Abstract
Prothioconazole is one of the most important commercially available demethylase inhibitors (DMIs) used to treat Mycosphaerella graminicola infection of wheat, but specific information regarding its mode of action is not available in the scientific literature. Treatment of wild-type M. graminicola (strain IPO323) with 5 μg of epoxiconazole, tebuconazole, triadimenol, or prothioconazole ml−1 resulted in inhibition of M. graminicola CYP51 (MgCYP51), as evidenced by the accumulation of 14α-methylated sterol substrates (lanosterol and eburicol) and the depletion of ergosterol in azole-treated cells. Successful expression of MgCYP51 in Escherichia coli enabled us to conduct spectrophotometric assays using purified 62-kDa MgCYP51 protein. Antifungal-binding studies revealed that epoxiconazole, tebuconazole, and triadimenol all bound tightly to MgCYP51, producing strong type II difference spectra (peak at 423 to 429 nm and trough at 406 to 409 nm) indicative of the formation of classical low-spin sixth-ligand complexes. Interaction of prothioconazole with MgCYP51 exhibited a novel spectrum with a peak and trough observed at 410 nm and 428 nm, respectively, indicating a different mechanism of inhibition. Prothioconazole bound to MgCYP51 with 840-fold less affinity than epoxiconazole and, unlike epoxiconazole, tebuconazole, and triadimenol, which are noncompetitive inhibitors, prothioconazole was found to be a competitive inhibitor of substrate binding. This represents the first study to validate the effect of prothioconazole on the sterol composition of M. graminicola and the first on the successful heterologous expression of active MgCYP51 protein. The binding affinity studies documented here provide novel insights into the interaction of MgCYP51 with DMIs, especially for the new triazolinethione derivative prothioconazole.
Mycosphaerella graminicola (anamorph: Septoria tritici) is a plant-pathogenic fungus causing septoria leaf blotch that is responsible for significant yield losses (13). The most widely used fungicides for the control of this disease are demethylase inhibitors (DMIs), which bind to the target cytochrome P450 (CYP51, also called Erg11p in yeast) that mediates sterol 14α-demethylation during ergosterol biosynthesis. The original observation in fungi of inhibition of sterol 14α-demethylation by DMIs was in the plant pathogen Ustilago maydis (31) and is also seen for azole drugs when treating Candida albicans infections (28). The 14α-demethylation step of sterol biosynthesis had been proposed to be a cytochrome P450 mediated activity (1) and the protein was first purified from Saccharomyces cerevisiae microsomal fraction (2). Using yeast genetics the gene encoding this ancient activity of the cytochrome P450 superfamily was isolated in 1987 (17), and all CYP51 genes encoding this activity in sterol biosynthesis in different Kingdoms of Life are classified to this family (19).
DMIs are generally imidazole or triazole compounds. The N-2 of imidazole and N-3 of triazole compounds form a sixth ligand with the heme of the CYP51 that is reflected in a type II binding spectrum formed when the azoles become ligands of low-spin CYP51 (14). The selectivity of DMIs is defined by the interaction of the N-1 substituent groups of the azole and the CYP51 structure. Recently, such interactions have been investigated by X-ray crystallography using trypanosomal and human CYP51 enzymes (24, 35). DMIs result in the depletion of ergosterol and the concomitant increase in 14α-methylated sterols. In contrast to S. cerevisiae and C. albicans, M. graminicola does not accumulate the 14-methyl-3,6-diol observed in these yeasts under azole treatment (15, 20), but the depletion of ergosterol and accumulation of other 14α-methylated sterols are indicative of CYP51 inhibition.
Resistance to antifungal compounds has developed in M. graminicola populations through mutations resulting in an altered CYP51 enzyme (9, 10). Similar mutations in CYP51 have also been observed in the clinical setting, firstly with C. albicans (26, 32). The introduction of new antifungals has allowed control of Septoria wheat blotch to be maintained. The most recently introduced DMI for the treatment of M. graminicola is the triazolinethione derivative prothioconazole.
We are interested in the development of resistance and the mode of action and efficacy of this class of antifungal. The difference in chemical structure of prothioconazole compared to triazole compounds led us to probe the biochemical basis of DMI fungicide affinity to the target protein. This requires a mechanism for heterologous production, and we report here the first purification of CYP51 from this economically important pathogen. We also present data which confirm that the triazole compounds epoxiconazole, tebuconazole, and triadimenol bind directly to the heme, as is observed with azole inhibitors of other CYP51s (14), and we have determined the affinities of these compounds for M. graminicola CYP51, which enables comparisons to be made with other species and compounds. Further investigation of the fungicide interaction with the target protein revealed that prothioconazole interacts with CYP51 in a novel way that is distinct from other azole fungicides. The mode of action of prothioconazole may therefore provide a new avenue of research for antifungals for the treatment of plant diseases and human infections such as candidiasis.
