocytogenes cell. These studies reported log10 r values of −9.93 based on data from Germany (2), −9.25 based on data from Sweden (21), and −9.75 based on the more extensive U.S. data (7). Using essentially the same approach but exposure data for RTE food categories from a 2001 draft risk assessment by the U.S. Food and Drug Administration/Food Safety and Inspection Service (FDA/FSIS) (37), a global exponential dose-response model was developed by a Food and Agriculture Organization/World Health Organization expert panel (11), which yielded a log10 r value of −11.97. In these studies, which used or were based on the exponential modeling approach, a simplifying assumption was made that all L. monocytogenes bacteria represent a single population. More recently, in an initial effort to quantify dose-response relationships for different L. monocytogenes subtypes, Chen et al. (6) showed that the two major L. monocytogenes genetic lineages and ribotypes within those lineages differ significantly in their log10 r values. A comprehensive approach was used in the FDA/FSIS risk assessment (37), where dose-response models were developed for different subpopulations such as the elderly and the perinatal population by using animal (mouse) data calibrated with human surveillance data in the United States.
In parallel efforts, several studies have developed dose-response relationships using animals that express an isoform of E-cadherin that binds InlA, including rhesus monkeys (32, 33) and guinea pigs (1, 39, 47). A log-logistic dose-response model fitted to data obtained from animal experiments with a clinical L. monocytogenes strain isolated from a rhesus monkey listeriosis outbreak resulted in an LD50 (i.e., 50% lethal dose for stillbirth) of 8.45 × 107 cells for pregnant monkeys (32) and 2.0 × 107 cells for pregnant guinea pigs (47). Recently, Van Stelten (39) and colleagues conducted experiments using a male juvenile guinea pig intragastric challenge model and showed ID50s (i.e., doses required for systemic infection in 50% of the subjects) of 2.57 × 107 and 5.50 × 108 for a human clinical epidemic clone strain from a 1998-1999 outbreak of listeriosis in the United States (4) and a food isolate carrying a naturally occurring PMSC mutation in inlA, respectively.
Recognizing that there are insufficient data to build a reliable dose-response model from either human outbreak data or animal experiment data alone, Ross et al. (30) chose the FAO/WHO exponential model as the most plausible dose-response model to conduct a risk assessment of L. monocytogenes in RTE meats in Australia. On the other hand, Williams et al. (48) reported that the risk of fetal mortality after exposure to a dose of 106 L. monocytogenes cells was 5.9 × 10−1 based on the log-logistic model developed from data collected in rhesus monkeys (32), approximately 6 orders of magnitude higher than that predicted using the corresponding dose-response model from the FDA/FSIS risk assessment (37). The investigators (48) applied the model for a clinical strain in monkeys directly to predict risk in humans without addressing factors essential for the extrapolation, such as differences in host susceptibility and variation in virulence among L. monocytogenes strains in RTE foods, which were considered in the FDA/FSIS risk assessment (37). Consequently, the model (48) estimated 733 stillbirths per annum in the U.S. population from the consumption of a single RTE product category, i.e., Mexican-style soft cheese, while the FDA/FSIS risk assessment (37) estimated less than 0.1 case of perinatal listeriosis per year attributable to the consumption of the same product category. Many unanswered questions remain on plausible dose-response relationships for L. monocytogenes, the extrapolation of dose-response curves from animal models to assessment of risk in humans, and whether there are distinct and quantitative differences among L. monocytogenes subtypes, including those with regard to virulence and dose response.
The primary objective of this study was to specifically investigate the hypothesis that dose responses for invasive listeriosis in humans vary among L. monocytogenes subtypes with or without a PMSC mutation in inlA. Furthermore, we made an attempt to quantify the influence of growth between retail and consumption on dose-response curves. We also attempted to derive a dose-response scaling factor by comparing dose-response models developed based on data relevant to humans in this study and those developed in another study using data from experiments in guinea pigs. Building upon published dose-response modeling approaches, findings from this study provide further quantitative evidence that variation exists in L. monocytogenes virulence with respect to humans. This study advances the knowledge of dose responses for L. monocytogenes by using, for the first time, molecular subtype classification based on a biologically meaningful genetic characteristic (i.e., allelic states of the gene encoding the virulence factor InlA) to characterize virulence differences between L. monocytogenes subtypes.
MATERIALS AND METHODS
L. monocytogenes database with enumeration data and inlA subtypes.
A database in Access (2003 version; Microsoft Corp., Redmond, WA), previously developed to tabulate L. monocytogenes presence/absence and enumeration data for 31,707 RTE samples from a 2-year food survey in two FoodNet sites (i.e., Maryland [MD] and northern California [CA]) (12), was further developed to include inlA allelic type data (i.e., presence/absence of 18 inlA PMSC mutations). While a total of 573 food samples were confirmed as L. monocytogenes positive, only 502 L. monocytogenes food isolates were available for characterization. As described in previous studies (6, 12, 40), 42 clinical isolates from sporadic listeriosis cases during the 2-year time frame of the food survey were obtained from the same FoodNet sites; these and an additional 465 human isolates from multiple other states (for a total of 507 human isolates) were used for this study.
Human and food L. monocytogenes isolates used here were previously characterized (40) by a multiplex single nucleotide polymorphism (SNP) genotyping assay to interrogate the presence/absence of 18 PMSC mutations in inlA that have been described worldwide to date. Seven of these 18 previously described PMSC mutations in inlA (i.e., PMSC mutations types 1, 2, 3, 4, 5, 6, and 12) were observed among the 502 food isolates and 507 clinical isolates. Based on these previously reported data, we created several tables in the Access database to further describe all isolates, including the presence/absence of a PMSC, the predicted length of the truncated InlA protein, the PMSC type as reported by Van Stelten et al. (40), and the isolate source. These new data tables were linked, through a common identification field, to presence/absence and enumeration data in previously created tables (6). The resulting Access database allowed for queries specific for data subsets based on inlA subtype data, e.g., the levels found in foods for L. monocytogenes isolates with a specific PMSC (such as PMSC type 4).
Subgrouping of L. monocytogenes isolates according to inlA allelic types.
inlA SNP genotyping data were also used to classify the isolates in two broad subgroups: (i) L. monocytogenes isolates with genes encoding a full-length InlA (without a PMSC in inlA) and (ii) L. monocytogenes isolates with genes encoding a truncated InlA (with a PMSC in inlA). Among the seven inlA PMSCs, PMSC type 4 is biologically unique because this is a frameshift mutation that occurs in a 5′ homopolymeric tract of adenine residues, which appears to represent a mutational hot spot that may facilitate a switch between inlA alleles encoding full-length and truncated InlA proteins, similar to phase shifting (29). As L. monocytogenes isolates carrying inlA PMSC type 4 may revert to an inlA allelic type encoding a full-length InlA (29), this inlA PMSC mutation was classified separately. Specifically, L. monocytogenes isolates containing an inlA PMSC were further divided into subgroups carrying (i) inlA PMSC type 4 or (ii) all other types of inlA PMSCs found among the isolates used here (i.e., PMSC types 1, 2, 3, 5, 6, and 12).
Mathematical considerations for inlA subtype-specific dose-response analysis.