MATERIALS AND METHODS
Chemicals, media, and strains.
Growth media, ampicillin, IPTG (isopropyl-β-d-thiogalactopyranoside), and 5-aminolevulenic acid were obtained from Foremedium, Ltd. (Hunstanton, United Kingdom). Eburicol was produced by David Nes. Enzymes for molecular biology were obtained from Promega (Madison, WI). All other chemicals were obtained from Sigma (Poole, United Kingdom). Escherichia coli DH5α (Stratagene, La Jolla, CA) was used for plasmid manipulation and protein expression.
Sterol composition of M. graminicola.
M. graminicola IPO323 was grown in YPD broth (1% [wt/vol] yeast extract, 2% [wt/vol] peptone, and 2% [wt/vol] glucose) at 25°C for 4 days. Cultures were diluted to an optical density at 600 nm of 1.0, and 500 μl was used to inoculate 10 ml of YPD, followed by treatment with 5 μg of fungicide ml−1. Azoles were diluted in dimethyl sulfoxide (adjusted to 1% [wt/vol] for azole solutions and the negative control). Cultures were grown for 48 h at 25°C, and the cells were harvested and washed twice with sterile water. Nonsaponifiable lipids were extracted as reported previously (18). Samples were dried in a vacuum centrifuge (Heto) and derivatized by the addition of 100 μl of 90% BSTFA-10% TMS (Sigma) and 50 μl of anhydrous pyridine (Sigma), followed by heating for 2 h at 80°C. TMS-derivatized sterols were analyzed and identified by using gas chromatography-mass spectrometry (GC-MS; Agilent 5975C Inert XL GC/MSD; Agilent Technologies, Ltd., Stockport, United Kingdom) with reference to retention times and fragmentation spectra for known standards. GC-MS data files were analyzed by using Agilent software (MSD Enhanced ChemStation) to determine the sterol profiles for all isolates and for integrated peak areas.
Heterologous expression of MgCYP51.
The M. graminicola CYP51 gene (MgCYP51 [GenBank AAU43734]) was synthesized by GeneCust (Evry, France). The nucleotide sequence was optimized for expression in E. coli (codon adaptation index of 0.85 compared to 0.63 for the wild-type MgCYP51 [34]) and engineered to contain 5′ NdeI and 3′ HindIII sites, a C-terminal hexahistidine tag, and the second residue, glycine, was replaced by alanine to aid overexpression (5). MgCYP51 was cloned into pCWori+ (5) using the NdeI and HindIII sites, and transformants were selected by using ampicillin. pCWori+::MgCYP51 transformants were grown in Terrific broth containing ampicillin at 30°C and 160 rpm for 24 h prior to induction with 1 mM IPTG and expression at 20°C and 140 rpm for 48 h in the presence of 1 mM 5-aminolevulenic acid.
MgCYP51 protein was isolated as described previously (4) using modified sonication buffer containing 2% (wt/vol) sodium cholate and no Tween 20. Solubilized MgCYP51 protein was purified by affinity chromatography using Ni2+-NTA agarose (Qiagen) and eluted using 1% (wt/vol) histidine in 0.1 M Tris-HCl (pH 8.1) containing 25% (wt/vol) glycerol. Purified protein was dialyzed against 4 liters of 0.1 M Tris-HCl (pH 8.1) overnight at 4°C using dialysis tubing with a 30-kDa molecular mass cutoff to remove histidine. Protein purity was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (21).
Cytochrome P450 spectral determinations.
Absolute spectra of the oxidized protein, the reduced protein (10 mM sodium dithionite), and the reduced carbon monoxide-P450 complex were determined between 300 and 700 nm by using 8 μM purified MgCYP51 in 0.1 M Tris-HCl (pH 8.1) and 25% (wt/vol) glycerol as previously described (8). Extinction coefficients of 125 mM−1 cm−1 for the oxidized heme Soret peak at 420 nm (12) and 91 mM−1 cm−1 at 445 nm (30) for the Soret peak of the red-shifted reduced carbon monoxide adduct were used. All UV-VIS spectrophotometry determinations were made by using a Hitachi U-3310 UV/VIS spectrophotometer (San Jose, CA) and quartz semi-micro cuvettes with a light path of 4.5 mm.
Azole-binding properties of MgCYP51.
Binding of azole to MgCYP51 were performed as previously described (22, 23) except that dimethylformamide (DMF) was also added to the cytochrome P450-containing compartment of the reference cuvette. Stock solutions of 0.1 mg of epoxiconazole, tebuconazole, and triadimenol ml−1 and 1 and 10 mg of prothioconazole ml−1 were prepared in DMF. Azoles were progressively titrated against 4 μM MgCYP51 in 0.1 M Tris-HCl (pH 8.1) and 25% (wt/vol) glycerol, with the difference spectra between 500 and 350 nm determined after each addition; azole binding determinations were performed in triplicate for each compound. Binding saturation curves were constructed from ΔApeak-trough versus the azole concentration. A rearrangement of the Morrison equation [ΔA = (ΔAmax × {[Et + [azole] + Kd] − [(Et + [azole] + Kd)^2 − (4 × Et × [azole])]^0.5}/{2 × Et})] (27, 29) (where Et is the total amount of CYP51 available to bind azole) was used to determine the dissociation constant (Kd) values when ligand binding was “tight.” Tight-binding is observed when the Kd for azole is similar or lower than the concentration of CYP51 present (11). The Michaelis-Menten equation ΔA = (ΔAmax × [azole])/(Kd + [azole]) was used when the ligand binding was not tight.
Substrate binding studies.
Lanosterol and eburicol (0.5 mg) were each dissolved in 0.1 ml of chloroform, to which 1 ml of acetone and 0.05 ml of Tween 80 were added. The solution was evaporated to dryness under nitrogen with constant vortexing. Sterol and Tween 80 residues were then dissolved in 1 ml of water to give 0.05% (vol/vol) stock sterol solutions. Lanosterol and eburicol were progressively titrated against 8 μM MgCYP51 in the sample cuvette with equivalent amounts of 5% (vol/vol) Tween 80 added to the reference cuvette also containing 8 μM MgCYP51. The absorbance difference spectrum between 500 and 350 nm was determined after each incremental addition of sterol up to 34 μM, and sterol saturation curves were constructed from the ΔA385-422 value. Eburicol binding determinations were performed in triplicate. The substrate binding constant (Ks) was determined by nonlinear regression (Levenberg-Marquardt algorithm) using the Michaelis-Menten equation: ΔA = (ΔAmax × [sterol])/(Ks + [sterol]).
Substrate binding studies were also performed with lanosterol and eburicol (3 to 80 μM) in the presence or absence of 0.35 mM prothioconazole or 6 μM epoxiconazole. Control determinations were made in the presence of 1.25% (vol/vol) DMF. Determinations were performed in triplicate and Lineweaver-Burk plots were constructed from resultant substrate binding spectra. To determine the modality of these two inhibitors the inhibitor constant (Kei) for the formation of the enzyme-prothioconazole complex was calculated by using the equation Ks.app = Ks × (1 + [I]/Kei) for pure competitive inhibition.
Data analysis.
Curve-fitting substrate and azole binding data were performed using the computer program ProFit 5.0.1 (QuantumSoft, Zurich, Switzerland). Protein targeting signal peptide prediction was performed using the Predotar (http://urgi.versailles.inra.fr/predotar/predotar.html), SignalP3.0 (http://www.cbs.dtu.dk/services/SignalP/), and TargetP1.1 (http://www.cbs.dtu.dk/services/TargetP/) programs.
RESULTS
Sterol composition of azole-treated M. graminicola.
The novel chemistry of prothioconazole led us to question whether the mode of action of inhibition of M. graminicola was through the inhibition of CYP51 activity. To investigate this initially, we first analyzed the sterol profiles of M. graminicola cultures treated with prothioconazole compared to those treated with epoxiconazole, tebuconazole, and triadimenol. It was expected that an increase in the CYP51 substrate, together with a depletion of ergosterol, would be observed upon treatment with azoles as seen previously. Table 1 shows the percentages of 14α-methylated sterols (eburicol and lanosterol) and 14α-demethylated sterols (ergosterol, ergosta-5,8,22-trienol, and ergosta-7,22-dienol) present in the treated samples. As expected, M. graminicola accumulated eburicol predominantly when treated with epoxiconazole, tebuconazole, and triadimenol in agreement with this being the main CYP51 substrate. Importantly, treatment with prothioconazole also resulted in an accumulation of eburicol and a depletion of the major 14α-demethylated sterols ergosterol and ergosta-5,8,22-trienol, confirming that prothioconazole does inhibit the activity of MgCYP51. In our previous work we had identified an ergosterol isomer as being detectable in M. graminicola, and we were able to confirm here that the isomer was ergosta-5,8,22-trienol present at lower levels in prior studies (6, 15).
TABLE 1.
Sterol profiles of azole-treated M. graminicola culturesa
| Treatment | Mean % content ± SD |
||||
|---|---|---|---|---|---|
| Ergosta-5,8,22-trienol | Ergosterol | Ergosta-7,22-dienol | Lanosterol | Eburicol | |
| Untreated | 4.5 ± 0.6 | 85.3 ± 0.5 | 6.4 ± 0.3 | ND | 3.0 ± 0.1 |
| Epoxiconazole | ND | ND | ND | 19.3 ± 0.2 | 80.7 ± 0.2 |
| Tebuconazole | ND | ND | ND | 17.5 ± 0.4 | 82.4 ± 0.4 |
| Triadimenol | ND | ND | ND | 17.5 ± 0.5 | 82.5 ± 0.4 |
| Prothioconazole | ND | ND | ND | 21.4 ± 1.8 | 78.6 ± 1.8 |
The percentage of different sterols in total sterol extracts of M. graminicola IPO323 treated with 5 μg of epoxiconazole, tebuconazole, triadimenol, or prothioconazole ml−1. ND, not detected.