To establish an inlA subtype-specific dose-response relationship, we used the inlA SNP typing data for food and clinical isolates as detailed above, along with food survey data on the prevalence and concentration of L. monocytogenes in eight food categories, epidemiological data on the incidence of human listeriosis, and consumption data (i.e., serving number and serving size) as reported in our previous study (7). The number of listeriosis cases was stratified and expressed as a mathematical function of exposure to inlA allelic type, as were the prevalence and concentration of L. monocytogenes in RTE foods. The food survey data generated in the previous retail survey (12) were used as a reasonable approximation of human exposure to L. monocytogenes in the sample population (i.e., the CA and MD FoodNet sites) and, by extrapolation, the U.S. population. In the food survey, sampling was weighted by the population within each of the two FoodNet sites and efforts had been made to sample products in proportion to consumption. To develop a dose-response relationship for inlA allelic types with or without a PMSC, we assumed that the number of human listeriosis cases was the measure of risk during the 2-year time period in which the food survey was conducted. Isolates representing specific inlA allelic types found in RTE foods and among clinical listeriosis cases, their prevalence and concentration in RTE foods, and the relative proportion of inlA allelic types associated with illness cases were determined using methods previously described (6), except that the subgrouping was based on classification into inlA allelic types.
Dose-response analysis was conducted using two approaches reported previously, including (i) an exponential modeling approach first described by Buchanan et al. (2) and (ii) a conditional probability approach that we described in our previous study (6). Briefly, for the exponential modeling approach, the probability that illness occurs given an exposure at dose d to subgroup or subtype k is denoted Pk,d. This probability can be expressed as
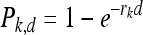 |
(1) |
where the virulence parameter rk is specific to inlA allelic type and reflects the sensitivity of the exposed population to inlA allelic type with or without a PMSC. A model previously developed to quantify the r value for L. monocytogenes regardless of subtype (7) was updated to quantify rk together with the attendant uncertainty using data specific to subtype k (i.e., subtype encoding a full-length or truncated InlA) as input. As part of the analysis, we also determined r (the r value for L. monocytogenes regardless of subtype) as an internal control to verify the algorithms and to generate PI, the probability of illness from exposure to L. monocytogenes regardless of subtype, for use in the analysis using the conditional probability approach.
For the conditional probability approach, we used the same method that we had reported previously for attributing risk to L. monocytogenes lineages and ribotypes. Briefly, based on various mathematical considerations, the probability of illness given exposure to inlA allelic type or subtype k, PI/k, can be expressed as a conditional probability
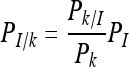 |
(2) |
where Pk/I, Pk, and PI are the probability of exposure to inlA allelic type or subtype k given that illness has occurred (i.e., the fraction of illness cases associated with the subgroup or subtype), the probability of exposure to subgroup or subtype k (i.e., fraction of exposures from foods attributed to the subgroup or subtype), and the probability of illness from exposure to L. monocytogenes regardless of subtype, respectively. These terms and attendant uncertainties were constructed from observed data. For L. monocytogenes, the exponential dose-response model is assumed to be linear over most of the range of exposures. Therefore, a simple approximation results in Pk,d ≅ rkd. Using this approximation together with the fact that PI/k can be obtained as the average of Pk,d over the exposure distribution for subgroup k yields the approximation PI/k ≅ rkμk, where μk is the average ingested dose for a subgroup. In this case, substitution for equation 2 resulted in
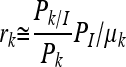 |
(3) |
where rk (essentially the probability of a single cell of a given subgroup or subtype causing illness) is the subtype-specific virulence parameter—a quantitative measure of the degree to which variations in virulence among inlA allelic types may exist. In classifying L. monocytogenes into two broad subgroups, i.e., subgroups with or without a PMSC in inlA, we made the assumption of homogeneous virulence for all isolates within the same subgroup. In the case of no subgrouping, this approximation reduced to r ≅ PI/μ. A model that we previously developed for lineages and ribotypes was updated to quantify rk together with the attendant uncertainty using data specific to inlA subgroups or subtypes associated with inlA PMSC mutations as input, where the approximation for r was included as an internal control to verify the algorithms. Estimation of confidence intervals for the individual inlA subgroups and the determination of the differences in virulence potential between two inlA subgroups were quantified using the conditional probability approach according to the equations and method that we reported previously (6).
Relative proportion of L. monocytogenes inlA subgroups in RTE foods or associated with listeriosis.
We determined the probability of exposure to a specific L. monocytogenes inlA allelic type given exposure to the pathogen in RTE foods based on observed prevalence in food samples collected at retail. For example, among the 502 food isolates, 227 isolates carried a PMSC in inlA (Table 1). This resulted in a fraction of 0.452 for Pk. The fraction values for the subgroups represent the parameters of a multinomial distribution, which through simulation was used to determine the uncertainties around prevalence estimates for inlA subgroups. Similarly, the proportions of human isolates associated with the inlA subgroups formed the basis for calculating Pk/I. The uncertainties about the estimates of Pk/I for the inlA subgroups associated with listeriosis cases were derived from the properties of a multinomial distribution through simulations, as described previously (6).
TABLE 1.
L. monocytogenes isolates found in RTE foods and in humans for different internalin A (InlA, encoded by inlA) subgroups
| Molecular subgroup | No. (%) of food isolates | No. (%) of human isolatesa |
|
|---|---|---|---|
| FoodNet sites | Multistate | ||
| inlA without PMSC | 275 (54.8) | 33 (78.6) | 481 (94.9) |
| inlA with PMSC | 227 (45.2) | 9 (21.4) | 26 (5.1) |
| inlA with PMSC4b | 33 (6.6) | 2 (4.8) | 4 (0.79) |
| inlA with PMSCnot4c | 194 (38.6) | 7 (16.7) | 22 (4.3) |
| Total no. of isolatesd | 502 | 42 | 507 |
The human isolates for two FoodNet sites were obtained from Maryland and northern California in 2000 and 2001; those from multiple states included the isolates from the two FoodNet sites as well as human clinical isolates obtained from New York, Connecticut, Ohio, Michigan, and a few other states primarily from 1997 through 2002 (see the work of Van Stelten et al. [40] for details).
PMSC4 represents one of the seven inlA subtypes found among the food and human isolates used in this study.
PMSCnot4 included isolates with PMSC types 1, 2, 3, 5, 6, and 12.
Total number of isolates (with and without premature stop codons [PMSCs] in inlA).
Concentration of L. monocytogenes subgroups in RTE foods.
We used two sets of concentration data as input: (i) the concentration of L. monocytogenes in RTE foods at retail and (ii) the concentration at consumption. Subgroup-specific concentration distributions were determined according to the method described in our study reported in 2006 (6). Briefly, food samples found to contain a specific L. monocytogenes subgroup were grouped into concentration ranges at 1-log intervals (Table 2). The average dose per serving (for individual subgroup, μk, or for L. monocytogenes regardless of subtypes, μ) was calculated based on the fraction of isolates in each interval, concentration corresponding to the midinterval value, and serving size. As shown in equation 3, the average dose per serving was used as an input to derive subtype-specific r values.
TABLE 2.
Number of food samples that contained different concentrations of L. monocytogenes separated by inlA subgroups
| Concn interval (log CFU/g) | No. of samples by subtype |
|||
|---|---|---|---|---|
| inlA without PMSC | inlA with PMSC | inlA with PMSC4 | All positive samplesa | |
| −2-−1 | 188 | 146 | 18 | 398 |
| −1-0 | 38 | 38 | 4 | 81 |
| 0-1 | 25 | 26 | 6 | 53 |
| 1-2 | 13 | 7 | 1 | 20 |
| 2-3 | 10 | 6 | 2 | 16 |
| 3-4 | 1 | 2 | 0 | 3 |
| 4-5 | 0 | 0 | 0 | 0 |
| 5-6 | 0 | 2 | 2 | 2 |
| Avg doseb (CFU/serving) | 1.76 × 103 | 2.00 × 105 | 1.36 × 106 | 8.03 × 104 |
The data for all the positive samples from the RTE food survey reported by Gombas et al. (12) are included for comparison. There were a total of 573 L. monocytogenes-positive samples among the 31,707 food samples tested; isolates from 502 samples were obtained and subjected to inlA subtyping.