Characterization of MgCYP51.
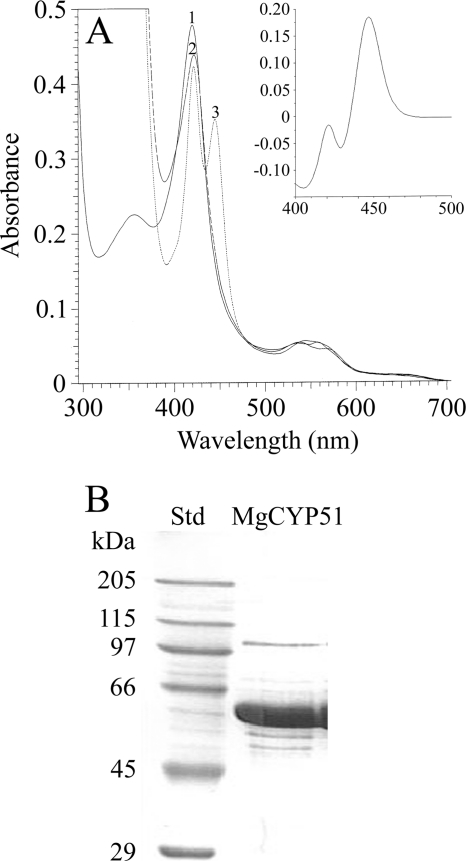
MgCYP51 was predicted to be a membrane bound protein localized in the endoplasmic reticulum (Predotar, SignalP3.0, and TargetP1.1), as is expected for eukaryotic CYPs, and sodium cholate was necessary to solubilize MgCYP51, confirming this prediction. Overexpression of MgCYP51 in E. coli yielded ∼300 nmol of MgCYP51 per liter of culture, as determined from the reduced carbon monoxide difference spectra. Purification by Ni2+-NTA agarose chromatography resulted in >90% purity when assessed by SDS-PAGE and an apparent molecular mass in agreement with the expected 62.177 kDa (Fig. 1 B).
FIG. 1.
Properties of purified MgCYP51. (A) Absolute spectra of 8 μM purified MgCYP51 were determined under oxidative conditions (line 1), indicating that MgCYP51 was isolated predominantly in the ferric low-spin state. Lines 2 and 3 show dithionite-reduced MgCYP51 and dithionite-reduced MgCYP51 in the presence of carbon monoxide, respectively. The reduced-CO difference spectrum (inset) was derived by subtracting line 3 from line 2 and confirmed that the protein was expressed in its active form. (B) SDS-PAGE was performed with 20 μg of purified protein. Std, standard.
The absolute spectra (Fig. 1A) and reduced carbon monoxide difference spectra (Fig. 1A, inset) of MgCYP51 were characteristic of a cytochrome P450 enzyme (8, 14), confirming that the protein was expressed in its active form. MgCYP51 was isolated predominantly in the ferric low-spin state with a heme Soret (γ) peak at 420 nm in addition to α, β, and δ peaks at 569, 537, and 356 nm, respectively. Dithionite one-electron reduction caused a small red-shift of the Soret peak to 422 nm with binding of carbon monoxide to the reduced ferrous form, resulting in a characteristic red-shift of the Soret peak from 420 to 445 nm. These studies confirmed the successful production of pure MgCYP51 for the first time and the ability to utilize this for in vitro investigation of inhibitors.
Fungicide binding studies to MgCYP51.
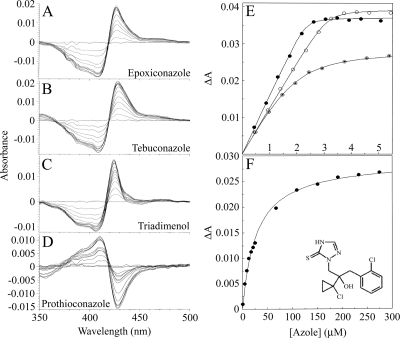
Binding of azole compounds to CYP51 enzymes has been investigated previously especially in the clinical setting (7). Purified MgCYP51 was used in spectrophotometric assessment of the interaction with epoxiconazole, tebuconazole, triadimenol, and prothioconazole. Epoxiconazole, tebuconazole, and triadimenol bound tightly to MgCYP51 (Fig. 2), producing strong type II difference spectra indicative of an azole-bound low-spin CYP51 complex (Fig. 2A, B, and C) with a peak at 423 to 429 nm and a trough at 406 to 409 nm. The Morrison equation was used to fit the saturation curve data (Fig. 2E) and that of the other two replicates for each azole. Mean Kd values of 0.0166, 0.0266, and 0.299 μM were obtained for epoxiconazole, tebuconazole, and triadimenol, respectively, with standard errors between replicates of <10%. In contrast, prothioconazole produced a novel spectrum on interaction with MgCYP51 with a peak at 410 nm and a trough at 428 nm (Fig. 2D). Azole-induced type II difference spectra reflect the direct coordination of an azole nitrogen atom to the heme as the sixth ligand, and the prothioconazole-induced spectrum indicated a difference in the mode of binding. The Michaelis-Menten equation was used to fit the prothioconazole-induced spectral changes to a saturation curve (Fig. 2F). Together with two replicates, this allowed a mean Kd of 14 ± 0.16 μM to be calculated, an affinity substantially less than for the other azole compounds.
FIG. 2.
Binding properties of azole fungicides to MgCYP51. Epoxiconazole (A), tebuconazole (B), triadimenol (C), and prothioconazole (D) bound to 4 μM MgCYP51. Each line represents the successive addition of antifungal, resulting in a progressive increase in absorbance at 423 to 429 nm and a decrease in absorbance at 406 to 409 nm with epoxiconazole, tebuconazole, and triadimenol and a progressive increase in absorbance at 410 nm and decrease in absorbance at 428 nm with prothioconazole, until saturation is reached. (E) The concentration of azole added to MgCYP51 and the resulting change in absorbance (ΔApeak-trough) were plotted to produce binding saturation profiles for epoxiconazole (•), tebuconazole (○), and triadimenol (⊙) using the Morrison equation. (F) A prothioconazole saturation profile was constructed by using the Michaelis-Menten equation. The data for one of three replicates are shown. The chemical structure of prothioconazole is also shown in panel F.
Fungicide interference with substrate binding.
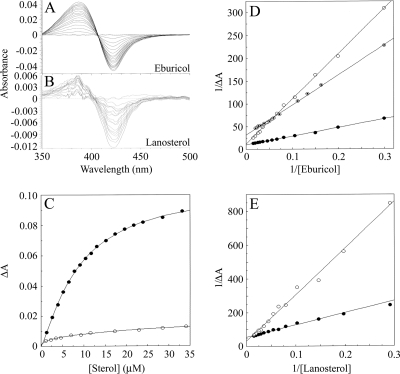
To gain further information on the MgCYP51 protein and mode of action, a comparison of the binding of two CYP51 substrates to MgCYP51 was undertaken, and then the ability of all four fungicides to interfere with these was measured. Progressive titration of MgCYP51 with lanosterol and eburicol gave type I binding spectra (Fig. 3 A and B) with peaks at 385 nm and troughs at 422 nm. This showed that the binding of sterol substrate caused a change in spin state from a low to a high spin by displacing the water molecule coordinated as the sixth ligand to the low-spin heme prosthetic group, causing the heme to adopt the high-spin pentacoordinated conformation (14). The type I binding spectrum obtained with eburicol was 9-fold more intense than that obtained with lanosterol, indicating that eburicol was more effective at displacing the water molecule coordinated as the sixth ligand to the heme than lanosterol. However, at saturating concentrations of eburicol, <10% of the MgCYP51 molecules changed spin state from low to high spin. This relatively low degree of spin state conversion induced by substrate binding has also been observed with other CYP51 enzymes (3, 7, 16, 25), with spin state changes usually not exceeding 10%.
FIG. 3.
Substrate binding properties of MgCYP51. Eburicol (A) and lanosterol (B) bound to 8 μM purified MgCYP51. Each line represents the successive addition of substrate and results in a progressive increase in absorbance at 385 nm and a decrease in absorbance at 422 nm, until saturation is reached. (C) The concentration of azole added to MgCYP51 and the resulting change in absorbance (ΔApeak-trough) were plotted to produce binding saturation profiles for eburicol (•) and lanosterol (○) using the Michaelis-Menten equation. The substrate binding affinities were determined to be 10.85 ± 0.92 and 13.4 μM, respectively. Lineweaver-Burk plots of eburicol (D) and lanosterol (E) binding to 8 μM purified MgCYP51 in the absence (•) or presence of 0.35 mM prothioconazole (○) or in the presence of 6 μM epoxiconazole (⊙ [eburicol only]) were constructed to determine the modality of inhibitor action on substrate binding.