The average dose per serving, given detection of L. monocytogenes in the RTE food survey, was determined from the discrete frequency data for each subgroup by using the mid-bin value for each interval and Weibull (1.40, 76.60) + 1.18 as the distribution for serving size.
Alternatively, we also used an exponential modeling approach to derive r values using the modeling framework that we reported in 2003 (7). A discrete concentration distribution was developed for the individual inlA subgroups by simulation and used as an input to derive concentration per serving and subsequently subtype-specific r values. As a control, we also derived an r value for L. monocytogenes regardless of subtype using the distribution beta (0.29, 2.68, −1.69, 6.10) as an input for concentration, as was done previously (6). We used the exponential modeling approach to determine an r value with or without considering L. monocytogenes growth between retail and consumption. To determine the concentration at consumption, the amount of growth (see growth model below) estimated between retail and consumption for each inlA subgroup and for all L. monocytogenes strains was added to the corresponding concentration at retail. Growth was estimated separately for two groups of isolates, i.e., L. monocytogenes with and without inlA PMSCs, as described in more detail below.
Estimating the growth between retail and consumption.
We implemented a growth model to estimate the amount of growth for L. monocytogenes with and without an inlA PMSC. This approach improves the dose-response analysis beyond the conservative assumption that we had made in previous studies (6, 7), that L. monocytogenes levels at retail represented levels to which the consumer was exposed. The amount of growth specific to inlA subtype was determined separately for each of the eight product categories: bagged leafy vegetable salads, fresh soft cheeses, soft ripened cheeses (soft mold-ripened cheeses), semisoft cheeses (blue-veined cheese), smoked seafood, deli salads, seafood salads, and luncheon meats.
Growth was determined using data reported in the literature on storage time, lag time, storage temperature, and growth rate. Storage time was obtained from a reported survey (3) to determine nationally representative consumer storage practices for refrigerated RTE foods. The combined storage times reported by Cates et al. (3) for smoked seafood, deli salads, deli meats, and soft cheeses were used for these product categories; “combined storage time” is the time for which a given product is stored unopened plus the additional time for which it is stored after opening. A distribution for storage time for the product categories other than bagged salads was obtained by simulation, which was designed to accommodate the format in which the combined (unopened and opened) storage time data were reported. Cates et al. (3) reported the weighted percentage of respondents who stored the product for ≤5, 6 to 7, 8 to 14, 15 to 21, or >22 days. Each time range (number of days) was represented by a uniform distribution in the growth model. A simulation was performed by sampling from all the bin ranges according to the reported percentage of respondents in the individual ranges. For bagged salads, we determined the combined (unopened and opened) storage time with the same method as that used for the other product categories (S. C. Cates, personal communication), using raw data in the “survey data” spreadsheet downloaded from the Joint Institute for Food Safety and Applied Nutrition website (http://www.foodrisk.org/exclusives/CSPRRTEF/index.cfm, accessed March 2010). Using a method previously described by Duffy and Schaffner (9) and Montville et al. (23), we found that the distribution [Weibull (1.43, 4.97) + 1.26] adequately described the storage time for bagged salads. To determine the time in which growth occurs, for all product categories, a lag time was subtracted from the storage time distributions. Lag time estimates were generated from simulations using lag time data reported by De Jesús and Whiting (8). The lag time (hour) distributions for inlA allelic types with or without a PMSC were represented by normal (1.61, 0.42) and normal (2.88, 0.79), which were derived from proxy data reported for lineage II and lineage I isolates, respectively. This approach is justified as lineage II isolates contain a high proportion of isolates with inlA PMSCs (58.5% of lineage II food isolates in the data set used here had inlA PMSCs), while lineage I isolates contain comparatively fewer isolates with inlA PMSCs (23.5% of lineage I food isolates in the data set used here had inlA PMSCs).
The temperature distribution was derived from EcoSure data for home refrigerator temperatures (available at http://foodrisk.org/exclusives/EcoSure/, accessed March 2010) and was represented by the distribution normal (3.55, 2.02) (temperature unit in °C). The exponential growth rate inputs were obtained from the 2003 FDA/FSIS L. monocytogenes risk assessment (37) and were represented by the distributions shown in Table 3. The same growth rates were applied regardless of subtype (i.e., L. monocytogenes with or without a PMSC). The 2003 FDA/FSIS risk assessment (37) included three choices for maximum growth, i.e., 5.0, 6.5, and 8.0 log10 CFU/g for scenarios with storage temperatures of <5.0, 5.0 to 7.0, and >7.0°C, respectively. In this study, although the storage temperature distribution included values above 5.0°C, we set the maximum amount of growth to 5.0 log10 CFU/g for all subgroups as a simplified approach. This allowed us to assess the potential impact of growth consideration on the dose-response analysis without the elaborate modeling of correlation between growth and temperatures that was done in the FDA/FSIS model (37).
TABLE 3.
Growth rate input variables for the growth modulea
| Product category | Exponential growth rate |
|---|---|
| Bagged salads | Uniform (−0.0374, 0.138) |
| Fresh soft cheeses | Beta (0.511, 0.742, −0.0816, 0.324) |
| Soft ripened cheeses | Uniform (−0.099, 0.11) |
| Semisoft cheeses | Normal (−0.043, 0.032) |
| Smoked seafood | Triangular (0.0519, 0.093, 0.27) |
| Deli salads | Uniform (0.1, 0.143) |
| Seafood salads | Uniform (0.1, 0.143) |
| Luncheon meats | Triangular (−0.0113, 0.0785, 0.755) |
Data obtained from the FDA/FSIS risk assessment (37).
Concentration distributions for the amount of growth between retail and consumption for the individual product categories were used to estimate growth for L. monocytogenes with or without a PMSC in inlA, as well as for L. monocytogenes regardless of inlA allelic type, according to the number of isolates belonging to each product category (Table 4). A concentration distribution for the increase in L. monocytogenes levels between retail and consumption for the subgroup comprised of L. monocytogenes with a PMSC in inlA was obtained by sampling the eight concentration distributions based on the number of isolates with a PMSC in inlA in a given RTE food product category. Similarly, a concentration distribution with growth was determined for the subgroup containing L. monocytogenes without a PMSC and for all L. monocytogenes isolates, regardless of subtype. Concentration distributions for the amount of growth between retail and consumption and the initial level of contamination for each subgroup were summed through simulation to yield the concentration at time of consumption.
TABLE 4.
L. monocytogenes inlA subtypes in various food categories
| Food category | No. of samples |
||
|---|---|---|---|
| inlA without PMSCs | inlA with PMSCs | Combined | |
| Bagged salads | 12 | 9 | 21 |
| Fresh soft cheeses | 2 | 2 | 4 |
| Soft ripened cheeses | 4 | 8 | 12 |
| Semisoft cheeses | 10 | 6 | 16 |
| Smoked seafood | 61 | 42 | 103 |
| Deli salads | 107 | 66 | 173 |
| Seafood salads | 48 | 56 | 104 |
| Luncheon meats | 31 | 38 | 69 |
| Total | 275 | 227 | 502 |
Dose-response models and simulations.