The Michaelis-Menten equation best fit for the sterol saturation curves (Fig. 3C) yielded Ks values of 10.85 μM (with standard errors between replicates of <10%) for eburicol and 13.4 μM (error of curve fit ± 2.0) for lanosterol, signifying that MgCYP51 bound both substrates with similar affinities. The presence of 0.35 mM prothioconazole inhibited the binding of both eburicol and lanosterol to MgCYP51 (Fig. 3D and E). The converging lines of the Lineweaver-Burk plots at the 1/ΔA axis are indicative of competitive inhibition, where the ΔAmax value remains constant and the apparent Ks value increases in response to the presence of inhibitor (33). In contrast, the presence of 6 μM epoxiconazole inhibited eburicol binding noncompetitively, as observed in the Lineweaver-Burk plot (Fig. 3D), where the epoxiconazole and control data sets converge to intersect on the 1/[eburicol] axis, indicating the apparent Ks remained unchanged, while the ΔAmax decreased. Due to the reduced quality of the lanosterol-induced spectra, the data obtained with this substrate were not included since reliable analysis was not possible.
DISCUSSION
There are no previous reports in the scientific literature of the effect of prothioconazole on sterols of treated fungi, and we demonstrated here that it behaves as an active inhibitor of sterol 14 α-demethylation and reduces ergosterol in M. graminicola cells. It is active against growing M. graminicola at concentrations equivalent to those of the azole fungicides epoxiconazole, tebuconazole, and triadimenol. The sterol profile of treated cultures showed a similar accumulation of eburicol and, to a lesser extent, lanosterol, which is consistent with the normal metabolic route suggested for filamentous fungi (eburicol rather than lanosterol being the CYP51 substrate). Since no other enzyme in fungi can fulfill the essential function of CYP51, the accumulation of 14 α-methyl sterols and the depletion of ergosterol confirms that CYP51 is the target of the prothioconazole-mediated growth inhibition of M. graminicola. The confirmation here that the ergosterol isomer observed in M. graminicola previously (6, 15) is ergosta-5,8,22-trienol suggests that C8 isomerization may be less efficient in M. graminicola than in other fungal species.
Successful heterologous expression of MgCYP51 was achieved with E. coli, and this allowed the interaction of DMIs, including prothioconazole, to be studied for the first time. The interaction of epoxiconazole, tebuconazole, and triadimenol indicated typical type II spectral interaction with high affinity for MgCYP51, confirming the formation of a low-spin complex with the compounds bound as a sixth ligand of the heme. Prothioconazole did not bind in this way, as reflected by the novel spectrum observed and the absence of type II spectral interaction. The binding affinity of MgCYP51 for prothioconazole was 840-fold less than for epoxiconazole. Epoxiconazole bound with greatest affinity to MgCYP51, followed closely by tebuconazole, and triadimenol bound with 18-fold less affinity than epoxiconazole. The ability to compare the affinities of different azole compounds for MgCYP51, as well as comparison to MgCYP51 strains harboring mutations implicated in resistance to azoles and with other CYP51s, may be of use in aiding the further understanding of the resistance to azoles and may aid in drug development.
The nature of the spectrum observed for prothioconazole is not currently clear and does not conform to inhibitor or substrate effects observed previously. The perturbation of prothioconazole on binding to MgCYP51 might be caused by a weak interaction between the electronegative sulfur atom on the azole ring of prothioconazole (Fig. 2F) and the ferric ion of the heme prosthetic group.
To compare the interaction of prothioconazole with MgCYP51 more closely, the interference with 14α-methyl substrate interaction was investigated. The presence of 0.35 mM prothioconazole caused 4.6- and 7.5-fold increases in the apparent Ks values for eburicol and lanosterol, respectively, resulting in estimates of the Kei for prothioconazole of 51 to 104 μM. This suggested that prothioconazole “competes” for the substrate binding site on MgCYP51. It is not surprising that epoxiconazole was a noncompetitive inhibitor of MgCYP51 since azole antifungal agents bind to the CYP51 molecule through direct coordination with the heme prosthetic group and not through interactions with the substrate binding site (8).
The treatment of whole cells revealed that prothioconazole has an efficacy comparable to that of epoxiconazole, tebuconazole, and triadimenol. However, the competitive inhibition observed with prothioconazole and the high Kd value for prothioconazole would not alone account for the effectiveness of prothioconazole in vivo as a CYP51 inhibitor if it were binding directly as a sixth ligand of the heme. Prothioconazole may therefore inhibit MgCYP51 activity in an additional capacity that does not result in the perturbation of the heme environment (and hence cannot be directly measured spectrophotometrically).