The dose-response models that integrated the various input variables were updated (for the parameters described in equations 1, 2, and 3 above) or newly developed (for the growth model) in Analytica (version 4.1; Lumina Decision Systems, Los Gatos, CA). The two modeling approaches were used as alternative approaches to quantify rk and r values. For the model with input variables for the exponential modeling approach, r value and confidence interval results were obtained by simulation runs, in which each run to determine rk and r values consisted of uncertainty samples of 30,000 iterations (10 trials). For the model with input variables for the conditional probability approach, results for r value, confidence interval, and the difference in r value between the subgroups were obtained by simulation runs in which each run to determine rk and r values consisted of uncertainty samples of 1,000 iterations (10 trials), using the standard random Latin Hypercube as part of the uncertainty sample option (41). In the simulations, we found very small variation in the results among the 10 trials obtained in each run, e.g., a 0.01-log difference among the r values from the 10 trials in each run for the subgroup of isolates carrying a PMSC in inlA (with growth considered using the exponential modeling approach).
RESULTS AND DISCUSSION
A better quantitative understanding of the degree to which L. monocytogenes subtypes differ in their abilities to cause human illness is critical to allow for accurate risk assessment of public health burden. In this study, we employed a conditional probability approach and built upon established modeling frameworks to enable the integration into a dose-response analysis of inlA molecular subtyping data for food and human isolates along with L. monocytogenes contamination levels and food survey data, epidemiologic data, and consumption data. We found that dose responses for invasive listeriosis in humans varied by several orders of magnitude among L. monocytogenes subtypes with or without a PMSC mutation in inlA.
L. monocytogenes inlA subtypes in RTE food and clinical isolates.
While the prevalence of L. monocytogenes isolates carrying an inlA PMSC was 45.2% among the RTE food isolates characterized here, the prevalence of isolates with an inlA PMSC was much lower among human clinical isolates: 21.4% of human isolates from CA and MD FoodNet sites and 5.1% of human isolates from the larger clinical isolate pool from multiple states were found to carry inlA PMSCs (Table 1) (40). The number of L. monocytogenes isolates with or without a PMSC among the RTE foods included in the food survey (12) varied depending on the individual food category, while there was no clear pattern for higher or lower prevalence of isolates with or without an inlA PMSC among the eight food categories (Table 4) (40). In our previous study (6), in which the same isolates were classified based on different subtyping methods (13), lineage II isolates represented 62.3% of the food isolates and 54.8% and 42.9% of the clinical isolates from the FoodNet sites and the multistate isolate pool, respectively. Interestingly, isolates with a PMSC in inlA are more common among lineage II isolates than among lineage I isolates, which is consistent with a recent study published by Ward et al. (42). This observation may at least partially explain why lineage II strains are less commonly found among human clinical isolates than expected based on their observed frequency among RTE foods (and hence based on the frequency of human exposure to lineage II isolates) (13). While lineage classification used in our previous work (6, 13) does not involve direct interrogation of genetic variation in loci that contribute to virulence, SNP genotyping to detect the presence/absence of 18 mutations leading to a PMSC in inlA (40) allowed us to classify L. monocytogenes isolates into subgroups that are mechanistically related to virulence. A dose-response analysis that uses classification of L. monocytogenes isolates based on inlA allelic types and the presence/absence of inlA PMSC in particular should thus lead to an improved understanding of virulence differences among L. monocytogenes subtypes and serve as a model for dose-response analyses for other food-borne pathogens where strain differences appear to be important (e.g., Shiga toxin-producing Escherichia coli).
The concentration of L. monocytogenes, given that it was detected in an RTE food sample, varied for isolates with and without a PMSC in inlA (Table 2). An average dose of L. monocytogenes per serving of RTE food for inlA subgroups was determined based on the number of isolates in the various concentration ranges using the method that we developed previously (6). The subgroup containing L. monocytogenes isolates with a PMSC in inlA showed an average dose of CFU/serving (2.00 × 105) 2 orders of magnitude higher than that of the subgroup of isolates without an inlA PMSC (1.76 × 103). In our previous study (6), we found that the concentration per serving differed by approximately 2 orders of magnitude between lineage II (1.47 × 105) and lineage I (6.92 × 102) isolates. Interestingly, isolates carrying inlA PMSC type 4 were characterized by an average dose of 1.36 × 106 CFU/serving, which was similar to the concentration found for isolates with ribotype DUP-1039C (1.29 × 106 CFU/serving) from our previous study and consistent with the finding that a considerable proportion (36.1%) of ribotype DUP-1039C isolates carry inlA PMSC type 4 (40).
Exponential model r value for L. monocytogenes inlA subtypes.
Using the conditional probability modeling approach, the average r value on a log scale for the subgroup without PMSCs was −8.13 (confidence interval [CI], −8.68 to −7.58). The r value for the subgroup with inlA PMSCs was −10.68 (CI, −11.06 to −10.30), more than 2 orders of magnitude lower (Table 5; illness and clinical isolate data from MD and CA as an input). When we further divided the subgroup with PMSC into two smaller subgroups, the subgroup with PMSC type 4 had an average log10 r value of −11.30 (Table 5). While inlA PMSC type 4 has the potential to revert to a wild-type inlA allelic type (as PMSC4 is a frameshift mutation in a homopolymeric tract in the signal sequence of InlA [29]), it also is the only inlA PMSC mutation that completely disrupts InlA production; all other PMSCs lead to the production of truncated and secreted forms of InlA (16, 24, 28, 40).
TABLE 5.
Virulence parameter for L. monocytogenes inlA subgroups using the conditional probability modeling approach
| Molecular subgroup | r value (log)b | Confidence interval (95%)b |
|---|---|---|
| inlA without PMSCs | −8.13 (−8.04) | −8.68-−7.58 (−8.59-−7.50) |
| inlA with PMSCs | −10.68 (−11.29) | −11.06-−10.30 (−11.59-−10.98) |
| inlA with PMSCnot4 | −8.88 (−9.45) | −10.01-−7.76 (−10.54-−8.36) |
| inlA with PMSC4 | −11.30 (−12.10) | −INF-NANc (−12.50-−11.70) |
| Combined modela | −9.94 | −9.66-−10.23 |
Values for L. monocytogenes regardless of subtype.
Values were obtained based on human illness and clinical isolate data from the MD and CA FoodNet sites (outside parentheses) and from multiple U.S. states (inside parentheses).
INF, infinite; NAN, not a number. These outputs were obtained in simulation because 0 was sometimes obtained in the distribution of the number of cases attributed to isolates with inlA PMSC4.
As shown in equation 3, the linear approximation to the dose-response function leads to an estimate of the r value for a subgroup or subtype k as a function of μk, the average ingested dose for a subgroup or subtype. Because of the large differences in μk among the subgroups (Table 2), this term is highly influential in determining differences in their r values (Table 5). It is important to consider sampling design and sources of variability in the interpretation of these results. For example, the large μk for the subgroup with inlA PMSCs was partly the result of two samples in which the L. monocytogenes level detected was 5 to 6 log10 CFU/g (Table 2). The magnitude of the two samples is unique in the survey. Given the size and extent of the survey (31,707 RTE food samples that spanned 2 years in CA and MD), the fact that the two samples occur in the same subgroup cannot be overlooked. Since there is no additional information that would lead to discounting these two samples, they were factored into the calculation of μk, which was used an input in equation 3 to derive the r value for the subgroup to which they belonged.