The possibility exists that in vivo metabolism of prothioconazole to the desthio metabolite (a triazole) in wheat may be important for the efficacy of this compound in the field, but our results show inhibition of sterol biosynthesis in M. graminicola cells grown in broth cultures and therefore indicate that the antifungal activity does not rely on in planta metabolism. Alternatively, it is possible that the in vivo metabolism of prothioconazole occurs in fungal cells and that this is necessary for the antifungal activity of the compound. Further work on prothioconazole-treated M. graminicola cultures will elucidate how prothioconazole itself is inhibiting CYP51 and whether biotransformation resulting in the desthio triazole compound contributes to CYP51 inhibition. In either case, prothioconazole presents a novel antifungal agent. The findings described here suggest an inhibition of CYP51 activity by binding to the enzyme in a manner other than direct coordination to the heme. Additionally, an eventual metabolism to the desthio triazole compound within the fungus would lead to an active antifungal. This mode of antifungal action therefore presents a possible basis for the development of new compounds both for agriculturally important pathogenic species and for the causative agents of human diseases, and we are currently examining other CYP51s of such pathogens for their interaction with this new fungicide.
Acknowledgments
We are grateful to the Biotechnology and Biological Science Research Council of the United Kingdom (projects BBE02257X1 and BBE0218321), the U.S. National Institutes of Health (NSF-MCB 0920212 [W.D.N.]), and the Welch Foundation (D-1276 [W.D.N.]) for supporting this work.
We are grateful also to N. Rolley for technical assistance and to the EPSRC National Mass Spectrometry Centre, Swansea University.
Footnotes
Published ahead of print on 17 December 2010.
REFERENCES
- 1.Alexander, K., M. Akhtar, R. B. Boar, J. F. McGhie, and D. H. R. Barton. 1972. The removal of the 32-carbon atom as formic acid in cholesterol biosynthesis. J. Chem. Soc. Chem. Commun. (Camb.) 1972(7):383-385. [Google Scholar]
- 2.Aoyama, Y., Y. Yoshida, and R. Sato. 1984. Yeast cytochrome P450 catalysing lanosterol 14α-demethylation. II. Lanosterol metabolism by purified P45014DM and by intact microsomes. J. Biol. Chem. 259:1661-1666. [PubMed] [Google Scholar]
- 3.Aoyama, Y., Y. Yoshida, Y. Sonoda, and Y. Sato. 1987. Metabolism of 32-hydroxy-24,25-dihydrolanosterol by purified cytochrome P-45014DM from yeast: evidence for contribution of the cytochrome to whole process of lanosterol 14α-demethylation. J. Biol. Chem. 262:1239-1243. [PubMed] [Google Scholar]
- 4.Arase, M., M. R. Waterman, and N. Kagawa. 2006. Purification and characterization of bovine steroid 21-hydroxylase (P450c21) efficiently expressed in Escherichia coli. Biochem. Biophys. Res. Commun. 344:400-405. [DOI] [PubMed] [Google Scholar]
- 5.Barnes, H. J., M. P. Arlotto, and M. R. Waterman. 1991. Expression and enzymatic activity of recombinant cytochrome P450 17α-hydroxylase in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 88:5597-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bean, T. P., et al. 2009. Sterol content analysis suggests altered eburicol 14α-demethylase (CYP51) activity in isolates of Mycosphaerella graminicola adapted to azole fungicides. FEMS Microbiol. Lett. 296:266-273. [DOI] [PubMed] [Google Scholar]
- 7.Bellamine, A., G. I. Lepesheva, and M. R. Waterman. 2004. Fluconazole binding and sterol demethylation in three CYP51 isoforms indicate differences in active site topology. J. Lipid Res. 45:2000-2007. [DOI] [PubMed] [Google Scholar]
- 8.Bellamine, A., A. T. Mangla, W. D. Nes, and M. R. Waterman. 1999. Characterization and catalytic properties of the sterol 14α-demethylase from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 96:8937-8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cools, H. J., and B. A. Fraaije. 2008. Are azole fungicides losing ground against Septoria wheat disease? Resistance mechanisms in Mycosphaerella graminicola. Pest Manag. Sci. 64:681-684. [DOI] [PubMed] [Google Scholar]
- 10.Cools, H. J., et al. 2010. Heterologous expression of mutated eburicol 14α-demethylase (CYP51) proteins of Mycosphaerella graminicola demonstrates effects on azole fungicide sensitivity and intrinsic protein function. Appl. Environ. Microbiol. 76:2866-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Copeland, R. A. 2005. Evaluation of enzyme inhibitors, p. 178-213. In Drug discovery: a guide for medicinal chemists and pharmacologists. Wiley-Interscience, New York, NY. [PubMed]
- 12.Correia, M. A. 2005. Table A.5, p. 644-646. In P. R. Ortiz de Montellano (ed.), Cytochrome P450: structure, mechanism and biochemistry, 3rd ed. Kluwer Academic/Plenum Press, Inc., New York, NY.