Keeping all other inputs the same and using data for a larger set of human illnesses across the United States and clinical isolates from multiple states, rather than data for human illnesses and clinical isolates from only the CA and MD FoodNet sites, the average r values, on a log10 scale, for the subgroups without inlA PMSCs, with inlA PMSCs, and with inlA PMSC type 4 were −8.04, −11.29, and −12.10, respectively (Table 5), further supporting a considerably (order of magnitude) lower likelihood of L. monocytogenes isolates with inlA PMSCs causing human disease. Interestingly, the r value for PMSC4 was substantially lower than the r value for the isolates with the other PMSCs (“PMSCnot4” in Table 5), suggesting that PMSC4 may show a particularly high level of virulence attenuation (e.g., due to complete lack of InlA expression, as the mutation leading to PMSC type 4 is localized early into the inlA open reading frame).
Similar r values were obtained when we applied the exponential modeling approach to the dose-response analysis. The r values obtained were, on a log10 scale, −8.12 and −10.67 for subgroups without and with inlA PMSCs, respectively (Table 6). Confidence intervals associated with the r values derived using the exponential modeling approach were not as wide as those derived using the conditional probability approach, which may likely be attributed to differences in the methods used to determine these intervals (6, 7). As an internal control to verify the algorithm for both modeling methods, average r values of −9.94 log (Table 5) and −9.44 log (Table 6) were obtained for L. monocytogenes regardless of subtype; these were similar to the values that we reported previously: −9.75 log using the exponential modeling approach (7) and −9.72 log using the conditional probability approach (6).
TABLE 6.
Virulence parameters for L. monocytogenes inlA subgroups using the exponential modeling approacha
| Molecular subgroup | No growth considered between retail and consumption |
Growth considered between retail and consumption |
||
|---|---|---|---|---|
| r value (log) | Confidence interval (95%) | r value (log) | Confidence interval (95%) | |
| inlA without PMSCs | −8.12 | −8.15-−8.06 | −10.44 | −10.53-−10.38 |
| inlA with PMSCs | −10.67 | −10.75-−10.61 | −13.76 | −13.84-−13.69 |
| Combined modelb | −9.44 | −9.52-−9.37 | −12.76 | −12.85-−12.69 |
Values were obtained based on human illness and clinical isolate data from the MD and CA FoodNet sites.
Values for L. monocytogenes regardless of subtypes for comparison.
Variation in virulence potential for L. monocytogenes inlA subgroups.
The exponential model parameter r value is a measure of virulence for L. monocytogenes, as this parameter essentially predicts the likelihood of a single cell causing listeriosis, i.e., a systemic infection (all dose-response analysis reported here was performed using systemic listeriosis as a disease outcome) (2, 6, 7). Differences in virulence for L. monocytogenes subgroups, as defined by the presence/absence of PMSC mutations in inlA, were thus further quantified by comparing the r values for the different subgroups, using the conditional probability modeling approach (6). There was a 2.55-log (95% confidence interval, 1.91 to 3.20) difference in the average r values for L. monocytogenes subgroups with and without inlA PMSCs; this difference was 3.24 logs (95% confidence interval, 2.62 to 3.86) when data for human illness and clinical isolates from multiple states rather than data from the CA and MD FoodNet sites were used as an input (Table 7). When the subgroup of L. monocytogenes isolates carrying inlA PMSC type 4 was compared to the subgroup of isolates with genes encoding a full-length InlA, the difference in the average r value was infinite when data for human illness and clinical isolates from the FoodNet sites were used as an input and 4.06 log when data for human illness and clinical isolates from multiple states were an input.
TABLE 7.
Comparison of virulence parameters
| Molecular subgroups compared | r value difference (log)a | Confidence interval (95%)a |
|---|---|---|
| inlA without PMSC vs inlA with PMSC | 2.55 (3.24) | 1.91-3.20 (2.62-3.86) |
| inlA without PMSC vs inlA with PMSCNot4 | 0.76 (1.40) | −0.48-1.99 (0.18-2.26) |
| inlA without PMSC vs inlA with PMSC4 | INFb (4.06) | NAN-INFb (3.38-4.73) |
Values were obtained based on human illness and clinical isolate data from the MD and CA FoodNet sites(not in parentheses) and from multiple U.S. states(in parentheses).
INF, infinite; NAN, not a number. These outputs were obtained in simulation because the number of cases could be 0 in the distribution of the number of cases attributed to isolates with inlA PMSC4.
Under the assumptions made, this study clearly demonstrated that there are quantifiable differences in virulence parameters among L. monocytogenes strains with or without PMSCs in inlA. These findings, which are based on a biological marker that can be mechanistically related to virulence (i.e., inlA premature stop codons) (25), are consistent with our previous findings that were based on broader genetic classification of L. monocytogenes, i.e., classification into lineages and ribotypes (6, 13). For example, the average differences in r values using MD and CA (or multistate) illness data as an input were 2.42 logs (or 2.65 logs) between lineage I and lineage II and 3.53 logs (or 4.19 logs) between ribotype DUP-1062A, with all isolates classified into this ribotype carrying inlA PMSCs (40), and DUP-1042B, a ribotype that has been linked to multiple human listeriosis outbreaks (13) and shows a low frequency (i.e., 3.3%) of inlA PMSCs (40). The overlapping results obtained from dose-response analyses using subgrouping data from the different subtyping methods support the biological relevance of the lineage and ribotype classification schemes. Furthermore, continuing improvement in the molecular subtyping tools for L. monocytogenes, moving toward more biologically meaningful classification based on virulence factors, such as those developed by Van Stelten and Nightingale (38) and Ward et al. (43), clearly increases the value of subtyping data in developing dose-response models. Collectively, results from this study and our previous study (6) suggest the existence of true quantifiable differences in pathogenic potential among L. monocytogenes subtypes.
Influence of growth consideration on r value.
The r values described above were derived under the assumption that L. monocytogenes levels found in the RTE samples in the food survey at retail were the levels at consumption that resulted in the illness cases reported. As expected, when postretail growth was considered in dose-response analysis, the r values obtained were lower than the r values derived when postretail growth was not considered (Table 6). Because exponential growth rate data were available for L. monocytogenes in general (37) and lag time data were available only for the common and broad L. monocytogenes genetic lineages (8), we modeled growth only for the two major subgroups with or without inlA PMSCs and did not further model growth for individual inlA subtypes. The data for lineages were used as proximate lag time because queries from our database, which included information on the lineage and inlA characteristics for individual isolates, showed that lineage I and II isolates contain a high proportion of isolates without inlA PMSCs (76.5%) and with inlA PMSCs (58.5%), respectively. As an initial attempt to quantify the impact of considering postretail growth on the r value, we chose the CA and MD FoodNet data subset for the analysis. When growth after retail was considered, the mean r values, on a log10 scale, were reduced by approximately 2 to 3 orders of magnitude, e.g., from −8.12 to −10.44 for the subgroup without inlA PMSCs, from −10.67 to −13.76 for the subgroup with inlA PMSCs, and from −9.44 to −12.76 for all L. monocytogenes strains regardless of subtype (Table 6). The difference in the r values with postretail growth consideration will influence risk-per-serving determination as well as the predicted degree of risk reduction when evaluating control measures that affect changes in L. monocytogenes concentration.
The median log10 r value obtained in this study for L. monocytogenes regardless of subtype, with growth considered between retail and consumption, was −12.75 logs, corresponding to an r value of 1.78 × 10−13 (by comparison, as described in Table 6, the mean log10 r value was −12.76). The median r value obtained from this study (i.e., the r value of 1.78 × 10−13) is essentially the same as the median r value of 1.34 × 10−13 for the population with increased susceptibility reported by an expert panel (11), in which growth was considered between retail and consumption, a “multiple-dose” approach was used to derive the r value, and the maximum dose was assumed to be 10.5 log CFU/serving. The FAO/WHO expert panel also reported a median r value of 1.06 × 10−12 for the population with increased susceptibility, which was derived assuming that all cases of listeriosis were due to ingestion of servings only at the highest dose level and assuming that the maximum dose varied from 7.5 to 10.5 log CFU/serving. Ross et al. (30) chose the median r value of 1.06 × 10−12 as a plausible dose-response model in their recently published risk assessment of L. monocytogenes in ready-to-eat meats in Australia.
The r values derived in this study and in the FAO/WHO study were both for the population with increased susceptibility, which we assumed to include 25% of the U.S. population (7) and the expert panel assumed to be 17.5% of the population (11). In this study, we quantified for the first time the r value with growth considered for two distinct molecular subgroups and found an average r value of 3.63 × 10−11 (i.e., log10 r value of −10.44) for the subgroup without inlA PMSCs and 1.74 × 10−14 (i.e., log10 r value of −13.76) for the subgroup with inlA PMSCs, further supporting a reduced likelihood of causing human disease for L. monocytogenes strains with PMSCs in inlA.
Emerging evidence for a dose-response scaling factor from guinea pigs to humans.
In this study and our previous studies, we developed a dose-response relationship through the use of data relevant to humans, including food survey data, epidemiologic data, and consumption data, as well as molecular subtyping data for food and human clinical isolates. In parallel efforts, dose-response models have been developed for L. monocytogenes using data from experiments in guinea pigs (32, 39). Generally, experiments in animals are conducted over a range of inoculum doses not typical of human exposures from RTE foods. Experiments with animals are designed to target dose levels of 105 to 108 log10 CFU, corresponding to a high probability of infection or mortality in the animals (e.g., 30 to 70% response rates). On the other hand, sporadic exposures of the human population to L. monocytogenes predominantly occur at low dose levels. For example, among the 573 positive samples obtained from the survey of 31,707 RTE food samples (12), 93% of the isolates had levels at or below 10 CFU/g. Difficulties are well known in extrapolating dose-response models derived from animal studies to low doses typical of human exposures from foods (11, 32, 34, 36, 47).
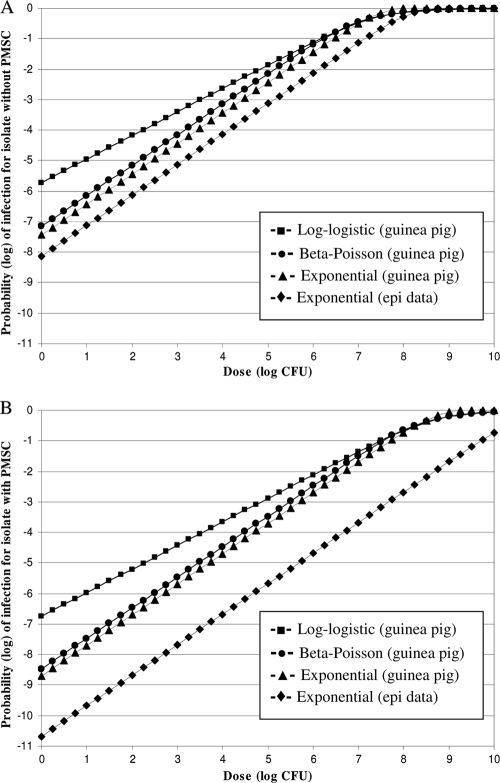
In an attempt to obtain insight into a dose-response scaling factor from guinea pigs to humans, we plotted and compared the inlA subtype-specific exponential dose response models derived from this study and models derived from data in guinea pigs (Fig. 1). The inlA subtype-specific dose-response models derived using data relevant to humans centered on the r value and the exponential model. The study by Van Stelten and colleagues using guinea pigs (39; A. Van Stelten, J. M. Simpson, Y. Chen, V. N. Scott, R. C. Whiting, W. H. Ross, and K. K. Nightingale, submitted for publication) showed that three dose-response models (log-logistic, beta-Poisson, and exponential) provided acceptable fit for the probability of spleen infection in response to an L. monocytogenes dose administered to guinea pigs; all three models predicted similar likelihoods of infection in the range of doses above 6 log CFU (Fig. 1). Similarly, Smith et al. (32) used three models to fit stillbirth mortality data in monkeys and reported that at a low dose with ≤1% response rates, the different models (including log-logistic, Weibull gamma, and exponential) diverged but the data were not sufficient to justify selection of a better fit among the models. In the dose-response study with guinea pigs by Van Stelten (39) and colleagues, the beta-Poisson and exponential models showed closely parallel probabilities of contracting a systemic infection at the same dose; however, a significant departure occurred for the log-logistic model when extrapolating to low doses (Fig. 1). A key difference in the log-logistic model, compared to the exponential and beta-Poisson models, is that it is not linear (on a log-log scale characterized by a slope that is not 1; for the data in Fig. 1, the slope is less than 1) at low doses, while the latter two are (characterized by a slope of 1 at a low dose such as <6 log CFU).
FIG. 1.
Dose-response models for L. monocytogenes systemic infection derived from human and food epidemiological data and models derived from guinea pig experiments. Guinea pig results were from the work of Van Stelten (39). Results from the exponential model obtained from epidemiological data (⧫) in the study reported here are shown for the subgroup without inlA PMSCs (A) and for the subgroup with inlA PMSCs (B). For comparison, three models (beta-Poisson, log-logistic, and exponential) that provided acceptable fit to L. monocytogenes numbers recovered from infected guinea pigs are shown; guinea pig dose-response curves for an L. monocytogenes isolate encoding a full-length InlA (isolate CUFSL N1-054, H7550; obtained from a human case associated with the 1998-1999 listeriosis outbreak in the United States reported by the CDC [4]) are shown in panel A, while guinea pig dose-response curves for an L. monocytogenes isolate with an inlA PMSC (isolate CUFSL N1-040) are shown in panel B. The models derived from guinea pig data were represented in both panels by the following symbols: ▪, log-logistic; •, beta-Poisson; and ▴, exponential.
Although further research is needed to determine which type of model (linear versus nonlinear at low doses) is mechanistically more plausible, the assumption of linearity at low doses is often made in dose-response analysis for microbial pathogens. The beta-Poisson model has been shown to fit experimental data from animal studies and human volunteer studies (35), such as for L. monocytogenes infections in mice (14), Salmonella infections in humans (18), Campylobacter jejuni infections in chickens (5), and infections with vaccine strains of influenza A virus in humans (44). The exponential model has been the choice for deriving a dose-response relationship from data relevant to humans (2, 7, 11, 21). Available knowledge does not preclude an assumption of models with linearity at low dose, e.g., the exponential model or the beta-Poisson model, for L. monocytogenes. A recent study (39; Van Stelten et al., submitted) showed that both the beta-Poisson and the exponential models provided an acceptable fit for L. monocytogenes infections in male guinea pigs and thus provided a gateway for comparing the models derived from guinea pigs and those from data relevant to humans (i.e., food survey, consumption, epidemiologic, and molecular subtyping data).