- 13.Eyal, Z. 1999. The Septoria tritici and Stagonospora nodorum blotch diseases of wheat. Eur. J. Plant Pathol. 105:629-641. [Google Scholar]
- 14.Jefcoate, C. R. 1978. Measurement of substrate and inhibitor binding to microsomal cytochrome P-450 by optical-difference spectroscopy. Methods Enzymol. 52:258-279. [DOI] [PubMed] [Google Scholar]
- 15.Joseph-Horne, T., D. Hollomon, N. Manning, and S. L. Kelly. 1996. Investigation of the sterol composition and azole resistance in field isolates of Septoria tritici. Appl. Environ. Microbiol. 62:184-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kahn, R. A., S. Bak, C. E. Olsen, I. Svendsen, and B. L. Moller. 1996. Isolation and reconstitution of the heme-thiolate protein obtusifoliol 14α-demethylase from Sorghum bicolor (L.) Moench. J. Biol. Chem. 271:32944-32950. [DOI] [PubMed] [Google Scholar]
- 17.Kalb, V. F., et al. 1987. Primary structure of the P450 lanosterol demethylase gene from Saccharomyces cerevisiae. DNA 6:529-537. [DOI] [PubMed] [Google Scholar]
- 18.Kelly, S. L., D. C. Lamb, A. J. Corran, B. C. Baldwin, and D. E. Kelly. 1995. Mode of action and resistance to azole antifungals associated with the formation of 14α-methylergosta-8,24(28)-dien-3β,6α-diol. Biochem. Biophys. Res. Commun. 207:910-915. [DOI] [PubMed] [Google Scholar]
- 19.Kelly, S. L., D. C. Lamb, C. J. Jackson, A. G. Warrilow, and D. E. Kelly. 2003. The biodiversity of microbial cytochromes P450. Adv. Microb. Physiol. 47:131-186. [DOI] [PubMed] [Google Scholar]
- 20.Kelly, S. L., D. C. Lamb, D. E. Kelly, J. Loeffler, and H. Einsele. 1996. Resistance to fluconazole and amphotericin in Candida albicans from AIDS patients. Lancet 348:1523-1524. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Lamb, D. C., et al. 1999. Generation of a complete, soluble, and catalytically active sterol 14α-demethylase-reductase complex. Biochemistry 38:8733-8738. [DOI] [PubMed] [Google Scholar]
- 23.Lamb, D. C., et al. 2002. The cytochrome P450 complement (CYPome) of Streptomyces coelicolor A3(2). J. Biol. Chem. 277:24000-24005. [DOI] [PubMed] [Google Scholar]
- 24.Lepesheva, G. I., et al. 2010. Crystal structures of Trypanosoma brucei sterol 14α-demethylase and implications for selective treatment of human infections. J. Biol. Chem. 285:1773-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lepesheva, G. I., C. Virus, and M. R. Waterman. 2003. Conservation in the CYP51 family: role of the B′ helix/BC loop and helices F and G in enzymatic function. Biochemistry 42:9091-9101. [DOI] [PubMed] [Google Scholar]
- 26.Löffler, J., et al. 1997. Molecular analysis of cyp51 from fluconazole-resistant Candida albicans strains. FEMS Microbiol. Lett. 151:263-268. [DOI] [PubMed] [Google Scholar]
- 27.Lutz, J. D., et al. 2009. Expression and functional characterization of cytochrome P450 26A1, a retinoic acid hydroxylase. Biochem. Pharmacol. 77:258-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marriott, M. S. 1980. Inhibition of sterol biosynthesis in Candida albicans by imidazole-containing antifungals. J. Gen. Microbiol. 117:253-255. [DOI] [PubMed] [Google Scholar]
- 29.Morrison, J. F. 1969. Kinetics of the reversible inhibition of enzyme-catalysed reactions by tight-binding inhibitors. Biochim. Biophys. Acta 185:269-286. [DOI] [PubMed] [Google Scholar]
- 30.Omura, T., and R. Sato. 1964. The carbon monoxide-binding pigment of liver microsomes. II. Solubilization, purification, and properties. J. Biol. Chem. 239:2379-2385. [PubMed] [Google Scholar]
- 31.Ragsdale, N. N., and H. D. Sisler. 1972. Inhibition of ergosterol synthesis in Ustilago maydis by the fungicide triarimol. Biochem. Biophys. Res. Commun. 46:2048-2053. [DOI] [PubMed] [Google Scholar]
- 32.Sanglard, D., F. Ischer, L. Koymans, and J. Bille. 1998. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob. Agents Chemother. 42:241-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Segel, I. H. 1975. Enzyme kinetics: behavior and analysis of rapid equilibrium and steady-state enzyme systems, p. 100-124. Wiley-Interscience, New York, NY.
- 34.Sharp, P. M., and W.-H. Li. 1987. The codon adaptation index: a measure of directional synonymous codon usage bias and its potential applications. Nucleic Acids Res. 15:1281-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strushkevich, N., S. A. Usanov, and H. W. Park. 2010. Structural basis of human CYP51 inhibition by antifungal azoles. J. Mol. Biol. 397:1067-1078. [DOI] [PubMed] [Google Scholar]