Just as the recent study by Van Stelten (39) and colleagues reported infectious dose curves, generated using guinea pigs, for both an L. monocytogenes isolate without an inlA PMSC and an isolate with an inlA PMSC, we were also able to compare the differences in probability of infection from 1 cell (i.e., the equivalent to the r value) between values calculated based on guinea pig data and values calculated based on human epidemiological data (i.e., the r values calculated here). There was an 0.69-log difference between the r value (based on the exponential model) derived from human epidemiological data for the subgroup without inlA PMSCs and the r value (based on the exponential model) derived from guinea pig data for the strain without an inlA PMSC (i.e., strain CUFSL N1-054, H7550; obtained from a human case associated with a 1998-1999 listeriosis outbreak in the United States [4]). The guinea pig data-derived r value for the isolate with an inlA PMSC was 1.98 logs higher than the r value for the subgroup with inlA PMSCs derived from human epidemiological data. When growth is considered in deriving the r value from the data relevant to humans, the differences in the log10 r value between the human data and guinea pig data were 2.91 for isolates without an inlA PMSC and 5.07 logs for isolates with an inlA PMSC. These results suggest that a scaling factor up to 3 logs for the L. monocytogenes subgroup without inlA PMSCs and a factor up to 5 logs for the subgroup with inlA PMSCs may be considered when extrapolating guinea pig dose-response models to predict illnesses in humans. Since virulence factors other than InlA play a role in L. monocytogenes infections, and available evidence does not favor the selection of a dose-response model with a particular shape at low doses (e.g., linear versus nonlinear at low doses), further research is needed to substantiate the scaling factor for extrapolation from dose-response models derived from experiments in guinea pigs to predict illness in humans.
Conclusions.
A number of in vitro studies (24, 25, 26, 27, 28, 31) as well as molecular subtyping and epidemiological studies of human listeriosis cases (15, 24, 25, 31) clearly support an important role of the L. monocytogenes surface protein InlA in virulence for human hosts. Interestingly, at least 18 different mutations in inlA that lead to a PMSC, which result either in production of truncated and secreted InlA or complete interruptions of InlA production (e.g., inlA PMSC type 4), have been identified (40). A number of studies (e.g., references 10, 24, 25, 26, 27, 28, and 31) have shown that selected strains with inlA PMSCs have significantly reduced abilities to invade intestinal epithelial cells and significantly reduced guinea pig virulence. Our data here provide, for the first time, quantitative estimates on the relative dose-response relationship for L. monocytogenes with and without inlA PMSCs. The average differences in the r values between the subgroup with PMSCs and the subgroup without PMSCs ranged from 1.4 to 4.1 log units, suggesting that L. monocytogenes isolates with inlA PMSCs may be as much as 10,000-fold less virulent than isolates that encode a full-length InlA. Continuing efforts to integrate molecular subtyping data into hazard characterization will enable better attribution of risk to L. monocytogenes subtypes in quantitative risk assessments. The use of subtype characterization approaches to target genetic variation biologically linked to virulence differences also holds considerable potential to improve risk assessment for other food-borne pathogens, such as Shiga toxin-producing E. coli.
Acknowledgments
This project was supported by the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service, grant 2005-35201-16266. We thank the North America Branch of the International Life Sciences Institute Technical Committee on Food Microbiology for a grant from which the framework of the growth model in this study was initially developed.
Footnotes
Published ahead of print on 17 December 2010.
REFERENCES
- 1.Bakardjiev, A. I., B. A. Stacy, S. J. Fisher, and D. A. Portnoy. 2004. Listeriosis in the pregnant guinea pig: a model of vertical transmission. Infect. Immun. 72:489-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchanan, R. L., W. G. Damert, R. C. Whiting, and M. van Schothorst. 1997. Use of epidemiologic and food survey data to estimate a purposefully conservative dose response relationship for Listeria monocytogenes levels and incidence of listeriosis. J. Food Prot. 60:918-922. [DOI] [PubMed] [Google Scholar]
- 3.Cates, S. C., K. M. Kosa, S. A. Karns, S. Godwin, and D. Chamers. 2007. Consumer storage practices for refrigerated ready-to-eat foods: results of a web-enabled survey. Food Prot. Trends 7:530-543. [Google Scholar]
- 4.Centers for Disease Control and Prevention. 1999. Update: multistate outbreak of listeriosis—United States, 1998-1999. MMWR Morb. Mortal. Wkly. Rep. 47:1117-1118. [PubMed] [Google Scholar]
- 5.Chen, L., H. Geys, S. Cawthraw, A. Havelaar, and P. Teunis. 2006. Dose response for infectivity of several strains of Campylobacter jejuni in chickens. Risk Anal. 26:1613-1621. [DOI] [PubMed] [Google Scholar]
- 6.Chen, Y., et al. 2006. Attributing risk to Listeria monocytogenes subgroup: dose response in relation to genetic lineages. J. Food Prot. 69:335-344. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Y., W. H. Ross, V. N. Scott, and D. E. Gombas. 2003. Listeria monocytogenes: low levels equal low risk. J. Food Prot. 66:570-577. [DOI] [PubMed] [Google Scholar]
- 8.De Jesús, A. J., and R. C. Whiting. 2003. Thermal inactivation, growth, and survival studies of Listeria monocytogenes strains belonging to three distinct genotypic lineages. J. Food Prot. 66:1611-1617. [DOI] [PubMed] [Google Scholar]
- 9.Duffy, S., and D. W. Schaffner. 2001. Modeling the survival of Escherichia coli O157:H7 in apple cider using probability distribution functions for quantitative risk assessment. J. Food Prot. 64:599-605. [DOI] [PubMed] [Google Scholar]
- 10.Felício, M. T. S., T. Hogg, P. Gibbs, P. Teixeira, and M. Wiedmann. 2007. Recurrent and sporadic Listeria monocytogenes contamination in alheiras represents considerable diversity including virulence attenuated isolates. Appl. Environ. Microbiol. 73:3887-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Food and Agriculture Organization of the United Nations, World Health Organization. 2004. Risk assessment of Listeria monocytogenes in ready-to-eat foods. Technical report. Microbiological risk assessment series 5. Food and Agriculture Organization (FAO) of the United Nations and World Health Organization (WHO), Rome, Italy. http://www.fao.org/docrep/010/y5394e/y5394e00.htm.
- 12.Gombas, D. E., Y. Chen, R. S. Clavero, and V. N. Scott. 2003. Survey of Listeria monocytogenes in ready-to-eat foods. J. Food Prot. 66:559-569. [DOI] [PubMed] [Google Scholar]
- 13.Gray, M. J., et al. 2004. Food and human isolates of Listeria monocytogenes form distinct but overlapping populations. Appl. Environ. Microbiol. 70:5833-5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haas, C. N., A. Thayyar-Madabusi, J. B. Rose, and C. P. Gerba. 1999. Development and validation of dose response relationship for Listeria monocytogenes. Quant. Microbiol. 1:89-102. [Google Scholar]
- 15.Jacquet, C., et al. 2004. A molecular marker for evaluating the pathogenic potential of foodborne Listeria monocytogenes. J. Infect. Dis. 189:2094-2100. [DOI] [PubMed] [Google Scholar]
- 16.Jonquieres, R., H. Bierne, J. Mengaud, and P. Cossart. 1998. The inlA gene of Listeria monocytogenes LO28 harbors a nonsense mutation resulting in release of internalin. Infect. Immun. 66:3420-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kathariou, S. 2002. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 65:1811-1829. [DOI] [PubMed] [Google Scholar]
- 18.Latimer, H. K., L.-A. Jaykus, R. A. Morales, P. Cowen, and D. Crawford-Brown. 2001. A weighted composite dose-response model for human salmonellosis. Risk Anal. 21:295-305. [DOI] [PubMed] [Google Scholar]
- 19.Lecuit, M., et al. 1999. A single amino acid in E-cadherin responsible for host specificity towards the human pathogen Listeria monocytogenes. EMBO J. 18:3956-3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lecuit, M., et al. 2001. A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science 292:1722-1725. [DOI] [PubMed] [Google Scholar]
- 21.Lindqvist, R., and A. Westöö. 2000. Quantitative risk assessment for Listeria monocytogenes in smoked or gravid salmon and rainbow trout in Sweden. Int. J. Food Microbiol. 58:181-196. [DOI] [PubMed] [Google Scholar]
- 22.Mead, P. S., et al. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. http://www.cdc.gov/ncidod/eid/vol5no5/mead.htm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montville, R., Y. Chen, and D. W. Schaffner. 2001. Glove barriers to bacterial cross-contamination between hands to food. J. Food Prot. 64:845-849. [DOI] [PubMed] [Google Scholar]
- 24.Nightingale, K. K., K. Windham, K. Martin, M. Yeung, and M. Wiedmann. 2005. Select Listeria monocytogenes subtypes commonly found in foods carry distinct nonsense mutations in inlA, leading to expression of truncated and secreted internalin A, and are associated with a reduced invasion phenotype for human intestinal epithelial cells. Appl. Environ. Microbiol. 71:8764-8772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nightingale, K. K., et al. 2008. inlA premature stop codons are common among Listeria monocytogenes isolated from foods and yield virulence-attenuated strains that confer protection against fully virulent strains. Appl. Environ. Microbiol. 74:6570-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olier, M., D. Garmyn, S. Rousseaux, J. P. Lemaitre, P. Piveteau, and J. Guzzo. 2005. Truncated internalin A and asymptomatic Listeria monocytogenes carriage: in vivo investigation by allelic exchange. Infect. Immun. 73:644-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olier, M., et al. 2002. Assessment of the pathogenic potential of two Listeria monocytogenes human faecal carriage isolates. Microbiology 148:1855-1862. [DOI] [PubMed] [Google Scholar]
- 28.Olier, M., et al. 2003. Expression of truncated internalin A is involved in impaired internalization of some Listeria monocytogenes isolates carried asymptomatically by humans. Infect. Immun. 71:1217-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orsi, R. H., B. M. Bowen, and M. Wiedmann. 2010. Homopolymeric tracts represent a general regulatory mechanism in prokaryotes. BMC Genomics 11:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ross, T., S. Rasmussen, A. Fazil, G. Paoli, and J. Sumner. 2009. Quantitative risk assessment of Listeria monocytogenes in ready-to-eat meats in Australia. Int. J. Food Microbiol. 131:128-137. [DOI] [PubMed] [Google Scholar]
- 31.Rousseaux, S., M. Olier, J. P. Lemaitre, P. Piveteau, and J. Guzzo. 2004. Use of PCR-restriction fragment length polymorphism of inlA for rapid screening of Listeria monocytogenes strains deficient in the ability to invade Caco-2 cells. Appl. Environ. Microbiol. 70:2180-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith, M. A., et al. 2008. Dose-response model for Listeria monocytogenes-induced stillbirths in non-human primates. Infect. Immun. 76:726-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, M. A., et al. 2003. A nonhuman primate model for Listeria monocytogenes-induced stillbirths. Infect. Immun. 71:1574-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teunis, P. F. M., and A. H. Havelaar. 2000. The Beta Poisson model is not a single-hit model. Risk Anal. 20:511-518. [DOI] [PubMed] [Google Scholar]
- 35.Teunis, P. F. M., N. J. D. Nagelkerke, and C. N. Haas. 1999. Dose response models for infectious gastroenteritis. Risk Anal. 19:1251-1260. [DOI] [PubMed] [Google Scholar]
- 36.Teunis, P. 2005. A reconsideration of the Campylobacter dose-response relation. Epidemiol. Infect. 133:583-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.U.S. Food and Drug Administration and USDA Food Safety and Inspection Service. 2003. Quantitative assessment of the relative risk to public health from foodborne Listeria monocytogenes among selected categories of ready-to-eat foods. Food Safety and Inspection Service, U.S. Food and Drug Administration, Washington, DC. http://www.foodsafety.gov/∼dma/lmr2-toc.html.
- 38.Van Stelten, A., and K. K. Nightingale. 2008. Development and implementation of a multiplex single-nucleotide polymorphism genotyping assay for detection of virulence-attenuating mutations in the Listeria monocytogenes virulence-associated gene inlA. Appl. Environ. Microbiol. 74:7365-7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Stelten, A. 2009. Characterization of Listeria monocytogenes with unique mutations in inlA. M.S. thesis. Colorado State University, Fort Collins, CO.
- 40.Van Stelten, A., J. M. Simpson, T. J. Ward, and K. K. Nightingale. 2010. Revelation by single-nucleotide polymorphism genotyping that mutations leading to a premature stop codon in inlA are common among Listeria monocytogenes isolates from ready-to-eat foods but not human listeriosis cases. Appl. Environ. Microbiol. 76:2783-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vose, D. 2000. Risk analysis: a quantitative guide, 2nd ed., p. 57-65. John Wiley & Sons, Chichester, England.
- 42.Ward, T. J., et al. 2010. Molecular and phenotypic characterization of Listeria monocytogenes from U.S. Department of Agriculture Food Safety and Inspection Service surveillance of ready-to-eat foods and processing facilities. J. Food Prot. 73:861-869. [DOI] [PubMed] [Google Scholar]
- 43.Ward, T. J., T. F. Ducey, T. Usgaard, K. A. Dunn, and J. P. Bielawski. 2008. Multilocus genotyping assays for single nucleotide polymorphism-based subtyping of Listeria monocytogenes isolates. Appl. Environ. Microbiol. 74:7629-7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watanabe, T., T. A. Bartrand, T. Omura, and C. N. Haas. 2009. Dose-response assessment for influenza A virus based on the datasets for its vaccine strains, abstr. T3-C10. Society for Risk Analysis annual meeting, Baltimore, MD, 6 to 9 December 2009.
- 45.Wiedmann, M. 2002. Molecular subtyping methods for Listeria monocytogenes. J. AOAC Int. 85:524-531. [PubMed] [Google Scholar]
- 46.Wiedmann, M., et al. 1997. Ribotyping and virulence gene polymorphisms suggested three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams, D., E. A. Irvin, R. A. Chmielewski, J. F. Frank, and M. A. Smith. 2007. Dose-response of Listeria monocytogenes after oral exposure in pregnant guinea pigs. J. Food Prot. 70:1122-1128. [DOI] [PubMed] [Google Scholar]
- 48.Williams, D., J. Castleman, C.-C. Lee, B. Mote, and M. A. Smith. 2009. Risk of fetal mortality after exposure to Listeria monocytogenes based on dose-response data from pregnant guinea pigs and primates. Risk Anal. 29:1495-1505. (Erratum, 30:710, 2010.) [DOI] [PubMed] [Google Scholar]